Mapping Natural Forest Remnants with Multi-Source and Multi-Temporal Remote Sensing Data for More Informed Management of Global Biodiversity Hotspots
Abstract
1. Introduction
2. Methods and Study Area
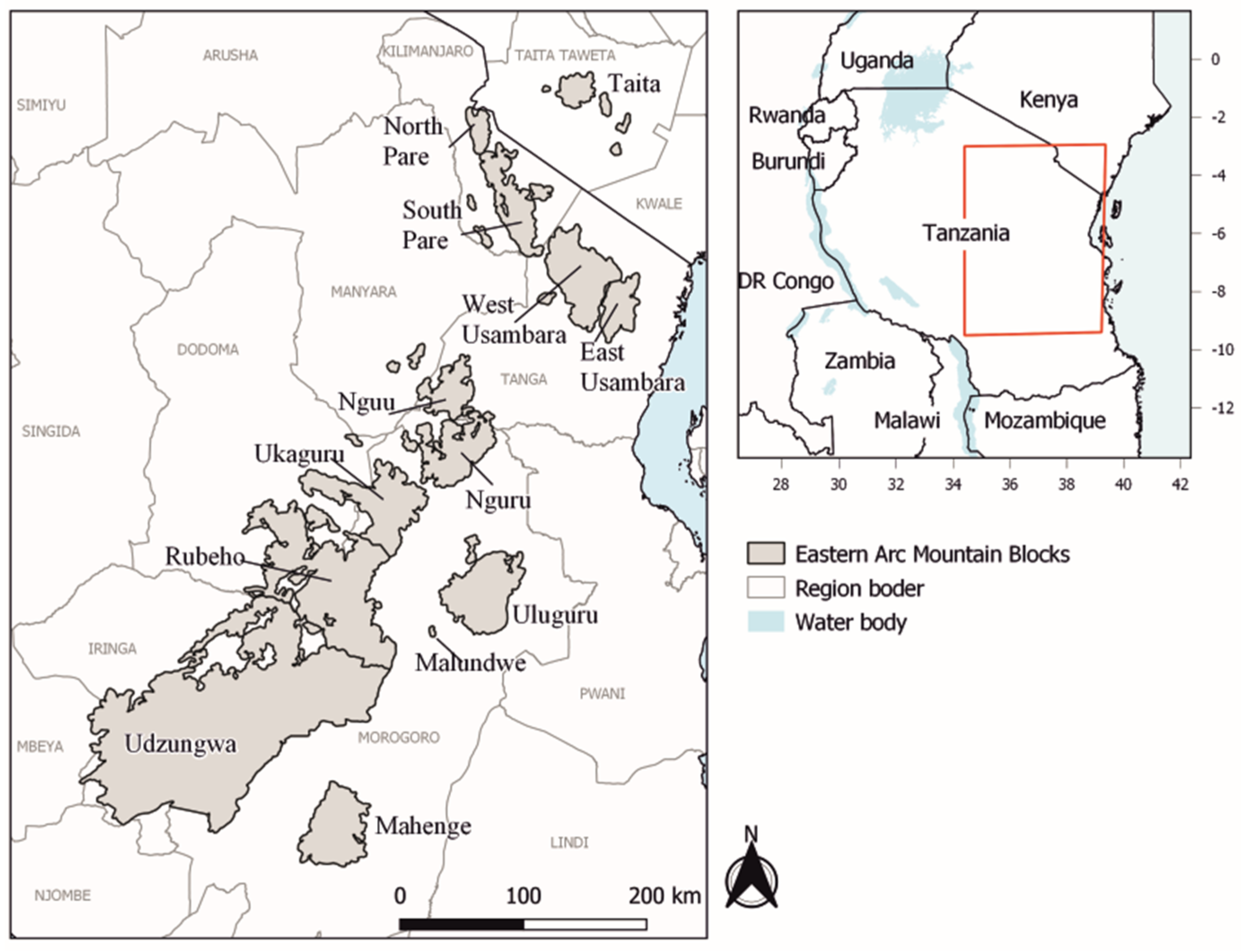
2.1. Study Area
2.2. Methods
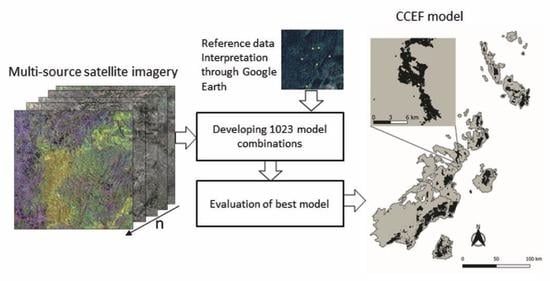
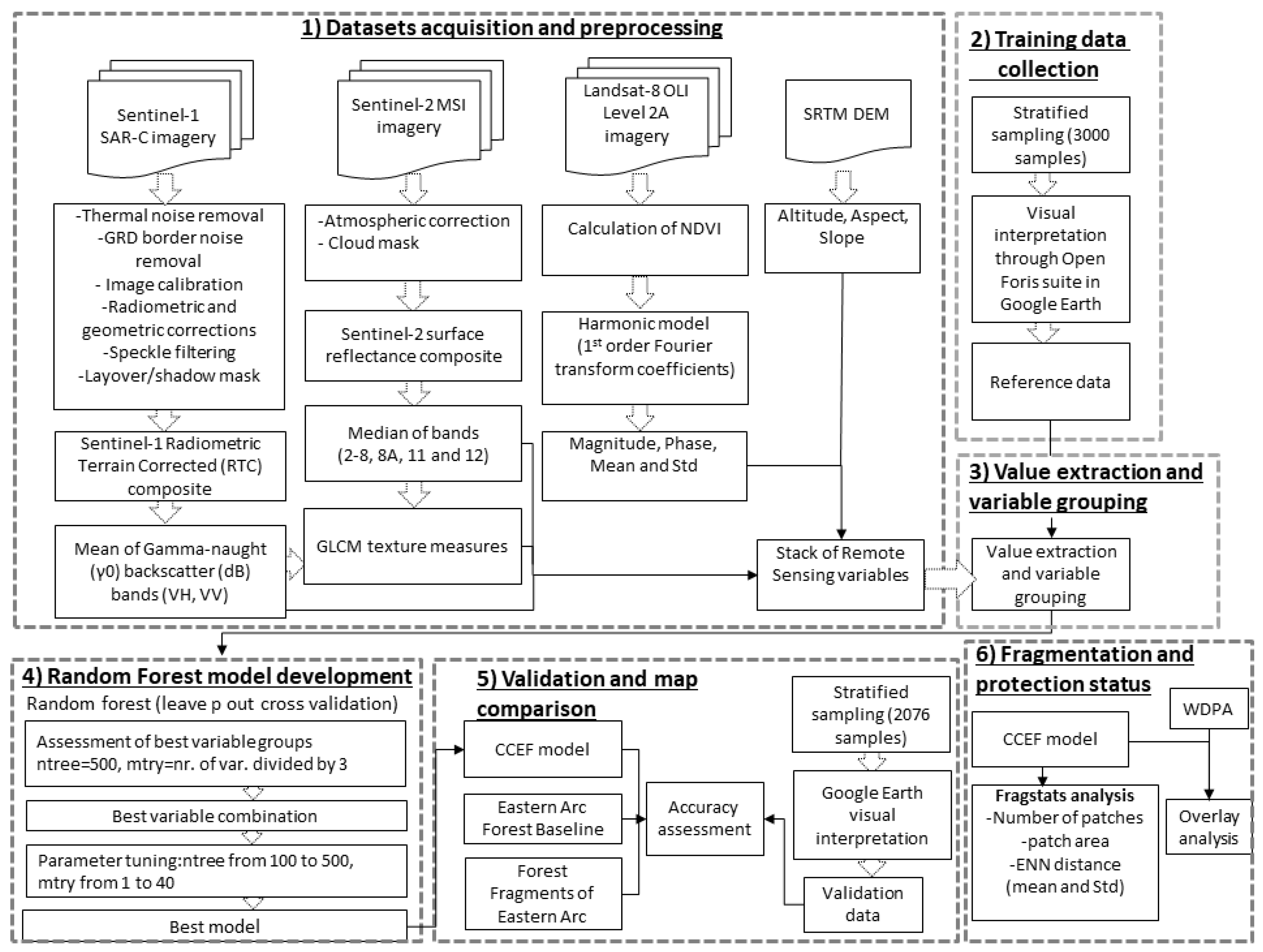
2.2.1. Methodological Design
2.2.2. Data Sets and Preprocessing
2.2.3. Training Data Collection
2.2.4. Value Extraction
2.2.5. Variable Grouping
2.2.6. Random Forest Model Development
2.2.7. Validation and Map Comparison
2.2.8. Fragmentation and Protection Status
3. Results
3.1. Variable Selection and Parameter Adjustment
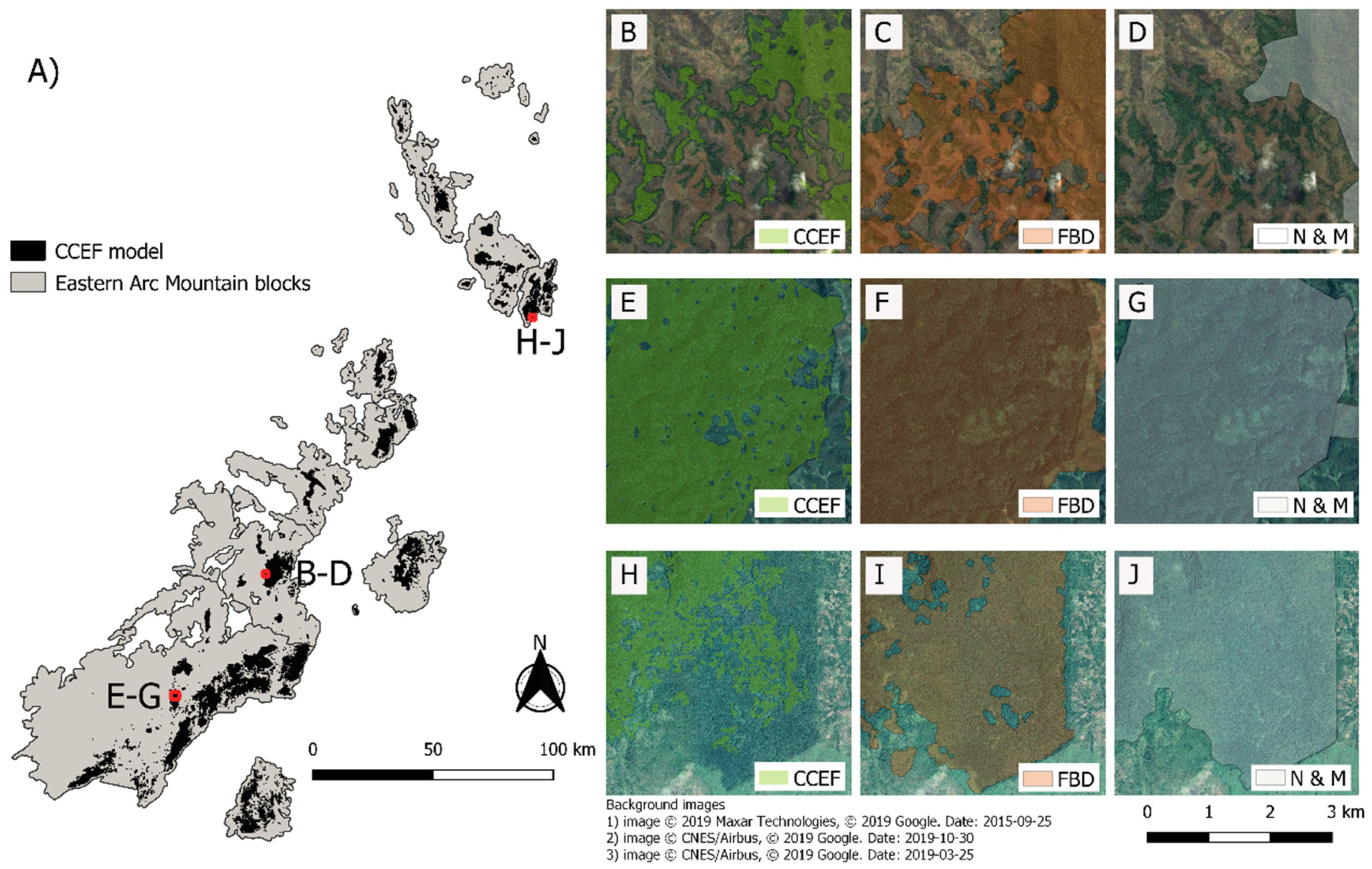
3.2. Coverage and Distribution of the Eastern Arc Forest
4. Discussion
4.1. Extent, Fragmentation, and Protection of Eastern Arc Forests
4.2. Accuracy of the CCEF Model
4.3. Optimal Modelling Setup for Detecting Closed Canopy Natural Forests
4.4. Global Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, C.; Sodhi, N.; Peh, K.; Brook, B. Global evidence that deforestation amplifies flood risk and severity in the developing world. Glob. Chang. Biol. 2007, 13, 2379–2395. [Google Scholar] [CrossRef]
- Brinck, K.; Fischer, R.; Groeneveld, J.; Lehmann, S.; Dantas De Paula, M.; Pütz, S.; Sexton, J.O.; Song, D.; Huth, A. High resolution analysis of tropical forest fragmentation and its impact on the global carbon cycle. Nat. Commun. 2017, 8, 14855. [Google Scholar] [CrossRef] [PubMed]
- Brook, B.W.; Sodhi, N.S.; Bradshaw, C.J.A. Synergies among extinction drivers under global change. Trends Ecol. Evol. 2008, 23, 453–460. [Google Scholar] [CrossRef]
- Bellard, C.; Bertelsmeier, C.; Leadley, P.; Thuiller, W.; Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 2012, 15, 365–377. [Google Scholar] [CrossRef]
- Garcia, R.A.; Cabeza, M.; Rahbek, C.; Araújo, M.B. Multiple Dimensions of Climate Change and Their Implications for Biodiversity. Science 2014, 344, 6183. [Google Scholar] [CrossRef]
- Cartwright, J. Ecological islands: Conserving biodiversity hotspots in a changing climate. Front. Ecol Environ. 2019, 17, 331–340. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef]
- Taubert, F.; Fischer, R.; Groeneveld, J.; Lehmann, S.; Müller, M.S.; Rödig, E.; Wiegand, T.; Huth, A. Global patterns of tropical forest fragmentation. Nature 2018, 554, 519–522. [Google Scholar] [CrossRef]
- Ganivet, E.; Bloomberg, M. Towards rapid assessments of tree species diversity and structure in fragmented tropical forests: A review of perspectives offered by remotely sensed and field-based data. For. Ecol. Manag. 2019, 432, 40–53. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Turner, W.R.; Larsen, F.W.; Brooks, T.M.; Gascon, C. Global biodiversity conservation: The critical role of hotspots. In Biodiversity Hotspots–Distribution and Protection of Conservation Priority Areas; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin, Germany, 2011; pp. 3–22. [Google Scholar]
- Hrdina, A.; Romportl, D. Evaluating global biodiversity hotspots–Very rich and even more endangered. J. Landsc. Ecol. 2017, 10, 108–115. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Decocq, G.; Andrieu, E.; Brunet, J.; Chabrerie, O.; De Frenne, P.; De Smedt, P.; Deconchat, M.; Diekmann, M.; Ehrmann, S.; Giffard, B.; et al. Ecosystem services from small forest patches in agricultural landscapes. Curr. For. Rep. 2016, 2, 30–44. [Google Scholar] [CrossRef]
- MacArthur, R.H.; Wilson, E.O. The Theory of Island Biogeography; Princeton University Press: Princeton, NJ, USA, 1967. [Google Scholar]
- Gibson, L.; Lynam, A.J.; Bradshaw, C.J.A.; He, F.; Bickford, D.P.; Woodruff, D.S.; Bumrungsri, S.; Laurance, W.F. Near-complete extinction of native small mammal fauna 25 years after forest fragmentation. Science 2013, 341, 1508–1510. [Google Scholar] [CrossRef]
- Pimm, S.L.; Brooks, T. Conservation: Forest Fragments, Facts, and Fallacies. Curr. Biol. 2014, 23, R1098–R1101. [Google Scholar] [CrossRef][Green Version]
- Tulloch, A.I.T.; Barnes, M.D.; Ringma, J.; Fuller, R.A.; Watson, J.E.M. Understanding the importance of small patches of habitat for conservation. J. Appl. Ecol. 2016, 53, 418–429. [Google Scholar] [CrossRef]
- Wintle, B.A.; Kujala, H.; Whitehead, A.; Cameron, A.; Veloz, S.; Kukkala, A.; Moilanen, A.; Gordon, A.; Lentini, P.E.; Cadenhead, N.C.R.; et al. Global synthesis of conservation studies reveals the importance of small habitat patches for biodiversity. Proc. Natl. Acad. Sci. USA 2019, 116, 909–914. [Google Scholar] [CrossRef]
- Sloan, S.; Jenkins, C.N.; Joppa, L.N.; Gaveau, D.L.A.; Lawrence, W.F. Remaining natural vegetation in the global biodiversity hotspots. Biol. Conserv. 2014, 177, 12–24. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 851–853. [Google Scholar] [CrossRef]
- Potapov, P.; Hansen, M.C.; Lastadius, L.; Turubanova, S.; Yaroshenko, A.; Thies, C.; Smith, W.; Zhuravleva, I.; Komarova, A.; Minnemeyer, S.; et al. The last frontiers of wilderness: Tracking loss of intact forest landscapes from 2017, 2000 to 2013. Sci. Adv. 2017, 3, e1600821. [Google Scholar] [CrossRef]
- Tuykavina, A.; Hansen, M.C.; Potapov, P.V.; Krylov, A.M.; Goetz, S.J. Pan-tropical hinterland forests: Mapping minimally disturbed forests. Glob. Ecol. Biogeogr. 2016, 25, 151–163. [Google Scholar] [CrossRef]
- Lechner, A.M.; Stein, A.; Jones, S.D.; Ferwerda, J.G. Remote sensing of small and linear features: Quantifying the effects of patch size and length, grid position and detectability on land cover mapping. Remote Sens. Environ. 2009, 113, 2194–2204. [Google Scholar] [CrossRef]
- Langner, A.; Miettinen, J.; Kukkonen, M.; Vancutsem, C.; Simonetti, D.; Vieilledent, G.; Verhegghen, A.; Gallego, J.; Stibig, H.-J. Towards Operational Monitoring of Forest Canopy Disturbance in Evergreen Rain Forests: A Test Case in Continental Southeast Asia. Remote Sens. 2018, 10, 544. [Google Scholar] [CrossRef]
- Lima, T.A.; Beuchle, R.; Lagner, A.; Grecchi, R.C.; Griess, V.C.; Achard, F. Comparing Sentinel-2 MSI and Landsat 8 OLI Imagery for Monitoring Selective Logging in the Brazilian Amazon. Remote Sens. 2019, 11, 961. [Google Scholar] [CrossRef]
- Laurin, G.V.; Puletti, N.; Hawthorne, W.; Liesenberg, V.; Corona, P.; Papale, D.; Chen, Q.; Valentini, R. Discrimination of tropical forest types, dominant species, and mapping of functional guilds by hyperspectral and simulated multispectral Sentinel-2 data. Remote Sens. Environ. 2016, 176, 163–176. [Google Scholar] [CrossRef]
- Banskota, A.; Kayastha, N.; Falkowski, M.J.; Wulder, M.A.; Froese, R.E.; White, J.C. Forest monitoring using landsat time series data: A review. Can. J. Remote Sens. 2014, 40, 362–384. [Google Scholar] [CrossRef]
- Laurin, G.V.; Balling, J.; Corona, P.; Mattioli, W.; Papale, D.; Puletti, N.; Rizzo, M.; Truckenbrodt, J.; Urban, M. Above-ground biomass prediction by sentinel-1 multitemporal data in central Italy with integration of alos2 and sentinel-2 data. J. Appl. Remote Sens. 2018, 12, 016008. [Google Scholar] [CrossRef]
- Vafaei, S.; Soosani, J.; Adeli, K.; Fadaei, H.; Naghavi, H.; Pham, T.D.; Bui, T.D. Improving accuracy estimation of forest aboveground biomass based on incorporation of alos-2 palsar-2 and sentinel-2a imagery and machine learning: A case study of the hyrcanian forest area (Iran). Remote Sens. 2018, 10, 172. [Google Scholar] [CrossRef]
- Marston, C.; Giraudoux, P. On the Synergistic Use of Optical and SAR Time-Series Satellite Data for Small Mammal Disease Host Mapping. Remote Sens. 2019, 11, 39. [Google Scholar] [CrossRef]
- Whyte, A.; Ferentinos, K.P.; Petropoulos, G.P. A new synergistic approach for monitoring wetlands using Sentinels-1 and 2 data with object-based machine learning algorithms. Environ. Model. Softw. 2018, 104, 40–54. [Google Scholar] [CrossRef]
- Torbick, N.; Ledoux, L.; Salas, W.; Zhao, M. Regional mapping of plantation extent using multisensor imagery. Remote Sens. 2016, 8, 236. [Google Scholar] [CrossRef]
- Ghosh, S.M.; Behera, M.D. Aboveground biomass estimation using multi-sensor data synergy and machine learning algorithms in a dense tropical forest. Appl. Geogr. 2018, 96, 29–40. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Schepaschenko, D.; See, L.; Lesiv, M.; Bastin, J.-F.; Mollicone, D.; Tsendbazar, N.E.; Bastin, L.; McCallum, I.; Laso Bayas, J.C.; Baklanov, A.; et al. Recent advances in forest observation with visual interpretation of very high-resolution imagery. Surv. Geophys. 2019, 40, 839–862. [Google Scholar] [CrossRef]
- Newmark, W.D. Forest Area, Fragmentation, and Loss in the Eastern Arc Mountains: Implications for the Conservation of Biological Diversity. J. East. Afr. Nat. Hist. 1998, 87, 29–36. [Google Scholar] [CrossRef]
- Burgess, N.D.; Butynski, T.M.; Cordeiro, N.J.; Doggart, N.H.; Fjeldsa, J.; Howell, K.M.; Kilahama, F.B.; Loader, S.P.; Lovett, J.C.; Mbilinyi, B.; et al. The biological importance of the Eastern Arc Mountains of Tanzania and Kenya. Biol. Conserv. 2007, 134, 209–231. [Google Scholar] [CrossRef]
- Newmark, W.D.; McNeally, P.B. Impact of habitat fragmentation on the spatial structure of the Eastern Arc forests in East Africa: Implications for biodiversity conservation. Biodivers. Conserv. 2018, 27, 1387–1402. [Google Scholar] [CrossRef]
- Balmford, A.; Moore, J.L.; Brooks, T.; Burgess, N.; Hansen, L.A.; Williams, P.; Rahbek, C. Conservation Conflicts Across Africa. Science 2001, 291, 2616–2619. [Google Scholar] [CrossRef]
- Brooks, T.M.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Rylands, A.B.; Konstant, W.R.; Flick, P.; Pilgrim, J.; Oldfield, S.; Magin, G.; et al. Habitat loss and extinction in the hotspots of biodiversity. Conserv. Biol. 2002, 16, 909–923. [Google Scholar] [CrossRef]
- Burgess, N.D.; Lovett, J.; Rodgers, A.; Kilahama, F.; Nashanda, E.; Davenport, T.; Butynski, T. Eastern Arc Mountains and Southern Rift. In Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Ecoregions, 2nd ed.; Mittermeier, R.A., Robles-Gil, P., Hoffmann, M., Pilgrim, J.D., Brooks, T.M., Mittermeier, C.G., Lamoreux, J.L., Fonseca, G.A.B., Eds.; Cemex: San Pedro Garza, Mexico, 2004; pp. 245–255. [Google Scholar]
- Hall, J.; Burgess, N.D.; Lovett, J.; Mbilinyi, B.; Gereau, R.E. Conservation implications of deforestation across an elevational gradient in the Eastern Arc Mountains, Tanzania. Biol. Conserv. 2009, 142, 2510–2521. [Google Scholar] [CrossRef]
- Newmark, W.D. Conserving Biodiversity in East African forests: A study of the Eastern Arc Mountains. In Ecological Studies; Springer: Heidelberg, Germany, 2002; Volume 155, p. 195. [Google Scholar]
- Forestry and Beekeeping Division. Forest Area Baseline for the Eastern Arc Mountains; Mbilinyi, B.P., Malimbwi, R.E., Shemwetta, D.T.K., Songorwa, Zahabu, E., Katani, J.Z., Kashaigili, J., Eds.; For Conservation and Management of the Eastern Arc Mountain Forests; Forestry and Beekeeping Division: Dar es Salaam, Tanzania, 2006. [Google Scholar]
- Lindenmayer, D. Small patches make critical contributions to biodiversity conservation. Proc. Natl. Acad. Sci. USA 2019, 116, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Platts, P.J.; Burgess, N.D.; Gereau, R.E.; Lovett, J.C.; Marshall, A.R.; McClean, C.J.; Pellikka, P.K.E.; Swetnam, R.D.; Marchant, R. Delimiting tropical mountain ecoregions for conservation. Environ. Conserv. 2011, 38, 312–332. [Google Scholar] [CrossRef]
- Lovett, J.C. Importance of the Eastern Arc Mountains for Vascular Plants. J. E. Afr. Nat. Hist. 1998, 87, 59–74. [Google Scholar] [CrossRef][Green Version]
- Davenport, T.R.B.; Stanley, W.T.; Sargis, E.J.; De Luca, D.W.; Mpunga, N.E.; Machaga, S.J.; Olson, L.E. A new genus of African Monkey, Rungwecebus: Morphology, ecology, and molecular phylogenetics. Science 2006, 312, 1378–1381. [Google Scholar] [CrossRef] [PubMed]
- Rovero, F.; Marshall, A.R.; Jones, T.; Perkin, A. The primates of Udzungwa mountains: Diversity, ecology and conservation. J. Anthropol. Sci. 2009, 87, 93–126. [Google Scholar] [PubMed]
- Swetnam, R.D.; Fisher, B.; Mbilinyi, B.; Munishi, P.K.T.; Willcock, S.; Ricketts, T.; Mwakalila, S.; Balmford, A.; Burgess, N.D.; Marshall, A.R. Mapping socio-economic scenarios of land cover change: A GIS method to enable ecosystem service modelling. J. Environ. Manag. 2011, 92, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Green, J.M.H.; Larrosa, C.; Burgess, N.D.; Balmford, A.; Johnston, A.; Mbilinyi, B.; Platts, P.J.; Coad, L. Deforestation in an African biodiversity hotspot: Extent, variation and the effectiveness of protected areas. Biol. Conserv. 2013, 164, 62–72. [Google Scholar] [CrossRef]
- Protected Planet, UNEP-WCMC and IUCN. The World Database on Protected Areas (WDPA) UNEP-WCMC and IUCN. 2019. Available online: www.protectedplanet.net (accessed on 12 December 2019).
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Kempeneers, P.; Soille, P. Optimizing Sentinel-2 image selection in a Big Data context. Big Earth Data 2017, 1, 145–148. [Google Scholar] [CrossRef]
- Martins, V.S.; Barbosa, C.C.F.; de Carvalho, L.A.S.; Jorge, D.S.F.; Lobo, F.D.L.; de Moraes Novo, E.M.L. Assessment of atmospheric correction methods for Sentinel-2 MSI images applied to Amazon floodplain lakes. Remote Sens. 2017, 9, 322. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Ottosen, T.-B.; Petch, G.; Hason, M.; Skjøth, C.A. Tree cover mapping based on Sentinel-2 images demonstrate high thematic accuracy in Europe. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101947. [Google Scholar] [CrossRef]
- Torres, R.; Snoeij, P.; Geudtner, D.; Bibby, D.; Davidson, M.; Attema, E.; Potin, P.; Rommen, B.; Floury, N.; Brown, M. Gmes sentinel-1 mission. Remote Sens. Environ. 2012, 120, 9–24. [Google Scholar] [CrossRef]
- Dostálová, A.; Wagner, W.; Milenković, M.; Hollaus, M. Annual seasonality in Sentinel-1 signal for forest mapping and forest type classification. Int. J. Remote Sens. 2018, 39, 7738–7760. [Google Scholar] [CrossRef]
- SNAP-ESA Sentinel Application Platform v6.0. Available online: http://step.esa.int (accessed on 7 September 2018).
- Filipponi, F. Sentinel-1 GRD Preprocessing Workflow. Proceedings 2019, 18, 11. [Google Scholar] [CrossRef]
- Gómez, C.; White, J.C.; Wulder, M.A. Optical remotely sensed time series data for land cover classification: A review. ISPRS J. Photogramm. Remote Sens. 2016, 116, 55–72. [Google Scholar] [CrossRef]
- Jakubauskas, M.E.; Legates, D.R.; Kastens, J.H. Harmonic analysis of time-series AVHRR NDVI data. Photogramm. Eng. Remote Sens. 2001, 67, 461–470. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Texture features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Hall-Beyer, M. Practical guidelines for choosing GLCM textures to use in landscape classification tasks over a range of moderate spatial scales. Int. J. Remote Sens. 2017, 38, 5. [Google Scholar] [CrossRef]
- Sesnie, S.E.; Gessler, P.E.; Finegan, B.; Thessler, S. Integrating Landsat TM and SRTM-DEM derived variables with decision trees for habitat classification and change detection in complex neotropical environments. Remote Sens. Environ. 2008, 112, 2145–2159. [Google Scholar] [CrossRef]
- Sexton, J.O.; Song, X.-P.; Feng, M.; Noojipady, P.; Anand, A.; Huang, C.; Kim, D.-H.; Collins, K.M.; Channan, S.; DiMiceli, C.; et al. Global, 30-m resolution continuous fields of tree cover: Landsat-based rescaling of MODIS vegetation continuous fields with lidar-based estimates of error. Int. J. Dig. Earth 2013, 6, 427–448. [Google Scholar] [CrossRef]
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-Filled Seamless SRTM Data (Online) V4. International Centre for Tropical Agriculture (CIAT). 2008. Available online: http://srtm.csi.cgiar.org (accessed on 7 January 2019).
- Saah, D.; Johnson, G.; Ashmall, B.; Tondapu, G.; Tenneson, K.; Patterson, M.; Poortinga, A.; Markert, K.; Quyen, N.H.; San Aung, K.; et al. Collect Earth: An online tool for systematic reference data collection in land cover and use applications. Environ. Model. Softw. 2019, 118, 166–171. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X.; Chen, B.; Torbick, N.; Jin, C.; Zhang, G.; Biradar, C. Mapping deciduous rubber plantations through integration of PALSAR and multitemporal Landsat imagery. Remote Sens. Environ. 2013, 134, 392–402. [Google Scholar] [CrossRef]
- Koskinen, J.; Leinonen, U.; Vollrath, A.; Ortmann, A.; Lindquist, E.; d’Annunzio, R.; Pekkarinen, A.; Käyhkö, N. Participatory mapping of forest plantations with Open Foris and Google Earth Engine. ISPRS J. Photogramm. Remote Sens. 2019, 148, 63–74. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.J.; Rigol-Sanchez, P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Lusk, C.H.; McGlone, M.S.; Overton, J.M. Climate predicts the proportion of divaricate plant species in New Zealand arborescent assemblages. J. Biogeogr. 2016, 43, 1881–1892. [Google Scholar] [CrossRef]
- Kukkonen, M.O.; Muhammad, M.J.; Käyhkö, N.; Luoto, M. Urban expansion in Zanzibar City, Tanzania: Analyzing quantity, spatial patterns and effects of alternative planning approaches. Land Use Policy 2018, 71, 554–565. [Google Scholar] [CrossRef]
- Korfanta, N.; Newmark, W.D.; Kaufman, M.J. Long-term demographic consequences of habitat fragmentation to a tropical understory bird community. Ecology 2012, 93, 2548–2559. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, N.J.; Borghesio, L.; Joho, M.P.; Monoski, T.J.; Mkongewa, V.J.; Dampf, C.J. Forest fragmentation in an African biodiversity hotspot impacts mixed-species bird flocks. Biol. Conserv. 2015, 188, 61–71. [Google Scholar] [CrossRef]
- McGarigal, K.; Cushman, S.A.; Ene, E. FRAGSTATS v4: Spatial Pattern Analysis Program for Categorical and Continuous Maps. Computer Software Program Produced by the Authors at the University of Massachusetts, Amherst. 2012. Available online: http://www.umass.edu/landeco/research/fragstats/fragstats.html (accessed on 20 September 2019).
- Doggart, N.; Perkin, A.; Kiure, J.; Fjeldså, J.; Poynton, J.; Burgess, N. Changing places: How the results of new field work in the Rubeho Mountains influence conservation priorities in the Eastern Arc Mountains of Tanzania. Afr. J. Ecol. 2006, 44, 134–144. [Google Scholar] [CrossRef]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good practices for estimating area and assessing accuracy of land change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Lu, D.; Li, G.; Moran, E.; Dutra, E.; Batistella, M. The roles of textural images in improving land-cover classification in the Brazilian Amazon. Int. J. Remote Sens. 2014, 35, 24. [Google Scholar] [CrossRef]
- Erinjery, J.J.; Singh, M.; Kent, R. Mapping and assessment of vegetation types in the tropical rainforests of the Western Ghats using multispectral Sentinel-2 and SAR Sentinel-1 satellite imagery. Remote Sens. Environ. 2018, 216, 345–354. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, X.; Chen, Y.; Liang, X. Land-cover mapping using Random Forest classification and incorporating NDVI time-series and texture: A case study of central Shandong. Int. J. Remote Sens. 2018, 39, 23. [Google Scholar] [CrossRef]
- Chiang, S.-H.; Valdez, M. Tree Species Classification by Integrating Satellite Imagery and Topographic Variables Using Maximum Entropy Method in a Mongolian Forest. Forests 2019, 10, 961. [Google Scholar] [CrossRef]
- Aguilar, C.; Herrero, J.; Polo, M. Topographic effects on solar radiation distribution in mountainous watersheds and their influence on reference evapotranspiration estimates at watershed scale. Hydrol. Earth Syst. Sci. 2010, 14, 2479–2494. [Google Scholar] [CrossRef]
- Crowson, M.; Warren-Thomas, E.; Hill, J.K.; Hariyadi, B.; Agu, F.; Saad, A.; Hamer, K.C.; Hodgson, J.A.; Kartika, W.D.; Lucey, J.; et al. A comparison of satellite remote sensing data fusion methods to map peat swamp forest loss in Sumatra, Indonesia. Remote Sens. Ecol. Conserv. 2018, 5, 247–258. [Google Scholar] [CrossRef]
- Barrett, B.; Raab, C.; Cawkwell, F.; Green, S. Upland vegetation mapping using Random Forests with optical and radar satellite data. Remote Sens. Ecol. Conserv. 2016, 2, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Mauya, E.W.; Koskinen, J.; Tegel, K.; Hämäläinen, J.; Kauranne, T.; Käyhkö, N. Modelling and Predicting the Growing Stock Volume in Small-Scale Plantation Forests of Tanzania Using Multi-Sensor Image Synergy. Forests 2019, 10, 279. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; Millán, V.E.G.; Strasser, T.; Borre, J.V.; Toon, S.; Förster, M. Remote sensing for mapping natural habitats and their conservation status−New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Schmidt, J.; Fassnacht, F.E.; Forster, M.; Schmidtlein, S. Synergetic use of Sentinel-1 and Sentinel-2 for assessments of heathland conservation status. Remote Sens. Ecol. Conserv. 2018, 4, 225–239. [Google Scholar] [CrossRef]
- Harris, G.M.; Jenkins, C.N.; Pimm, S.L. Refining biodiversity conservation priorities. Conserv. Biol. 2005, 19, 1957–1968. [Google Scholar] [CrossRef]
- Murray-Smith, C.; Brummitt, N.A.; Oliveira-Filho, A.T.; Bachman, S.; Moat, J.; Lughadha, E.M.N.; Lucas, E.J. Plant diversity hotspots in the Atlantic Coastal Forests of Brazil. Conserv. Biol. 2009, 23, 151–163. [Google Scholar] [CrossRef]
- Noroozi, J.; Talebi, A.; Doostmohammadi, M.; Rumpf, S.B.; Linder, H.P.; Schneeweiss, G.M. Hotspots within a global biodiversity hotspot-areas of endemism are associated with high mountain ranges. Sci. Rep. 2018, 8, 10345. [Google Scholar] [CrossRef]
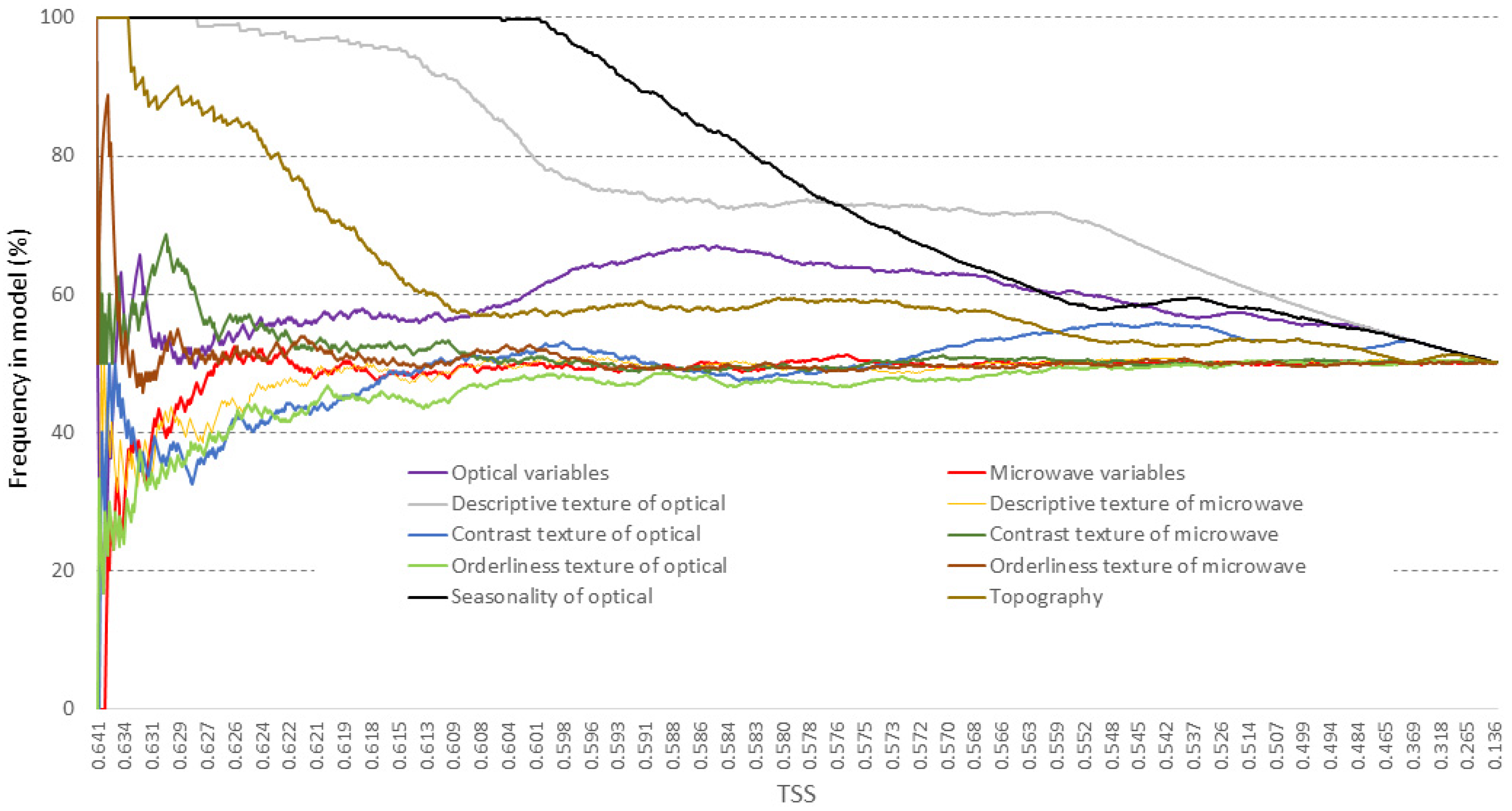

| Group | Variables within Group | Number of Variables |
|---|---|---|
| Optical variables | Sentinel-2 bands (B2–B8A, B11–B12) | 10 |
| Microwave variables | Sentinel-1 bands (VH, VV) | 2 |
| Descriptive texture of optical | Descriptive group haralicks (mean, variance, correlation) of Sentinel-2 bands | 30 |
| Descriptive texture of microwave | Descriptive group haralicks (mean, variance, correlation) of Sentinel-1 bands | 6 |
| Contrast texture of optical | Contrast group haralicks (contrast, dissimilarity, homogeneity) of Sentinel-2 bands | 30 |
| Contrast texture of microwave | Contrast group haralicks (contrast, dissimilarity, homogeneity) of Sentinel-1 bands | 6 |
| Orderliness texture of optical | Orderliness group haralicks (energy, entropy) of Sentinel-2 bands | 20 |
| Orderliness texture of microwave | Orderliness group haralicks (energy, entropy) of Sentinel-1 bands | 4 |
| Seasonality of optical | Seasonality of Landsat-8 derived NDVI (magnitude, phase, mean, std) | 4 |
| Topography | Topography (altitude, slope, aspect) | 3 |
| CCEF Model | FBD [45] | Newmark and McNeally [39] | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimated Area (ha) | Map Area (ha) | Map OA | Map TSS | Map Area (ha) | Map OA | Map TSS | Map Area (ha) | Map OA | Map TSS | |
| Taita | 607 ± 73 | 623 | 0.76 | 0.36 | - | - | - | 590 | 0.76 | 0.34 |
| North Pare | 2287 ± 295 | 2648 | 0.7 | 0.39 | 2700 | 0.8 | 0.59 | 1546 | 0.8 | 0.61 |
| South Pare | 11,174 ± 834 | 10,942 | 0.87 | 0.72 | 13,800 | 0.75 | 0.34 | 11,380 | 0.78 | 0.47 |
| West Usambara | 23,116 ± 1352 | 23,578 | 0.83 | 0.62 | 31,900 | 0.66 | 0.17 | 28,213 | 0.76 | 0.39 |
| East Usambara | 25,995 ± 1603 | 21,779 | 0.72 | 0.55 | 26,300 | 0.59 | 0.09 | 30,110 | 0.86 | 0.57 |
| Nguu | 12,472 ± 766 | 11,799 | 0.89 | 0.82 | 18,800 | 0.69 | 0.23 | 16,170 | 0.78 | 0.48 |
| Nguru | 21,897 ± 1340 | 22,666 | 0.84 | 0.63 | 29,700 | 0.69 | 0.23 | 29,927 | 0.73 | 0.28 |
| Uluguru | 22,704 ± 1291 | 25,243 | 0.83 | 0.56 | 26,698 | 0.74 | 0.38 | 25,963 | 0.87 | 0.68 |
| Ukaguru | 12,993 ± 678 | 13,700 | 0.93 | 0.87 | 17,271 | 0.6 | 0.01 | 17,742 | 0.8 | 0.50 |
| Rubeho | 35,212 ± 1519 | 32,394 | 0.88 | 0.73 | 47,131 | 0.69 | 0.21 | 42,274 | 0.76 | 0.38 |
| Malundwe | 803 ± 68 | 344 | 0.9 | 0.77 | 174 | 0.52 | 0.18 | 1141 | 0.58 | 0 |
| Mahenge | 11,789 ± 1235 | 11,214 | 0.77 | 0.48 | 1918 | 0.42 | 0.17 | 5474 | 0.3 | −0.38 |
| Udzungwa | 150,254 ± 5000 | 119,279 | 0.75 | 0.50 | 135,276 | 0.6 | 0.20 | 195,321 | 0.66 | 0.20 |
| Total | 333,978 ± 5621 | 296,209 | 0.8 | 0.57 | 351,668 | 0.63 | 0.21 | 405,851 | 0.72 | 0.41 |
| Fragmentation and Isolation | Protection Status | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Num.of Patches | Num.of Patches (<10 ha) | Forest Area (<10 ha) (%) | ENN Mean (m) | ENN StD (m) | Mean Forest Patch Area (ha) | Small Forest Patches (1–10 ha) Unprotected (%) | Forests Unprotected (%) | Forest Protected with IUCN (%) | Forests Protected (%) | |
| Taita | 64 | 58 | 26.8 | 224 | 477 | 10 | 100 | 100 | - | - |
| North Pare | 92 | 74 | 7 | 268 | 486 | 29 | 66 | 41.1 | 53.9 | 58.9 |
| South Pare | 226 | 195 | 5.1 | 260 | 569 | 48 | 62 | 13.4 | 3 | 86.6 |
| West Usambara | 411 | 342 | 3.9 | 191 | 507 | 57 | 63 | 23.3 | 75.6 | 76.7 |
| East Usambara | 491 | 433 | 4.9 | 120 | 170 | 44 | 53 | 27.8 | 72.2 | 72.2 |
| Nguu | 167 | 147 | 3.5 | 416 | 1683 | 71 | 35 | 13.3 | 70.8 | 86.7 |
| Nguru | 298 | 262 | 2.8 | 199 | 298 | 76 | 92 | 77.7 | 21.8 | 22.3 |
| Uluguru | 732 | 691 | 6.5 | 204 | 463 | 34 | 87 | 19.3 | 1.1 | 80.7 |
| Ukaguru | 177 | 152 | 3 | 371 | 895 | 77 | 79 | 15.6 | 31.8 | 84.4 |
| Rubeho | 570 | 496 | 4 | 250 | 797 | 57 | 35 | 38 | 54.7 | 62 |
| Malundwe | 29 | 28 | 22.9 | 171 | 272 | 12 | 0 | 0 | 100 | 100 |
| Mahenge | 1569 | 1446 | 33 | 204 | 351 | 7 | 97 | 89.5 | 0.2 | 10.5 |
| Udzungwa | 5259 | 4739 | 9.8 | 190 | - | 22 | 19 | 6.4 | 44.8 | 93.6 |
| Total/Mean | 10,085 | 9063 | 7.7 | 203 | 581 | 29 | 88 | 23.9 | 42.1 | 76.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koskikala, J.; Kukkonen, M.; Käyhkö, N. Mapping Natural Forest Remnants with Multi-Source and Multi-Temporal Remote Sensing Data for More Informed Management of Global Biodiversity Hotspots. Remote Sens. 2020, 12, 1429. https://doi.org/10.3390/rs12091429
Koskikala J, Kukkonen M, Käyhkö N. Mapping Natural Forest Remnants with Multi-Source and Multi-Temporal Remote Sensing Data for More Informed Management of Global Biodiversity Hotspots. Remote Sensing. 2020; 12(9):1429. https://doi.org/10.3390/rs12091429
Chicago/Turabian StyleKoskikala, Joni, Markus Kukkonen, and Niina Käyhkö. 2020. "Mapping Natural Forest Remnants with Multi-Source and Multi-Temporal Remote Sensing Data for More Informed Management of Global Biodiversity Hotspots" Remote Sensing 12, no. 9: 1429. https://doi.org/10.3390/rs12091429
APA StyleKoskikala, J., Kukkonen, M., & Käyhkö, N. (2020). Mapping Natural Forest Remnants with Multi-Source and Multi-Temporal Remote Sensing Data for More Informed Management of Global Biodiversity Hotspots. Remote Sensing, 12(9), 1429. https://doi.org/10.3390/rs12091429