Hierarchical Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing with Spectral Variability
Abstract
1. Introduction
- It introduces the new assumption that endmember variability can be represented by the conical hulls of prototypal endmember spectra.
- It derives the resulting mixing model, which, coupled with a hierarchically structured group-sparsity constraint, allow for the prototypal endmember spectra to be automatically extracted and clustered from the data.
- It develops the associated unmixing algorithm that jointly estimates the prototypal endmembers and the abundances within an unsupervised framework.
2. Related Works
2.1. LMM with Fixed Endmember Spectra
2.2. LMM with Pixel-Wise Endmember Spectra
2.2.1. Spectral Library-Free Mixing Models
2.2.2. Bundle-Based Mixing Models
2.2.3. Towards Library-Free Bundle-Based Models
- which is able to propose a bundle-driven description of the spectral variability to ensure physically sound interpretation of this variability; and,
- which does not require the prior knowledge of a spectral library, akin to the unsupervised methods.
3. Proposed Model
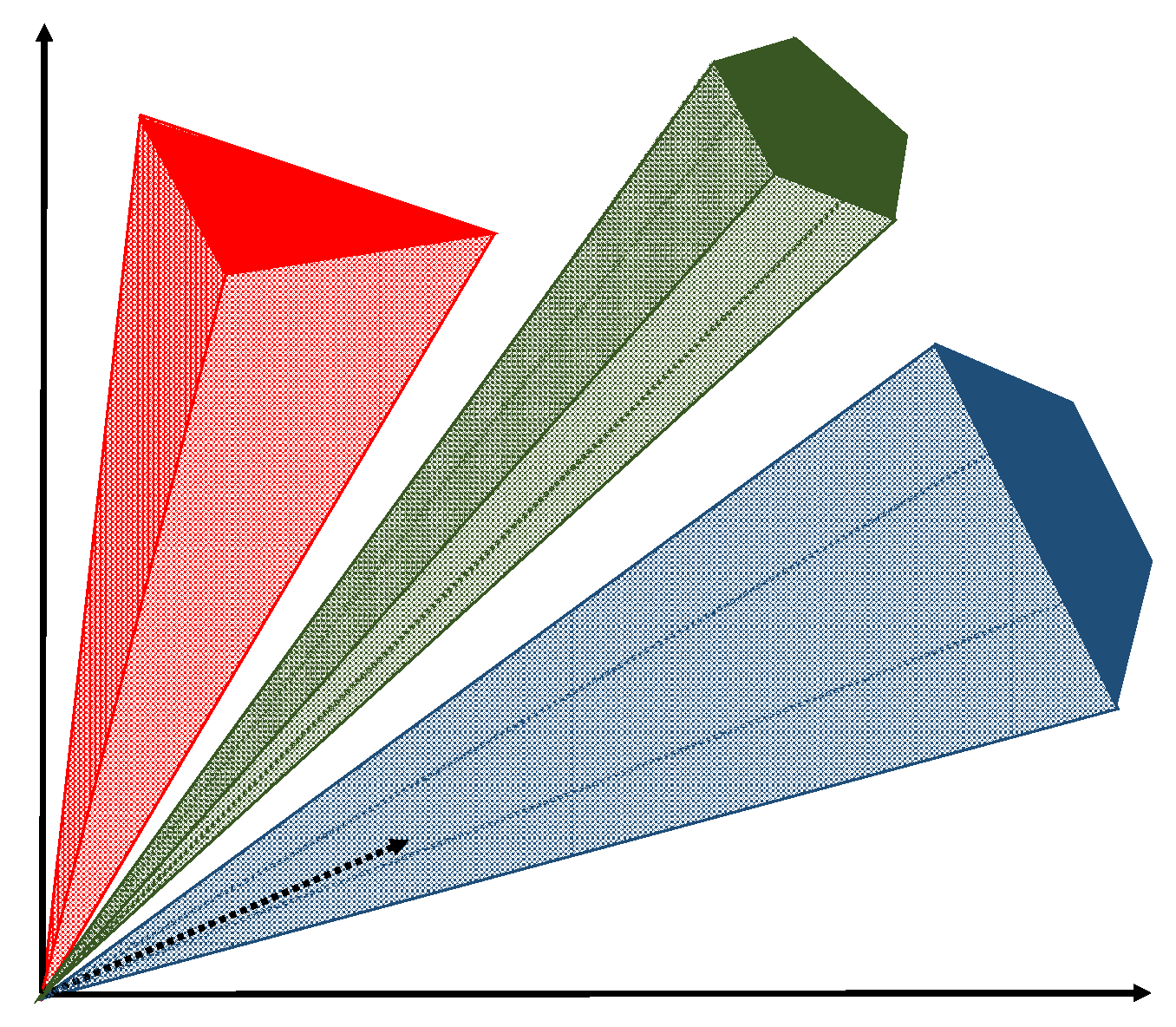
3.1. A New Paradigm of Endmember Variability
3.2. A New Spectral Library-Free Bundle-Based Model
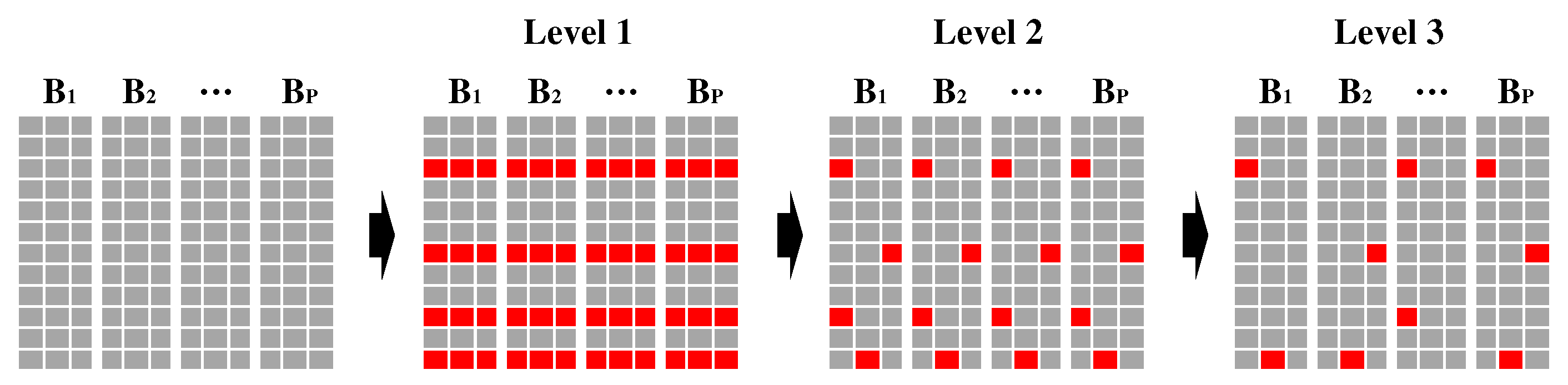
3.3. Hierarchical Group-Sparsity
3.4. Further Relationships with Existing Models
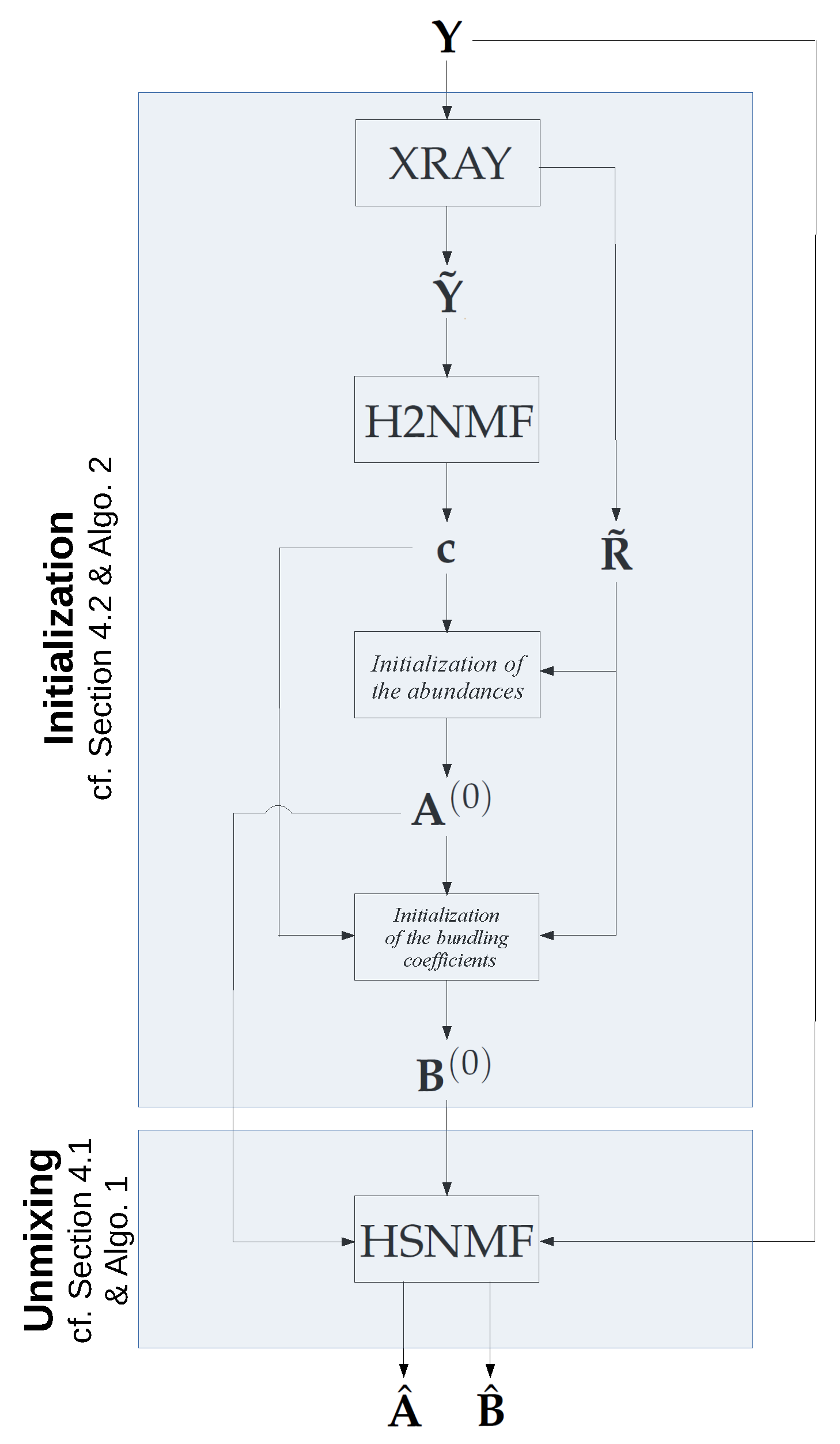
4. HSNMF Unmixing Algorithm
4.1. PALM Algorithm
| Algorithm 1 HSNMF algorithm |
|
4.1.1. Updating
4.1.2. Updating
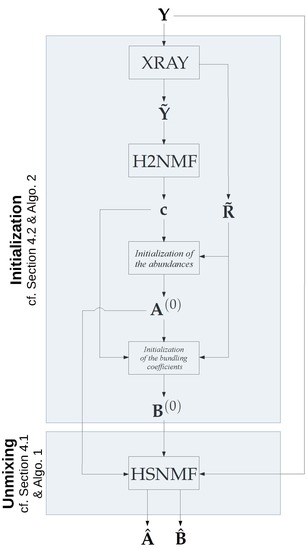
4.2. Initialization and Computational Cost Reduction
| Algorithm 2 Initialization |
|
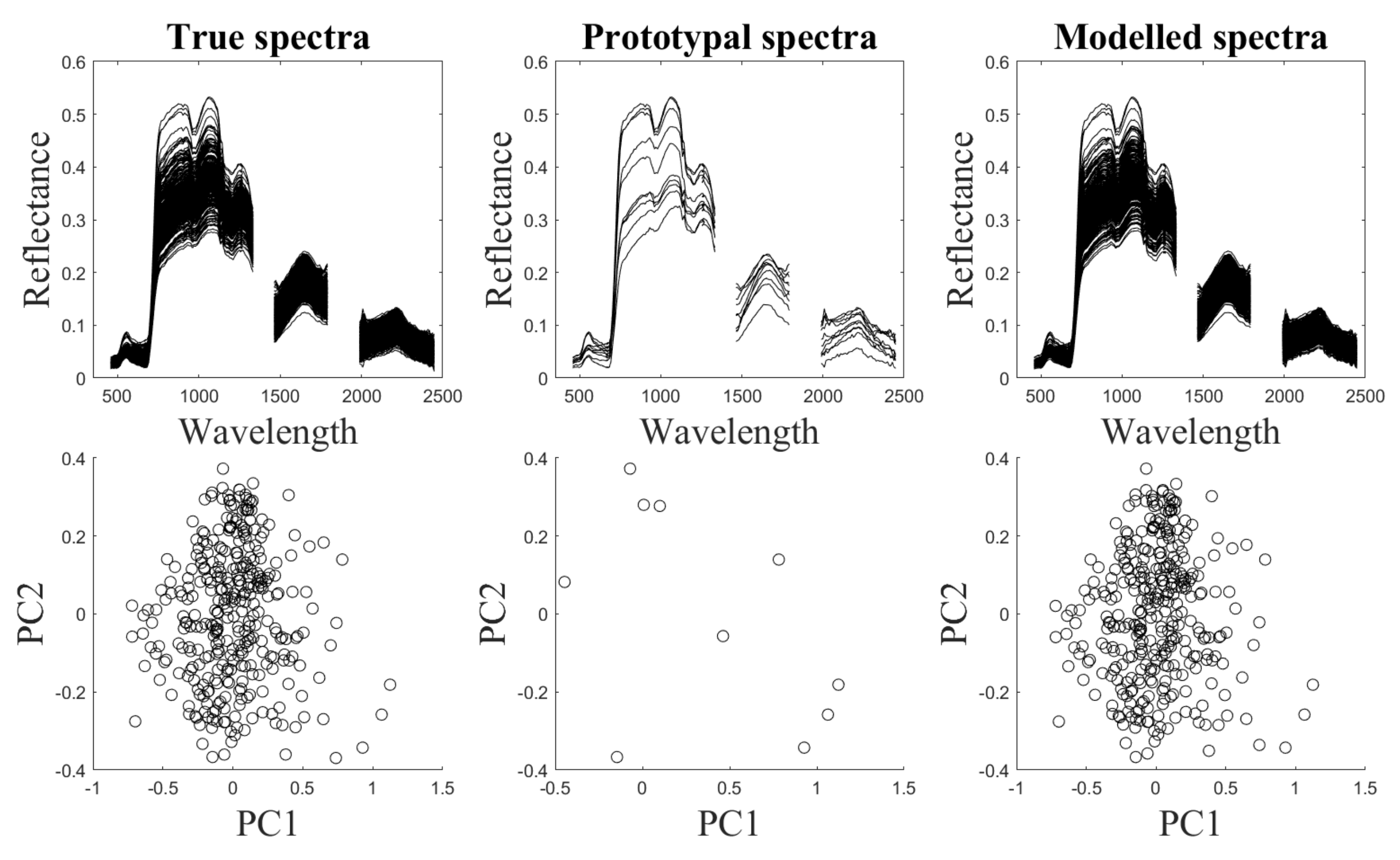
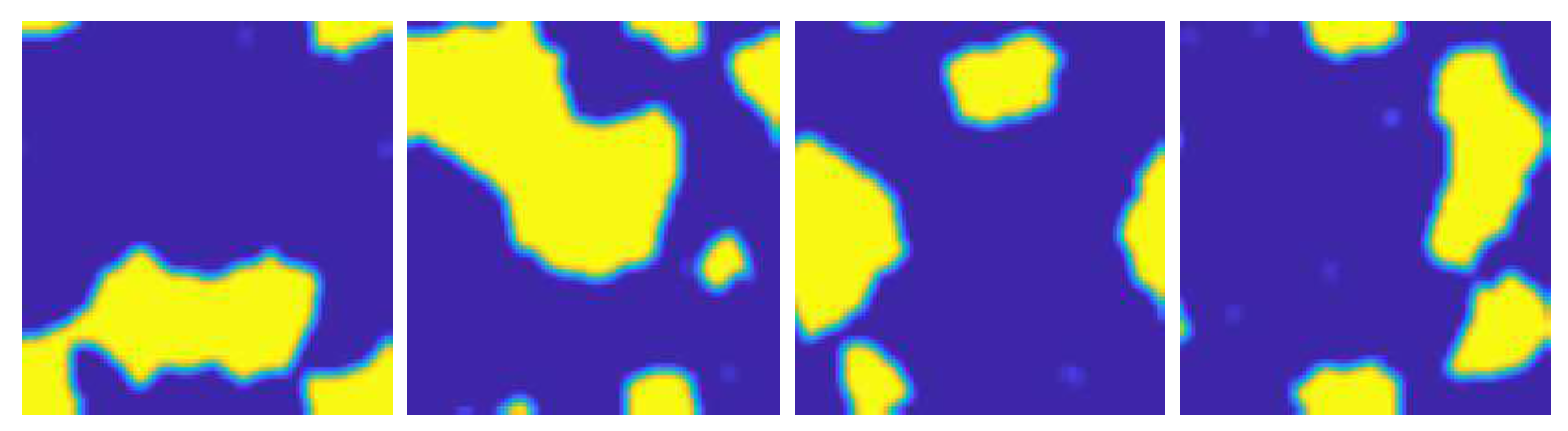
5. Experiments Using Simulated Data
5.1. Simulated Data Sets
5.2. Compared Methods
5.3. Performance Criterion
5.4. Results
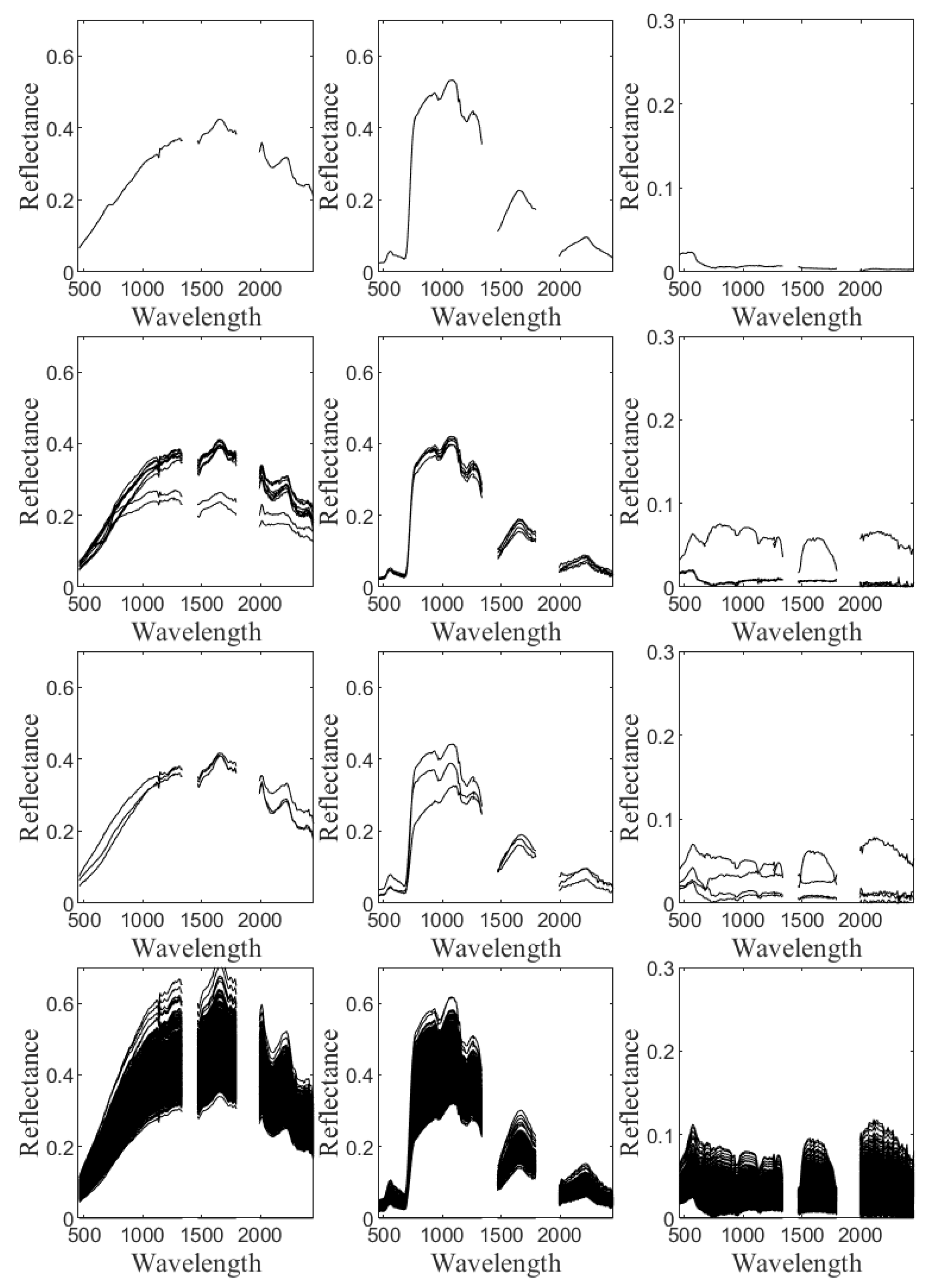
6. Experiments Using Real Data
6.1. Real Datasets
6.1.1. MUESLI Image
6.1.2. AVIRIS Image
6.2. Initialization
6.3. Results
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, 110–122. [Google Scholar] [CrossRef]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Uezato, T.; Fauvel, M.; Dobigeon, N. Hyperspectral image unmixing with LiDAR data-aided spatial regularization. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4098–4108. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Dobigeon, N.; Parente, M.; Qian, D.; Gader, P.; Chanussot, J. Hyperspectral Unmixing Overview: Geometrical, Statistical, and Sparse Regression-Based Approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 354–379. [Google Scholar] [CrossRef]
- Murphy, R.J.; Monteiro, S.T.; Schneider, S. Evaluating Classification Techniques for Mapping Vertical Geology Using Field-Based Hyperspectral Sensors. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3066–3080. [Google Scholar] [CrossRef]
- Uezato, T.; Murphy, R.J.; Melkumyan, A.; Chlingaryan, A. A Novel Spectral Unmixing Method Incorporating Spectral Variability Within Endmember Classes. IEEE Trans. Geosci. Remote Sens. 2016, 54, 2812–2831. [Google Scholar] [CrossRef]
- Zare, A.; Ho, K.C. Endmember Variability in Hyperspectral Analysis: Addressing Spectral Variability During Spectral Unmixing. IEEE Signal Process. Mag. 2014, 31, 95–104. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P.; Tits, L.; Coppin, P. Endmember variability in Spectral Mixture Analysis: A review. Remote Sens. Environ. 2011, 115, 1603–1616. [Google Scholar] [CrossRef]
- Bateson, C.A.; Asner, G.P.; Wessman, C.A. Endmember bundles: A new approach to incorporating endmember variability into spectral mixture analysis. IEEE Trans. Geosci. Remote Sens. 2000, 38, 1083–1094. [Google Scholar] [CrossRef]
- Roberts, D.A.; Gardner, M.; Church, R.; Ustin, S.; Scheer, G.; Green, R.O. Mapping Chaparral in the Santa Monica Mountains Using Multiple Endmember Spectral Mixture Models. Remote Sens. Environ. 1998, 65, 267–279. [Google Scholar] [CrossRef]
- Iordache, M.; Bioucas-Dias, J.; Plaza, A. Hyperspectral unmixing with sparse group lasso. In Proceedings of the 2011 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Vancouver, BC, Canada, 24–29 July 2011; pp. 3586–3589. [Google Scholar]
- Drumetz, L.; Meyer, T.R.; Chanussot, J.; Bertozzi, A.L.; Jutten, C. Hyperspectral image unmixing with endmember bundles and group sparsity inducing mixed norms. IEEE Trans. Image Process. 2019, 28, 3435–3450. [Google Scholar] [CrossRef] [PubMed]
- Goenaga, M.A.; Torres-Madronero, M.C.; Velez-Reyes, M.; Van Bloem, S.J.; Chinea, J.D. Unmixing Analysis of a Time Series of Hyperion Images Over the Guanica Dry Forest in Puerto Rico. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 329–338. [Google Scholar] [CrossRef]
- Uezato, T.; Murphy, R.J.; Melkumyan, A.; Chlingaryan, A. A Novel Endmember Bundle Extraction and Clustering Approach for Capturing Spectral Variability within Endmember Classes. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6712–6731. [Google Scholar] [CrossRef]
- Somers, B.; Zortea, M.; Plaza, A.; Asner, G.P. Automated Extraction of Image-Based Endmember Bundles for Improved Spectral Unmixing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 396–408. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, L.; Du, B. An Image-Based Endmember Bundle Extraction Algorithm Using Both Spatial and Spectral Information. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 6, 2607–2617. [Google Scholar] [CrossRef]
- Eches, O.; Dobigeon, N.; Mailhes, C.; Tourneret, J.Y. Bayesian Estimation of Linear Mixtures Using the Normal Compositional Model. Application to Hyperspectral Imagery. IEEE Trans. Image Process. 2010, 19, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Rangarajan, A.; Gader, P.D. A Gaussian mixture model representation of endmember variability in hyperspectral unmixing. IEEE Trans. Image Process. 2018, 27, 2242–2256. [Google Scholar] [CrossRef]
- Drumetz, L.; Veganzones, M.A.; Henrot, S.; Phlypo, R.; Chanussot, J.; Jutten, C. Blind Hyperspectral Unmixing Using an Extended Linear Mixing Model to Address Spectral Variability. IEEE Trans. Image Process. 2016, 25, 3890–3905. [Google Scholar] [CrossRef]
- Thouvenin, P.A.; Dobigeon, N.; Tourneret, J.Y. Hyperspectral Unmixing with Spectral Variability Using a Perturbed Linear Mixing Model. IEEE Trans. Signal Process. 2016, 64, 525–538. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An Augmented Linear Mixing Model to Address Spectral Variability for Hyperspectral Unmixing. IEEE Trans. Image Process. 2018, 28, 1923–1938. [Google Scholar] [CrossRef]
- Imbiriba, T.; Borsoi, R.A.; Bermudez, J.C.M. Generalized linear mixing model accounting for endmember variability. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 1862–1866. [Google Scholar]
- Meyer, T.R.; Drumetz, L.; Chanussot, J.; Bertozzi, A.L.; Jutten, C. Hyperspectral unmixing with material variability using social sparsity. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 2187–2191. [Google Scholar]
- Henrot, S.; Chanussot, J.; Jutten, C. Dynamical spectral unmixing of multitemporal hyperspectral images. IEEE Trans. Image Process. 2016, 25, 3219–3232. [Google Scholar] [CrossRef] [PubMed]
- Thouvenin, P.A.; Dobigeon, N.; Tourneret, J.Y. Online Unmixing of Multitemporal Hyperspectral Images Accounting for Spectral Variability. IEEE Trans. Image Process. 2016, 25, 3979–3990. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, J.; Ulfarsson, M.O.; Sveinsson, J.R.; Bioucas-Dias, J.M. Sparse distributed multitemporal hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6069–6084. [Google Scholar] [CrossRef]
- Thouvenin, P.A.; Dobigeon, N.; Tourneret, J.Y. Partially asynchronous distributed unmixing of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2009–2021. [Google Scholar] [CrossRef]
- Uezato, T.; Fauvel, M.; Dobigeon, N. Hyperspectral unmixing with spectral variability using adaptive bundles and double sparsity. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3980–3992. [Google Scholar] [CrossRef]
- Cutler, A.; Breiman, L. Archetypal analysis. Technometrics 1994, 36, 338–347. [Google Scholar] [CrossRef]
- Sun, W.; Jiang, M.; Li, W.; Liu, Y. A symmetric sparse representation based band selection method for hyperspectral imagery classification. Remote Sens. 2016, 8, 238. [Google Scholar] [CrossRef]
- Drees, L.; Roscher, R.; Wenzel, S. Archetypal Analysis for Sparse Representation-based Hyperspectral Sub-pixel Quantification. Photogramm. Eng. Remote Sens. 2018, 84, 279–286. [Google Scholar] [CrossRef]
- Sun, W.; Yang, G.; Wu, K.; Li, W.; Zhang, D. Pure endmember extraction using robust kernel archetypoid analysis for hyperspectral imagery. ISPRS J. Photogramm. Remote Sens. 2017, 131, 147–159. [Google Scholar] [CrossRef]
- Sun, W.; Ma, J.; Yang, G.; Du, B.; Zhang, L. A Poisson nonnegative matrix factorization method with parameter subspace clustering constraint for endmember extraction in hyperspectral imagery. ISPRS J. Photogramm. Remote Sens. 2017, 128, 27–39. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, D.; Xu, Y.; Tian, L.; Yang, G.; Li, W. A probabilistic weighted archetypal analysis method with Earth mover’s distance for endmember extraction from hyperspectral imagery. Remote Sens. 2017, 9, 841. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, G.; Jia, X. Hyperspectral Image Unmixing Based on Fast Kernel Archetypal Analysis. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 2016, 10, 331–346. [Google Scholar] [CrossRef]
- Akhtar, N.; Mian, A. RCMF: Robust constrained matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3354–3366. [Google Scholar] [CrossRef]
- Zhao, G.; Jia, X.; Zhao, C. Multiple endmembers based unmixing using archetypal analysis. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 5039–5042. [Google Scholar]
- Xu, M.; Zhang, G.; Fan, Y.; Du, B.; Li, J. Archetypal analysis for endmember bundle extraction considering spectral variability. In Proceedings of the IEEE GRSS Workshop Hyperspectral Image SIgnal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 23–26 September 2018; pp. 1–4. [Google Scholar]
- Revel, C.; Deville, Y.; Achard, V.; Briottet, X. Inertia-Constrained Pixel-by-Pixel Nonnegative Matrix Factorisation: A Hyperspectral Unmixing Method Dealing with Intra-class Variability. Remote Sens. 2018, 10, 1706. [Google Scholar] [CrossRef]
- Dobigeon, N.; Tourneret, J.Y.; Richard, C.; Bermudez, J.C.M.; McLaughlin, S.; Hero, A.O. Nonlinear unmixing of hyperspectral images: Models and algorithms. IEEE Signal Process. Mag. 2014, 31, 82–94. [Google Scholar] [CrossRef]
- Burazerović, D.; Heylen, R.; Geens, B.; Sterckx, S.; Scheunders, P. Detecting the adjacency effect in hyperspectral imagery with spectral unmixing techniques. IEEE J. Sel. Top. Signal Process. 2013, 6, 1070–1078. [Google Scholar] [CrossRef]
- Halimi, A.; Bioucas-Dias, J.M.; Dobigeon, N.; Buller, G.S.; McLaughlin, S. Fast hyperspectral unmixing in presence of nonlinearity or mismodelling effects. IEEE Trans. Comput. Imag. 2017, 3, 146–159. [Google Scholar] [CrossRef]
- Stein, D. Application of the normal compositional model to the analysis of hyperspectral imagery. In Proceedings of the IEEE Workshop on Advances in Techniques for Analysis of Remotely Sensed Data, Greenbelt, MD, USA, 27–28 October 2003. [Google Scholar]
- Halimi, A.; Dobigeon, N.; Tourneret, J. Unsupervised Unmixing of Hyperspectral Images Accounting for Endmember Variability. IEEE Trans. Image Process. 2015, 24, 4904–4917. [Google Scholar] [CrossRef]
- Zhou, Y.; Rangarajan, A.; Gader, P.D. A Spatial Compositional Model for Linear Unmixing and Endmember Uncertainty Estimation. IEEE Trans. Image Process. 2016, 25, 5987–6002. [Google Scholar] [CrossRef] [PubMed]
- Woodbridge, Y.; Okun, U.; Elidan, G.; Wiesel, A. Unmixing K-Gaussians With Application to Hyperspectral Imaging. IEEE Trans. Geosci. Remote Sens. 2019, 57, 7281–7293. [Google Scholar] [CrossRef]
- Xiaoxiao, D.; Zare, A.; Gader, P.; Dranishnikov, D. Spatial and Spectral Unmixing Using the Beta Compositional Model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1994–2003. [Google Scholar] [CrossRef]
- Hong, D.; Zhu, X.X. SULoRA: Subspace unmixing with low-rank attribute embedding for hyperspectral data analysis. IEEE J. Sel. Top. Signal Process. 2018, 12, 1351–1363. [Google Scholar] [CrossRef]
- Uezato, T.; Murphy, R.J.; Melkumyan, A.; Chlingaryan, A. Incorporating Spatial Information and Endmember Variability Into Unmixing Analyses to Improve Abundance Estimates. IEEE Trans. Image Process. 2016, 25, 5563–5575. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.; Roberts, D.A.; Halligan, K.; Menz, G. Hierarchical Multiple Endmember Spectral Mixture Analysis (MESMA) of hyperspectral imagery for urban environments. Remote Sens. Environ. 2009, 113, 1712–1723. [Google Scholar] [CrossRef]
- Winter, M.E. N-FINDR: An algorithm for fast autonomous spectral end-member determination in hyperspectral data. Proc. SPIE 1999, 3753, 266–275. [Google Scholar]
- Nascimento, J.M.; Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Gillis, N. Successive nonnegative projection algorithm for robust nonnegative blind source separation. SIAM J. Imaging Sci. 2014, 7, 1420–1450. [Google Scholar] [CrossRef]
- Ammanouil, R.; Ferrari, A.; Richard, C.; Mary, D. Blind and fully constrained unmixing of hyperspectral images. IEEE Trans. Image Process. 2014, 23, 5510–5518. [Google Scholar] [CrossRef]
- Kowalski, M. Sparse regression using mixed norms. Appl. Comput. Harmon. Anal. 2009, 27, 303–324. [Google Scholar] [CrossRef]
- Shi, Z.; Shi, T.; Zhou, M.; Xu, X. Collaborative Sparse Hyperspectral Unmixing Using l0 Norm. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5495–5508. [Google Scholar] [CrossRef]
- Fu, X.; Ma, W.K.; Chan, T.H.; Bioucas-Dias, J.M. Self-dictionary sparse regression for hyperspectral unmixing: Greedy pursuit and pure pixel search are related. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 9, 1128–1141. [Google Scholar] [CrossRef]
- Giampouras, P.V.; Themelis, K.E.; Rontogiannis, A.A.; Koutroumbas, K.D. Simultaneously Sparse and Low-Rank Abundance Matrix Estimation for Hyperspectral Image Unmixing. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4775–4789. [Google Scholar] [CrossRef]
- Jenatton, R.; Mairal, J.; Obozinski, G.; Bach, F.R. Proximal Methods for Hierarchical Sparse Coding. J. Mach. Learn. Res. 2011, 12, 2297–2334. [Google Scholar]
- Cohen, J.E.; Gillis, N. A new approach to dictionary-based nonnegative matrix factorization. In Proceedings of the 2017 25th European Signal Processing Conference (EUSIPCO), Kos, Greece, 28 August–2 September 2017; pp. 493–497. [Google Scholar]
- Gillis, N.; Luce, R. A Fast Gradient Method for Nonnegative Sparse Regression With Self-Dictionary. IEEE Trans. Image Process. 2018, 27, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Bolte, J.; Sabach, S.; Teboulle, M. Proximal alternating linearized minimization for nonconvex and nonsmooth problems. Math. Program. 2014, 146, 459–494. [Google Scholar] [CrossRef]
- Yu, Y. On Decomposing the Proximal Map. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–8 December 2013; pp. 91–99. [Google Scholar]
- Kyrillidis, A.; Becker, S.; Cevher, V.; Koch, C. Sparse projections onto the simplex. In Proceedings of the International Conference on Machine Learning (ICML), Atlanta, GA, USA, 16 June–21 June 2013. [Google Scholar]
- Kumar, A.; Sindhwani, V.; Kambadur, P. Fast conical hull algorithms for near-separable non-negative matrix factorization. In Proceedings of the International Conference on Machine Learning (ICML), Atlanta, GA, USA, 16 June–21 June 2013. [Google Scholar]
- Gillis, N.; Kuang, D.; Park, H. Hierarchical Clustering of Hyperspectral Images Using Rank-Two Nonnegative Matrix Factorization. IEEE Trans. Geosci. Remote Sens. 2015, 53, 2066–2078. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Figueiredo, M.A.T. Alternating direction algorithms for constrained sparse regression: Application to hyperspectral unmixing. In Proceedings of the IEEE GRSS Workshop Hyperspectral Image SIgnal Processing: Evolution in Remote Sensing (WHISPERS), Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Chan, T.H.; Ma, W.K.; Ambikapathi, A.; Chi, C.Y. A Simplex Volume Maximization Framework for Hyperspectral Endmember Extraction. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4177–4193. [Google Scholar] [CrossRef]
- Besson, O.; Dobigeon, N.; Tourneret, J.Y. Minimum mean square distance estimation of a subspace. IEEE Trans. Signal Process. 2011, 59, 5709–5720. [Google Scholar] [CrossRef]
- Févotte, C.; Dobigeon, N. Nonlinear Hyperspectral Unmixing With Robust Nonnegative Matrix Factorization. IEEE Trans. Image Process. 2015, 24, 4810–4819. [Google Scholar] [CrossRef]
- Wright, J.; Ganesh, A.; Rao, S.R.; Peng, Y.; Ma, Y. Robust Principal Component Analysis: Exact Recovery of Corrupted Low-Rank Matrices via Convex Optimization. In Advances in Neural Information Processing Systems; Vancouver, BC, Canada, 2010. [Google Scholar]

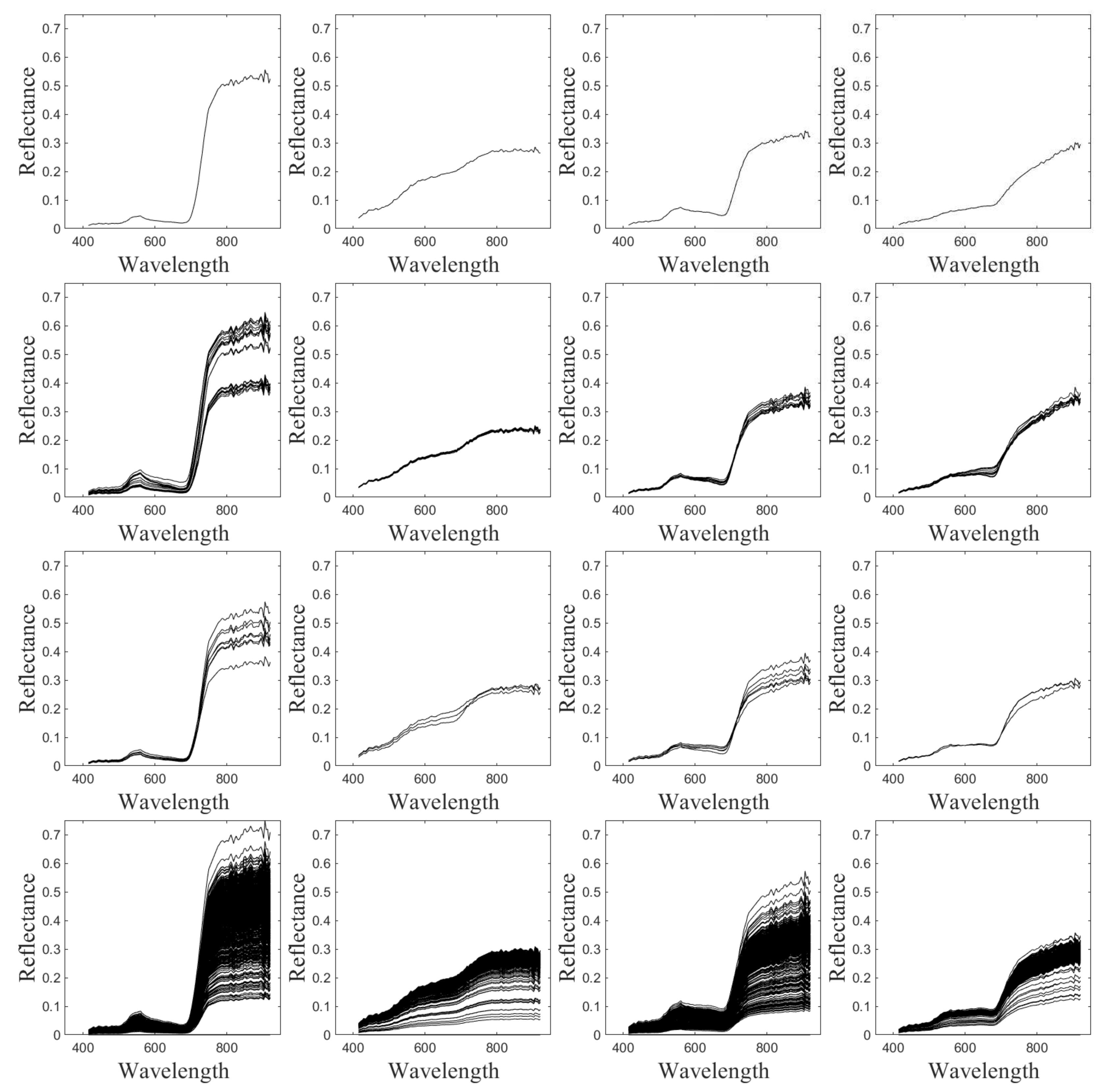
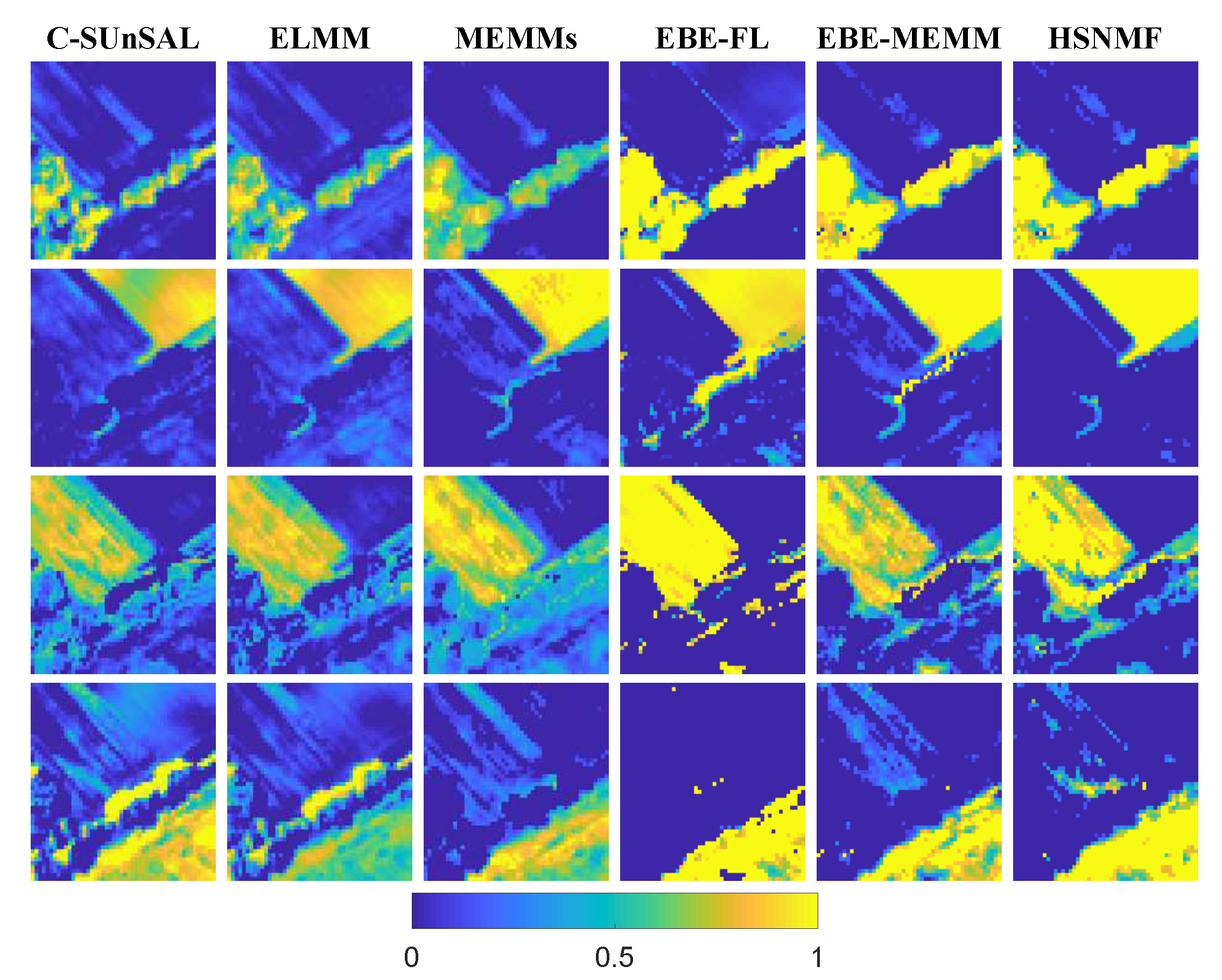
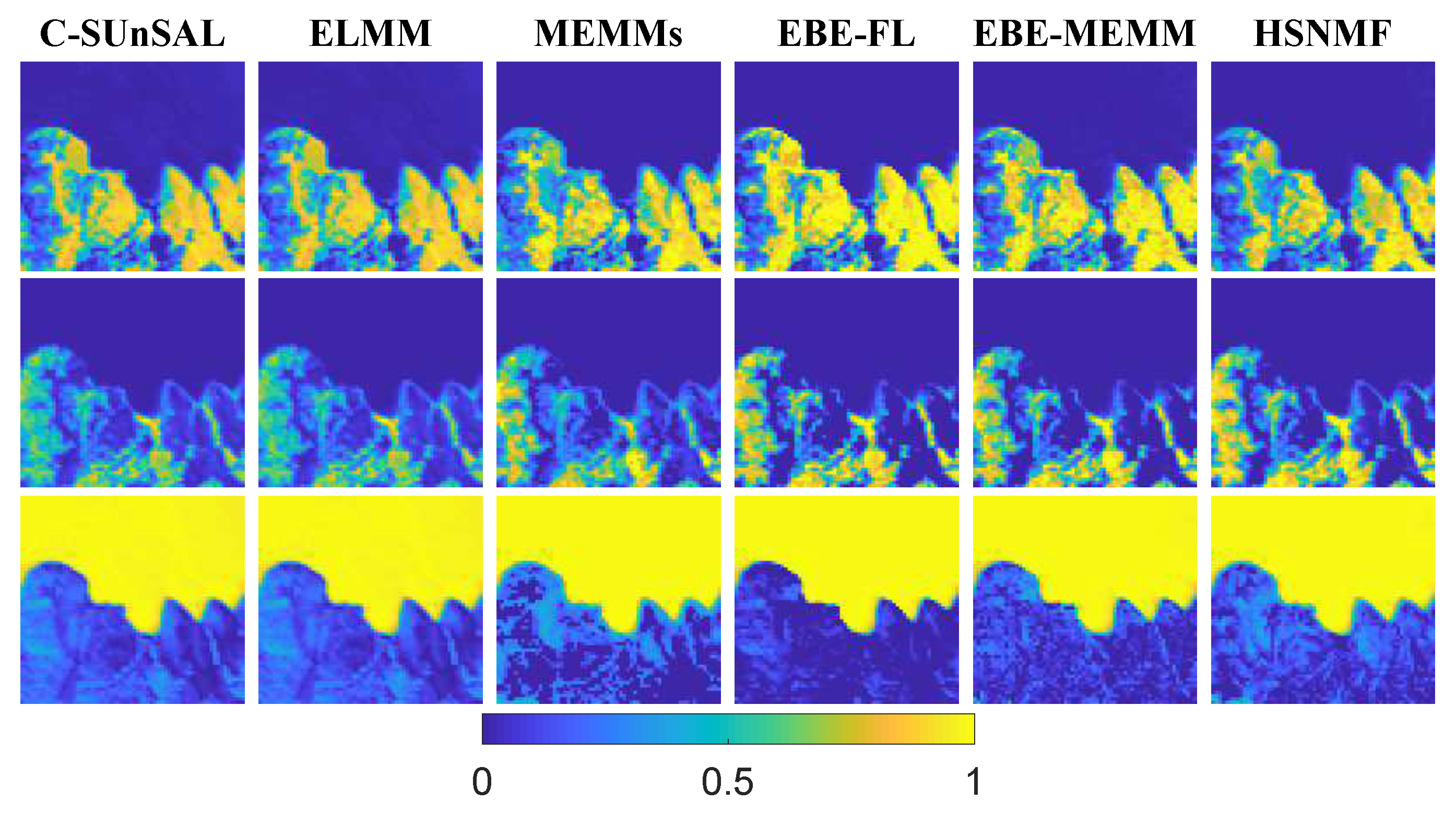
| C-SUnSAL | ELMM | MEMMs | EBE-FL | EBE-MEMM | HSNMF | |
|---|---|---|---|---|---|---|
| RMSE | 0.0724 | 0.0408 | 0.0218 | 0.0378 | 0.0154 | 0.0105 |
| RE | 0.0220 | 0.0093 | 0.0205 | 0.0116 | 0.0106 | 0.0107 |
| SAM | 0.0174 | 0.0141 | 0.0173 | 0.0092 | 0.0086 | 0.0081 |
| JD | 0.6671 | 0.647 | 0.1109 | 0.6219 | 0.0905 | 0.0729 |
| Time | 0.0646 | 97.0853 | 2.5008 | 5.3933 | 11.0964 | 71.18 |
| C-SUnSAL | ELMM | MEMMs | EBE-FL | EBE-MEMM | HSNMF | |
|---|---|---|---|---|---|---|
| RMSE | 0.2001 | 0.1588 | 0.1183 | 0.1822 | 0.1134 | 0.027 |
| RE | 0.0410 | 0.0193 | 0.0328 | 0.0462 | 0.0295 | 0.0127 |
| SAM | 0.0506 | 0.0404 | 0.0502 | 0.0483 | 0.0442 | 0.0159 |
| JD | 0.5247 | 0.5027 | 0.1660 | 0.5059 | 0.1680 | 0.0584 |
| Time | 0.2563 | 206.3764 | 21.4221 | 4.4775 | 73.3072 | 599.6684 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uezato, T.; Fauvel, M.; Dobigeon, N. Hierarchical Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing with Spectral Variability. Remote Sens. 2020, 12, 2326. https://doi.org/10.3390/rs12142326
Uezato T, Fauvel M, Dobigeon N. Hierarchical Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing with Spectral Variability. Remote Sensing. 2020; 12(14):2326. https://doi.org/10.3390/rs12142326
Chicago/Turabian StyleUezato, Tatsumi, Mathieu Fauvel, and Nicolas Dobigeon. 2020. "Hierarchical Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing with Spectral Variability" Remote Sensing 12, no. 14: 2326. https://doi.org/10.3390/rs12142326
APA StyleUezato, T., Fauvel, M., & Dobigeon, N. (2020). Hierarchical Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing with Spectral Variability. Remote Sensing, 12(14), 2326. https://doi.org/10.3390/rs12142326