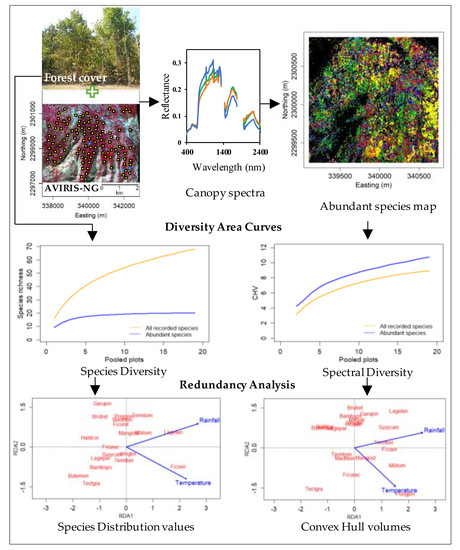
Inferring Species Diversity and Variability over Climatic Gradient with Spectral Diversity Metrics
Abstract
1. Introduction
- How species diversity correlates with spectral diversity calculated from remote sensing;
- Can measured CHVs assist in elucidating the intra- and inter-species variability of abundant species;
- To what extent climatic gradient impacts the distribution of species and the associated spectral diversity.
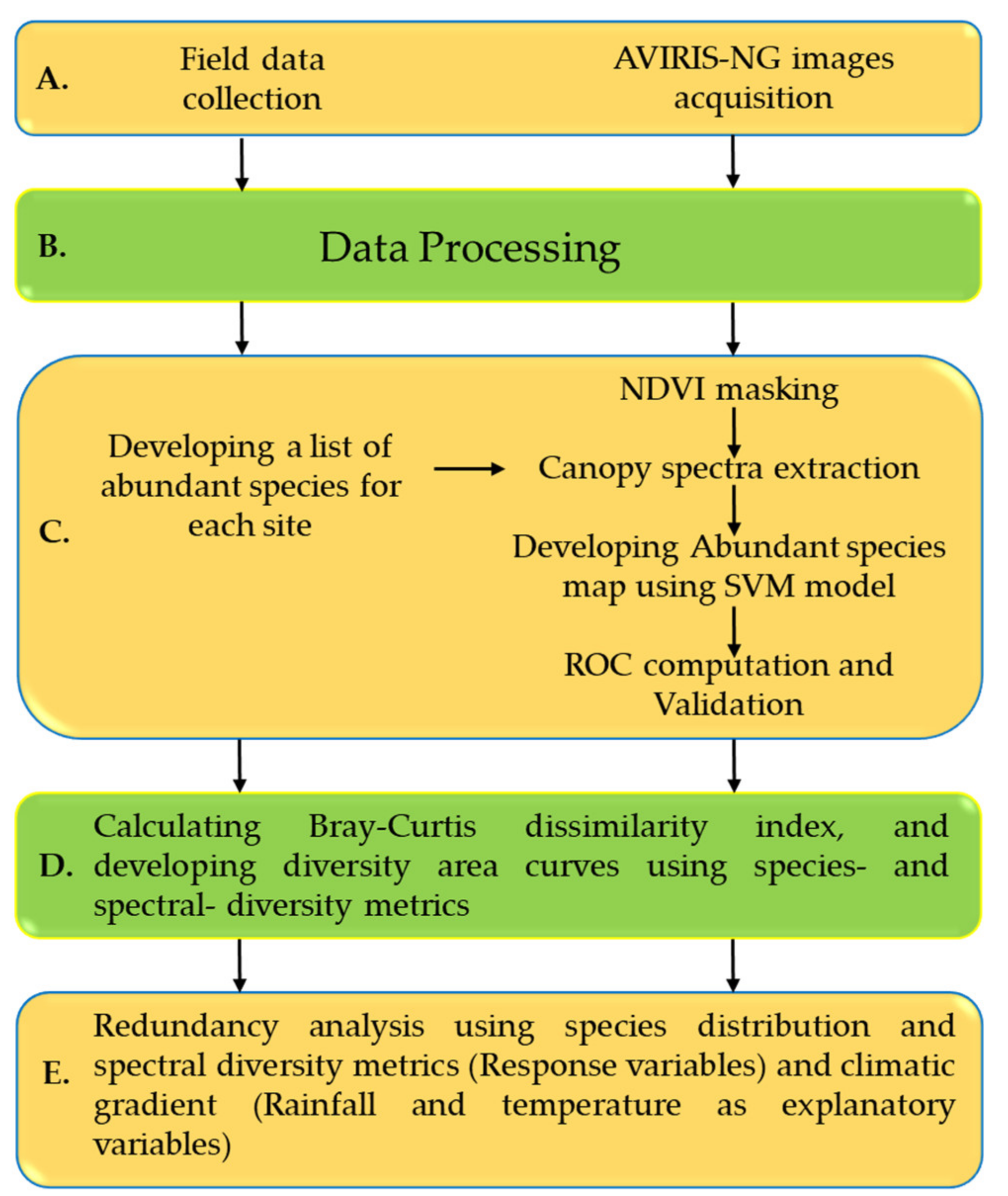
2. Materials and Methods
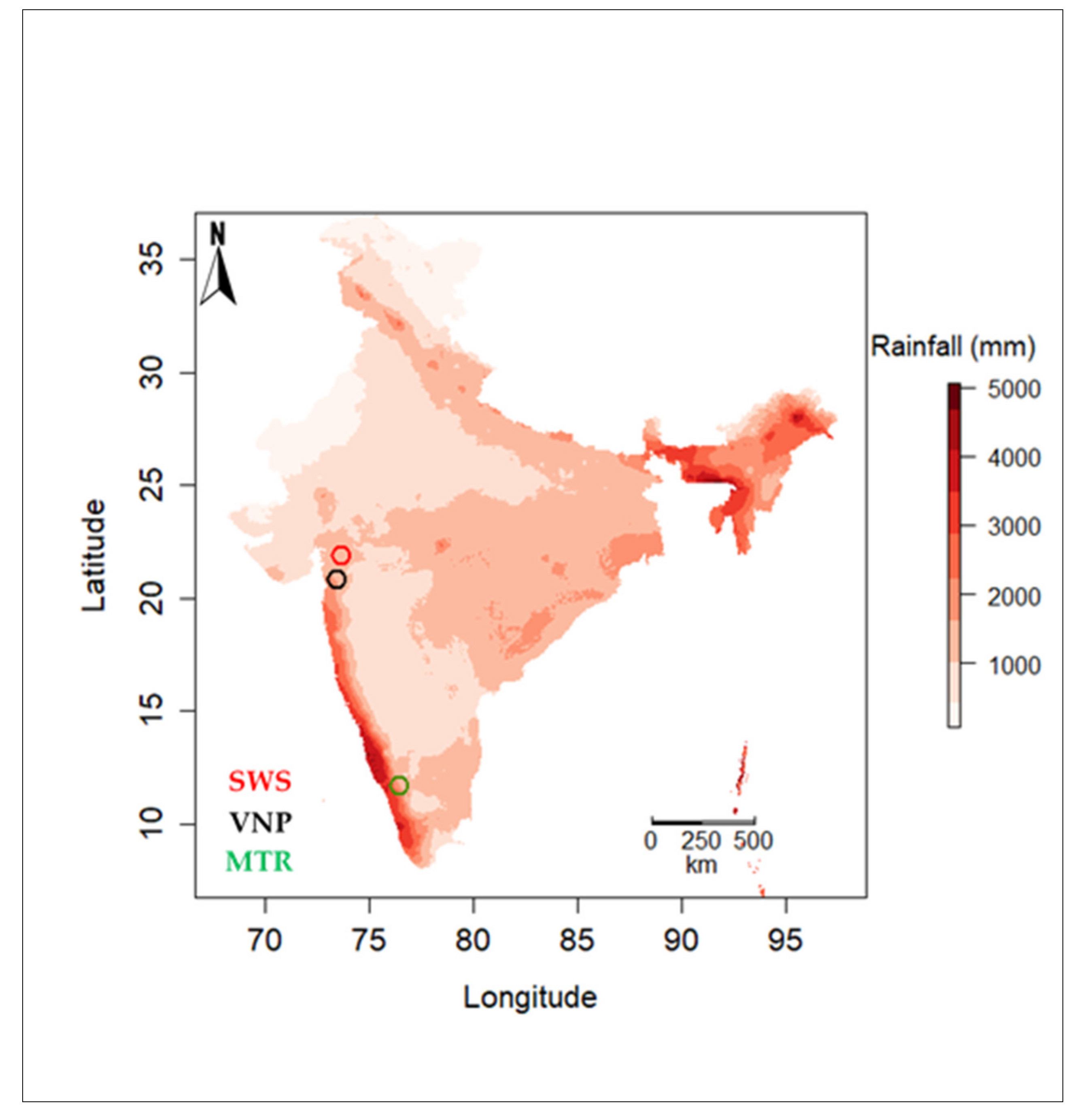
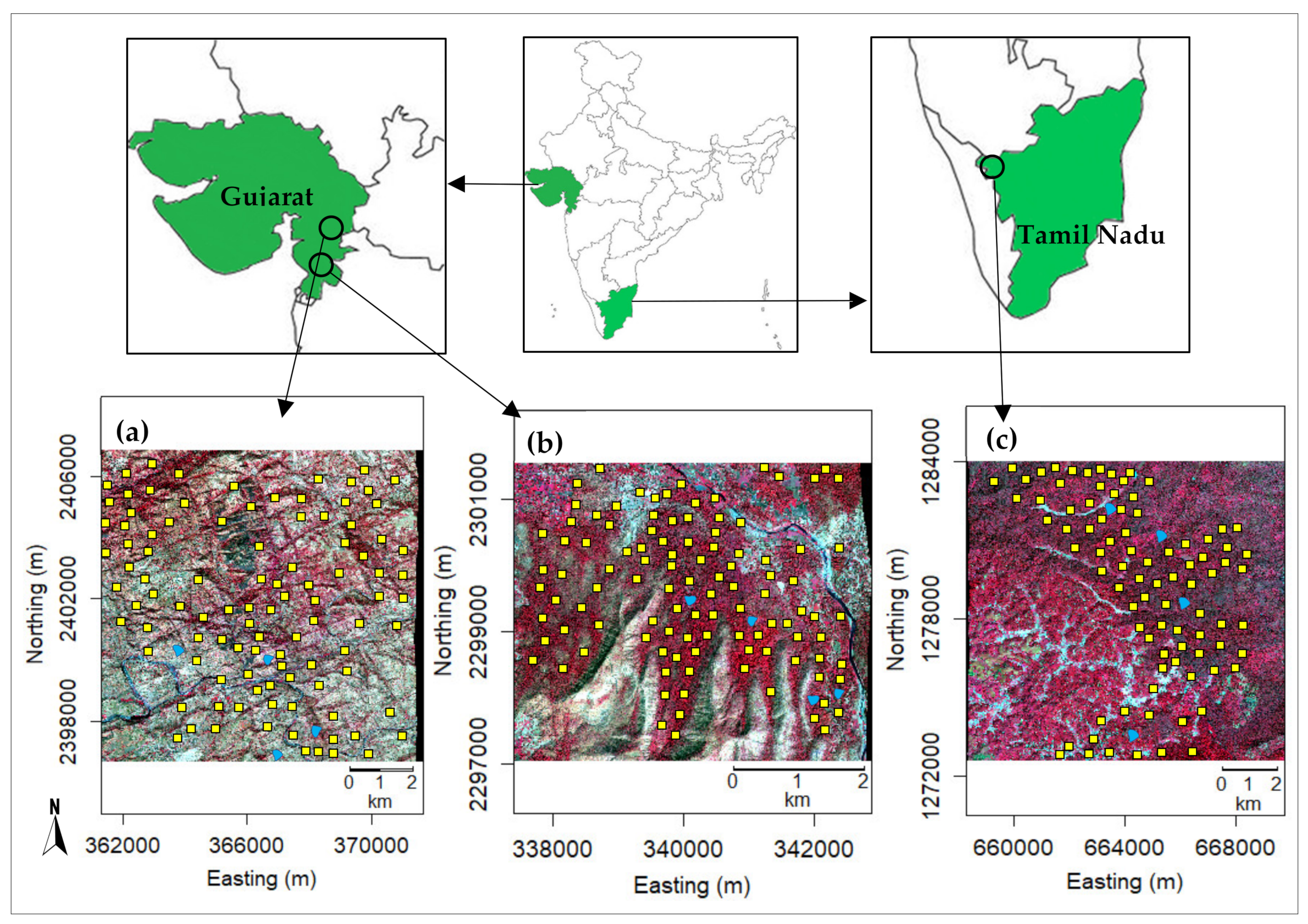
2.1. Site Description
2.2. Vegetation Characteristics
2.3. Field Data Collection
2.4. Climatic Data
2.5. AVIRIS-NG Data Acquisition and Processing
2.6. Data Analysis
2.6.1. Field Data Analysis
2.6.2. Abundant Species Mapping with the Support Vector Machine (SVM) Model
2.6.3. Spectral Diversity Area Curves and Metrics
2.6.4. Intra- and Inter-Species Variability, CHV Values
2.6.5. Redundancy Analysis
3. Results
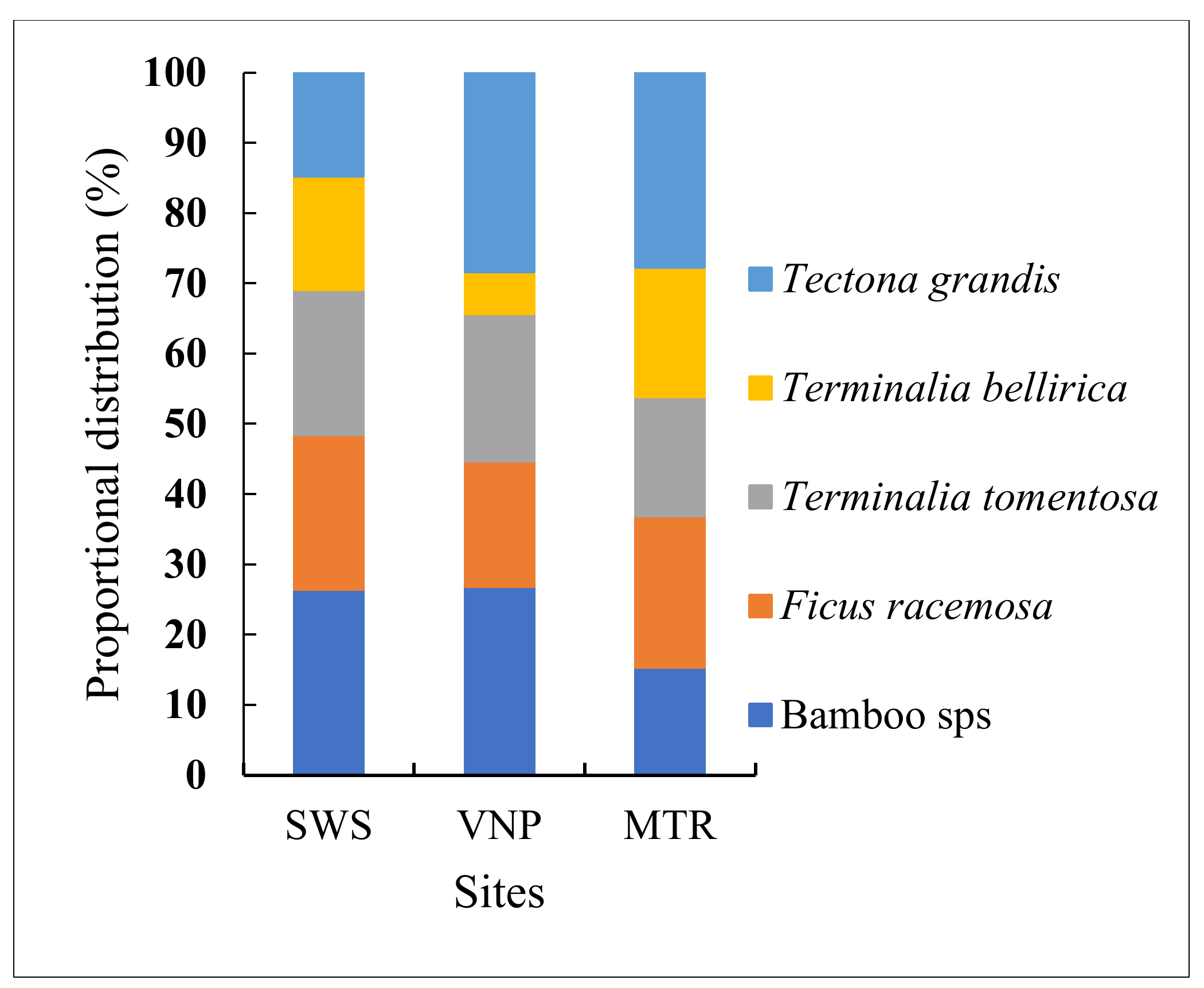
3.1. Tree Species Diversity, Distribution
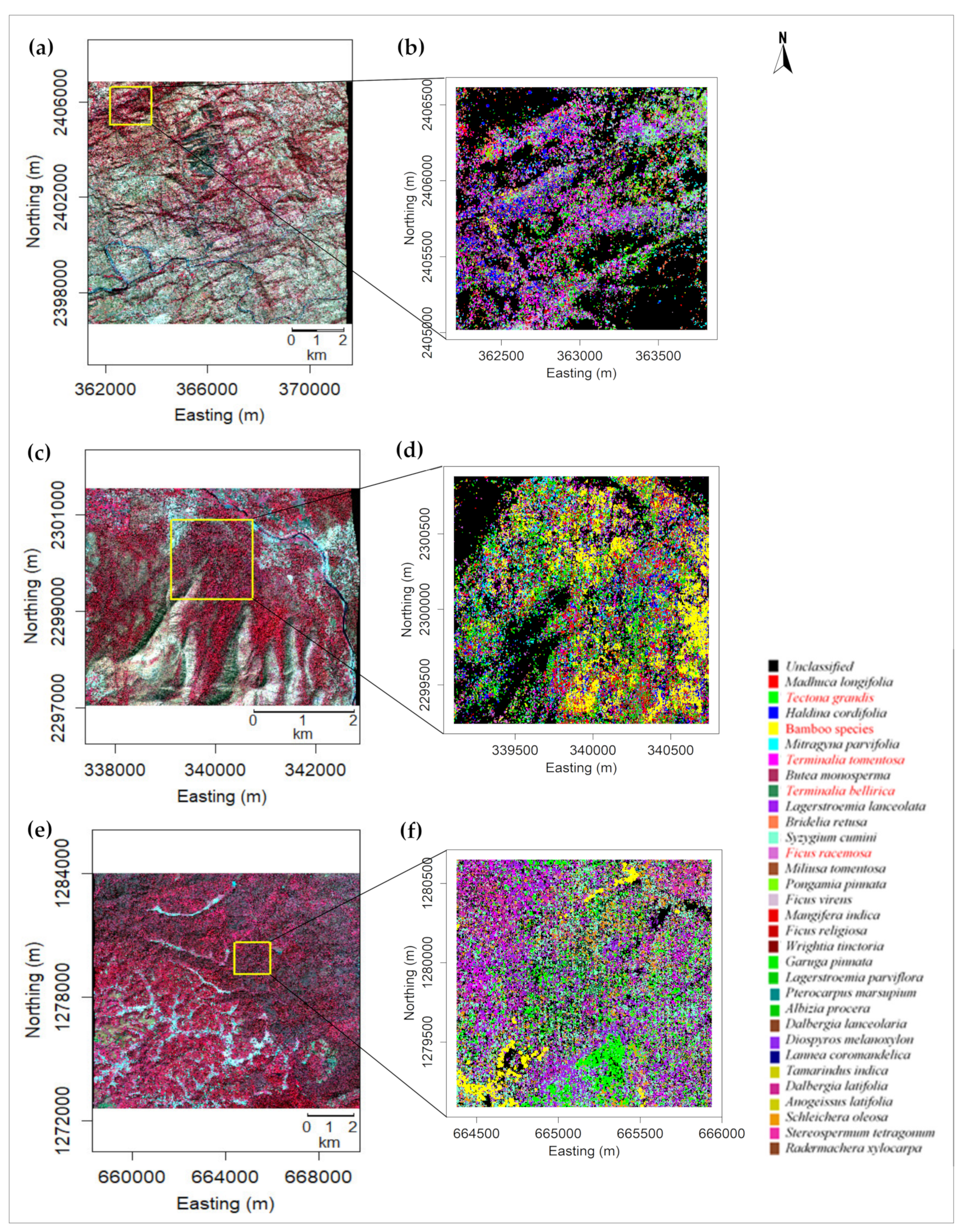
3.2. Abundant Species Maps
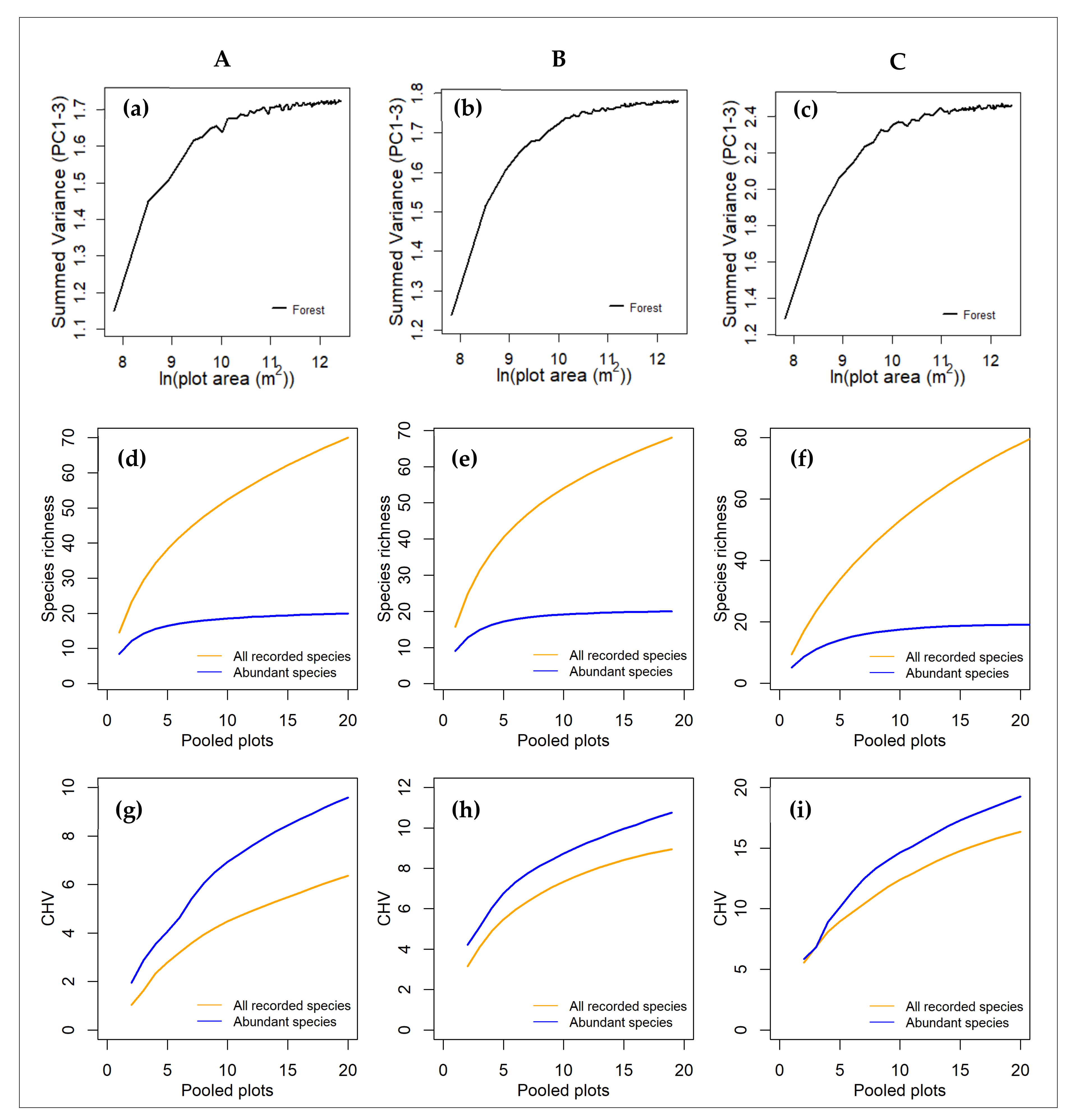
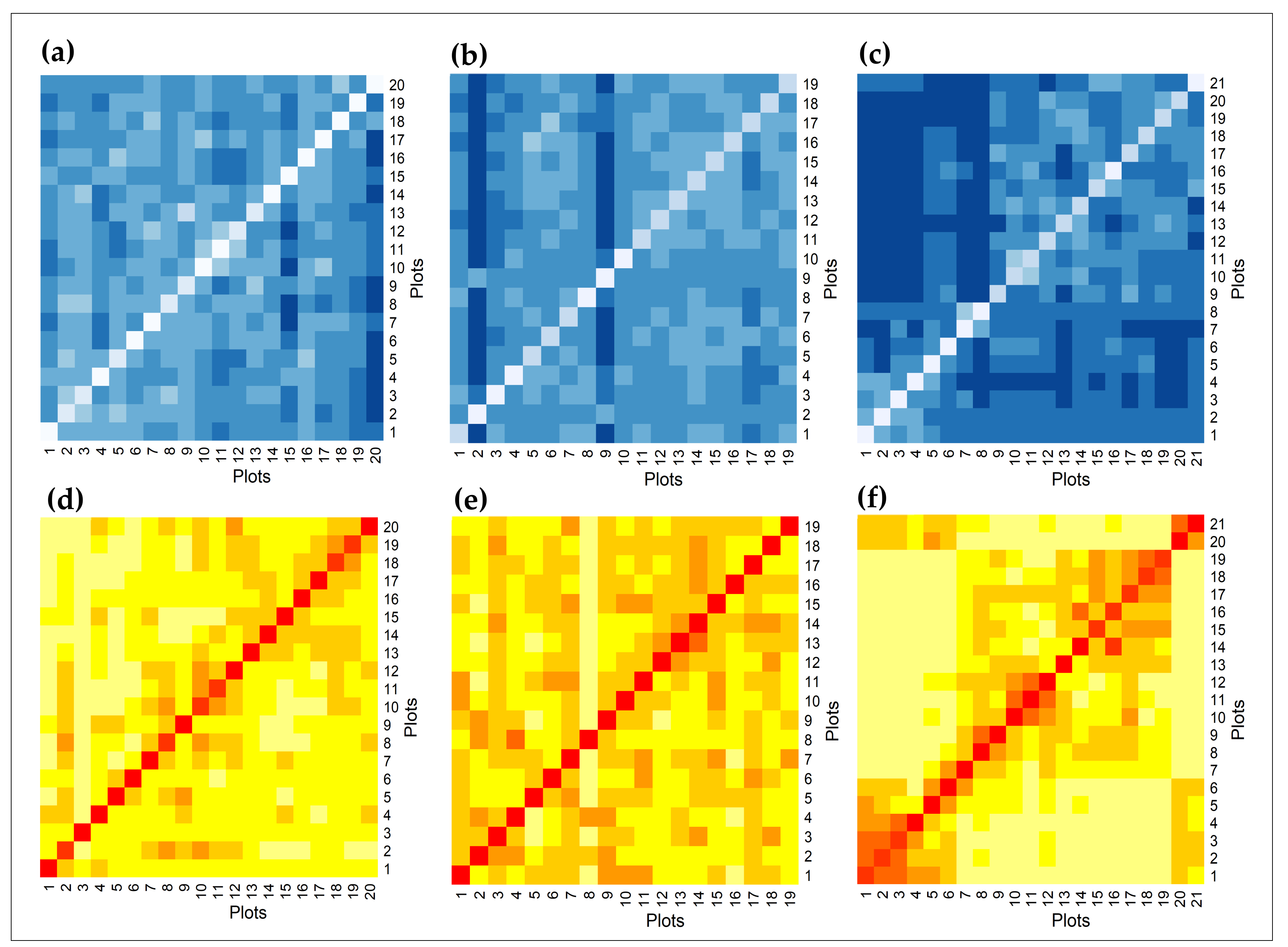
3.3. Diversity–Area Curves
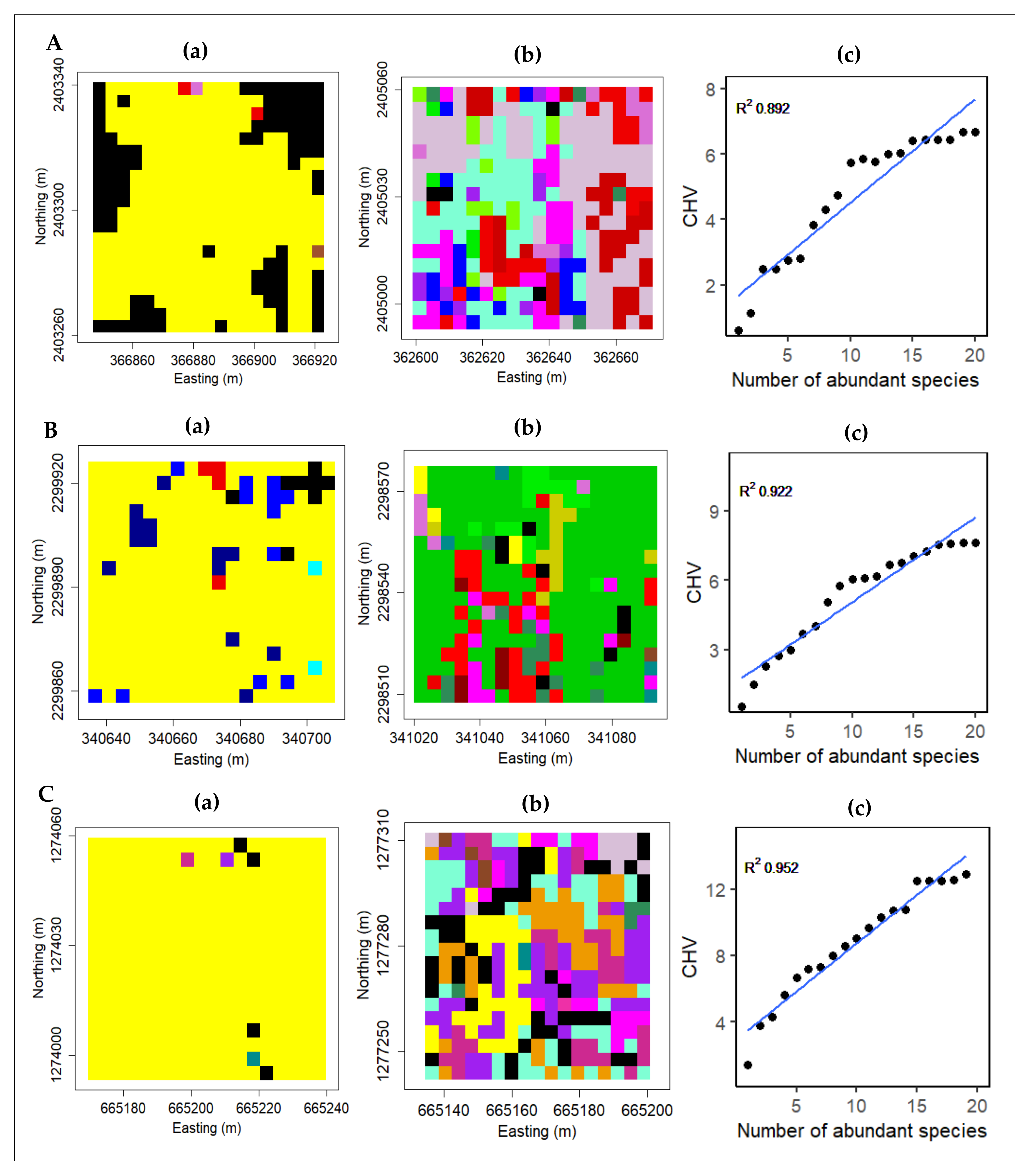
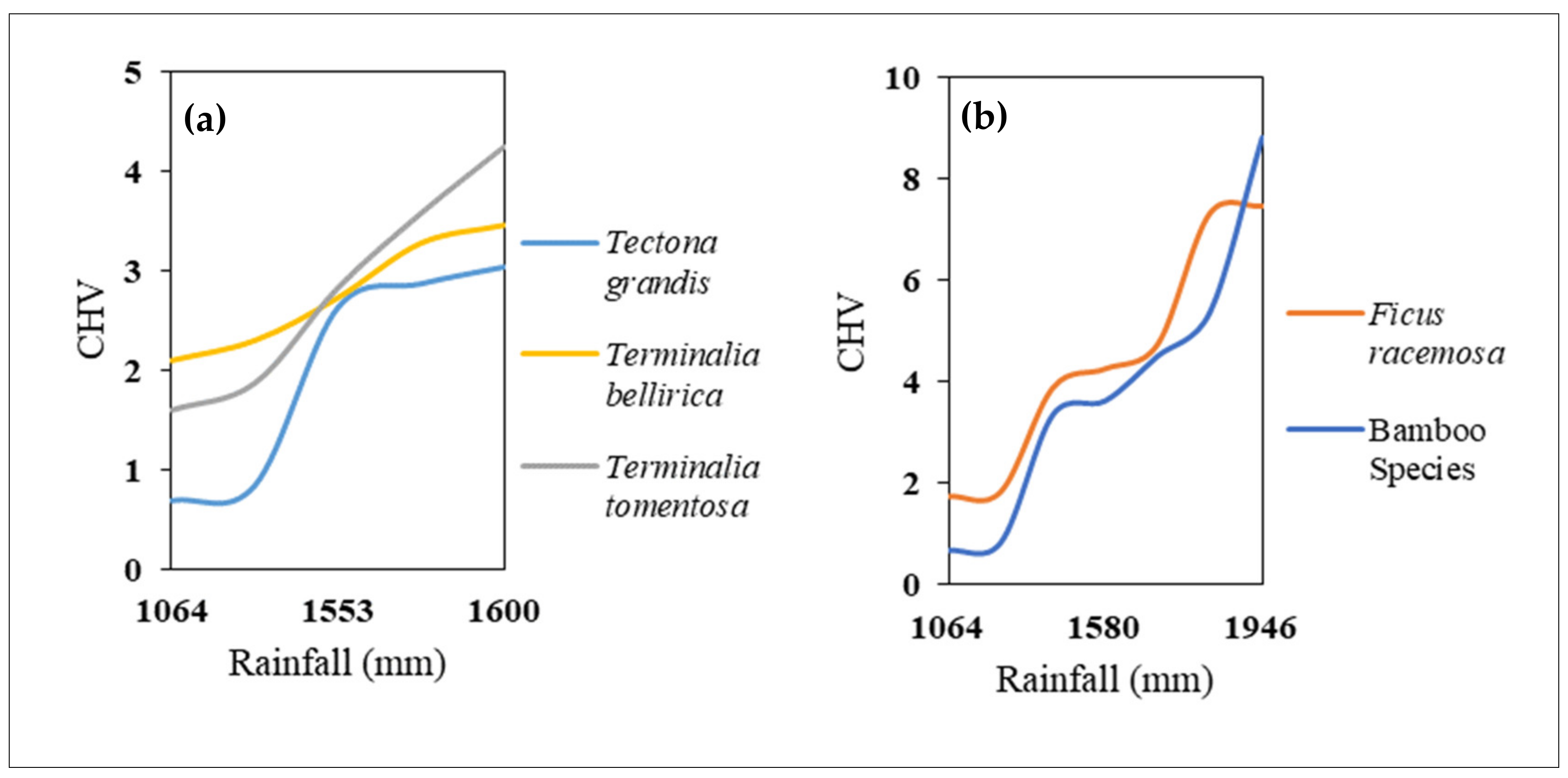
3.4. Intra- and Inter-Species Spectral Diversity, Rainfall Gradient
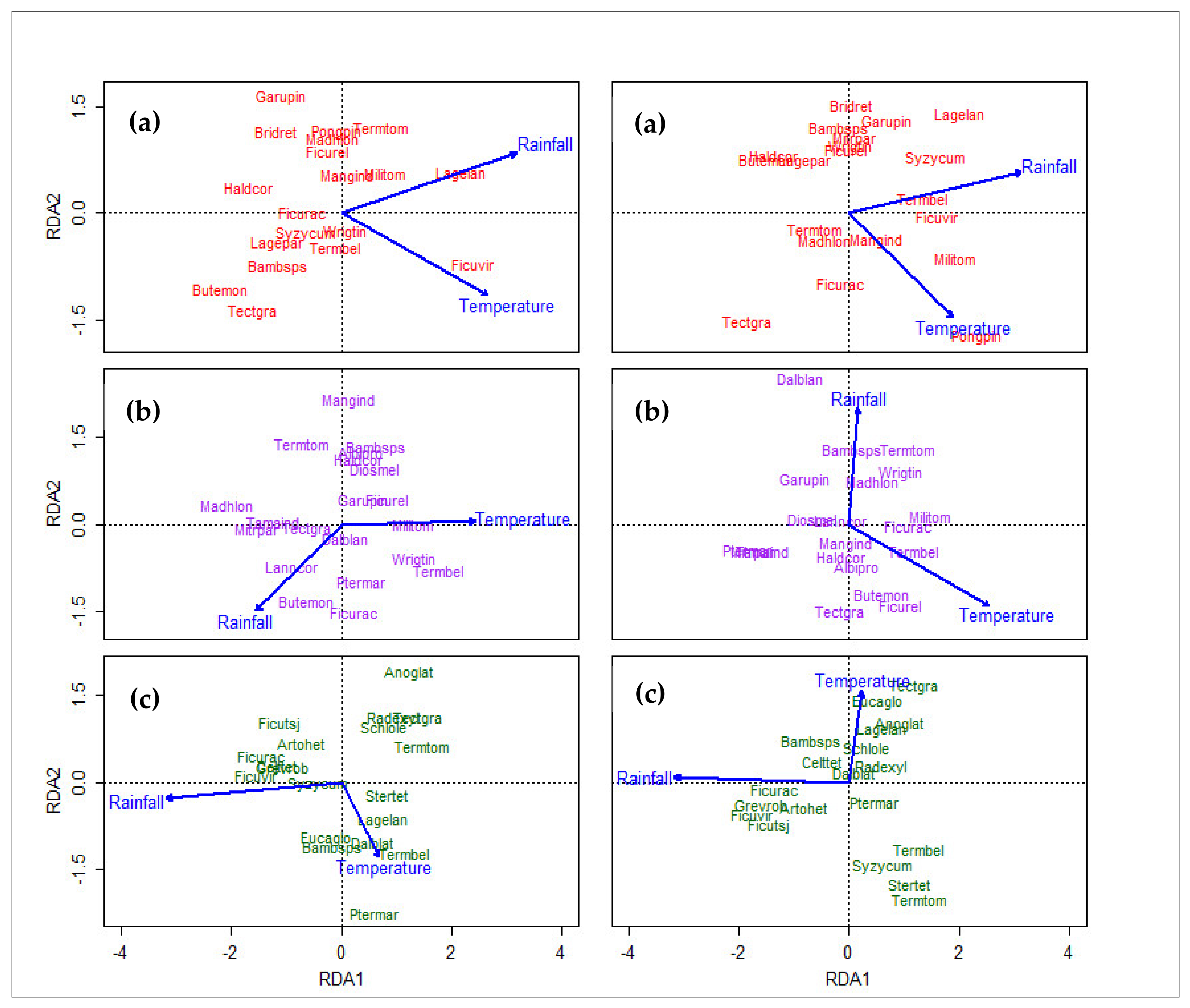
3.5. Redundancy Analysis
3.6. Caveats
4. Discussion
4.1. Tree Species Diversity Metrics
4.2. Abundant Species Maps
4.3. Intra- and Inter-Species Spectral Variability
4.4. Climatic Gradient and Species Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Esquivel-Muelbert, A.; Baker, T.R.; Dexter, K.G.; Lewis, S.L.; ter Steege, H.; Lopez-Gonzalez, G.; Mendoza, A.M.; Brienen, R.; Feldpausch, T.R.; Pitman, N.; et al. Seasonal drought limits tree species across the Neotropics. Ecography 2017, 40, 618–629. [Google Scholar] [CrossRef]
- Kraft, N.J.B.; Valencia, R.; Ackerly, D.D. Functional Traits and Niche-Based Tree Community Assembly in an Amazonian Forest. Science 2008, 322, 580–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Niinemets, Ü.; Sheffield, J.; Lichstein, J.W. Shifts in tree functional composition amplify the response of forest biomass to climate. Nature 2018, 556, 99–102. [Google Scholar] [CrossRef]
- Knapp, A.K.; Ciais, P.; Smith, M.D. Reconciling inconsistencies in precipitation–productivity relationships: implications for climate change. New Phytol. 2017, 214, 41–47. [Google Scholar] [CrossRef]
- Babst, F.; Poulter, B.; Trouet, V.; Tan, K.; Neuwirth, B.; Wilson, R.; Carrer, M.; Grabner, M.; Tegel, W.; Levanic, T.; et al. Site- and species-specific responses of forest growth to climate across the European continent. Glob. Ecol. Biogeogr. 2013, 22, 706–717. [Google Scholar] [CrossRef]
- Kapos, V. Seeing the forest through the trees. Science 2017, 355, 347–349. [Google Scholar] [CrossRef]
- Poorter, L.; Wright, S.J.; Paz, H.; Ackerly, D.D.; Condit, R.; Ibarra-Manríquez, G.; Harms, K.E.; Licona, J.C.; Martínez-Ramos, M.; Mazer, S.J.; et al. Are Functional Traits Good Predictors of Demographic Rates? Evidence from Five Neotropical Forests. Ecology 2008, 89, 1908–1920. [Google Scholar] [CrossRef]
- Pélissier, R. Tree spatial patterns in three contrasting plots of a southern Indian tropical moist evergreen forest. J. Trop. Ecol. 1998, 14, 1–16. [Google Scholar] [CrossRef]
- Swenson, N.G. The assembly of tropical tree communities—The advances and shortcomings of phylogenetic and functional trait analyses. Ecography 2013, 36, 264–276. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A. Remote sensing of terrestrial plant biodiversity. Remote Sens. Environ. 2019, 231, 111218. [Google Scholar] [CrossRef]
- Díaz, S.; Kattge, J.; Cornelissen, J.H.C.; Wright, I.J.; Lavorel, S.; Dray, S.; Reu, B.; Kleyer, M.; Wirth, C.; Colin Prentice, I.; et al. The global spectrum of plant form and function. Nature 2016, 529, 167–171. [Google Scholar] [CrossRef]
- Schepaschenko, D.; See, L.; Lesiv, M.; Bastin, J.-F.; Mollicone, D.; Tsendbazar, N.-E.; Bastin, L.; McCallum, I.; Laso Bayas, J.C.; Baklanov, A.; et al. Recent Advances in Forest Observation with Visual Interpretation of Very High-Resolution Imagery. Surv. Geophys. 2019, 40, 839–862. [Google Scholar] [CrossRef]
- Hill, J.; Buddenbaum, H.; Townsend, P.A. Imaging Spectroscopy of Forest Ecosystems: Perspectives for the Use of Space-borne Hyperspectral Earth Observation Systems. Surv. Geophys. 2019, 40, 553–588. [Google Scholar] [CrossRef]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Miraglio, T.; Adeline, K.; Huesca, M.; Ustin, S.; Briottet, X. Monitoring LAI, Chlorophylls, and Carotenoids Content of a Woodland Savanna Using Hyperspectral Imagery and 3D Radiative Transfer Modeling. Remote Sens. 2020, 12, 28. [Google Scholar] [CrossRef]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-Bares, J. Mapping foliar functional traits and their uncertainties across three years in a grassland experiment. Remote Sens. Environ. 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Kunstler, G.; Falster, D.; Coomes, D.A.; Hui, F.; Kooyman, R.M.; Laughlin, D.C.; Poorter, L.; Vanderwel, M.; Vieilledent, G.; Wright, S.J.; et al. Plant functional traits have globally consistent effects on competition. Nature 2016, 529, 204–207. [Google Scholar] [CrossRef]
- Palmer, M.W.; Wohlgemuth, T.; Earls, P.; Arévalo, J.R.; Thompson, S.D. Opportunities for long-term ecological research at the Tallgrass Prairie Preserve, Oklahoma. In Proceedings of the ILTER Regional Workshop: Cooperation in Long Term Ecological Research in Central and Eastern Europe, Budapest, Hungary, 22–25 June 1999; Volume 2229, p. 123128. [Google Scholar]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Rocchini, D.; Chiarucci, A.; Loiselle, S.A. Testing the spectral variation hypothesis by using satellite multispectral images. Acta Oecologica 2004, 26, 117–120. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing. Glob. Ecol. Conserv. 2016, 8, 212–219. [Google Scholar] [CrossRef]
- Dahlin, K.M. Spectral diversity area relationships for assessing biodiversity in a wildland–agriculture matrix. Ecol. Appl. 2016, 26, 2758–2768. [Google Scholar] [CrossRef]
- Gholizadeh, H.; Gamon, J.A.; Zygielbaum, A.I.; Wang, R.; Schweiger, A.K.; Cavender-Bares, J. Remote sensing of biodiversity: Soil correction and data dimension reduction methods improve assessment of α-diversity (species richness) in prairie ecosystems. Remote Sens. Environ. 2018, 206, 240–253. [Google Scholar] [CrossRef]
- Bello, F.D.; Lepš, J.; Sebastià, M.-T. Predictive value of plant traits to grazing along a climatic gradient in the Mediterranean. J. Appl. Ecol. 2005, 42, 824–833. [Google Scholar] [CrossRef]
- Díaz, S.; Cabido, M.; Zak, M.; Carretero, E.M.; Araníbar, J. Plant functional traits, ecosystem structure and land-use history along a climatic gradient in central-western Argentina. J. Veg. Sci. 1999, 10, 651–660. [Google Scholar] [CrossRef]
- Aguirre-Gutiérrez, J.; Oliveras, I.; Rifai, S.; Fauset, S.; Adu-Bredu, S.; Affum-Baffoe, K.; Baker, T.R.; Feldpausch, T.R.; Gvozdevaite, A.; Hubau, W.; et al. Drier tropical forests are susceptible to functional changes in response to a long-term drought. Ecol. Lett. 2019, 22, 855–865. [Google Scholar] [CrossRef]
- Féret, J.-B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef]
- Martin, M.E.; Plourde, L.C.; Ollinger, S.V.; Smith, M.-L.; McNeil, B.E. A generalizable method for remote sensing of canopy nitrogen across a wide range of forest ecosystems. Remote Sens. Environ. 2008, 112, 3511–3519. [Google Scholar] [CrossRef]
- Wieczynski, D.J.; Boyle, B.; Buzzard, V.; Duran, S.M.; Henderson, A.N.; Hulshof, C.M.; Kerkhoff, A.J.; McCarthy, M.C.; Michaletz, S.T.; Swenson, N.G.; et al. Climate shapes and shifts functional biodiversity in forests worldwide. Proc. Natl. Acad. Sci. USA 2019, 116, 587–592. [Google Scholar] [CrossRef]
- Lewis, S.L. Tropical forests and the changing earth system. Philos. Trans. R. Soc. B: Biol. Sci. 2006, 361, 195–210. [Google Scholar] [CrossRef]
- Enquist, B.J.; Enquist, C.A.F. Long-term change within a Neotropical forest: assessing differential functional and floristic responses to disturbance and drought. Glob. Chang. Biol. 2011, 17, 1408–1424. [Google Scholar] [CrossRef]
- Lewis, S.L.; Sonké, B.; Sunderland, T.; Begne, S.K.; Lopez-Gonzalez, G.; van der Heijden, G.M.F.; Phillips, O.L.; Affum-Baffoe, K.; Baker, T.R.; Banin, L.; et al. Above-ground biomass and structure of 260 African tropical forests. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120295. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Roy, P.S. Biodiversity information in India: Status and future scope. In Biodiversity in Tropical Ecosystems; Springer: Berlin, Germany, 2015; pp. 9–13. ISBN 1-55528-370-5. [Google Scholar]
- UNNI, N.V.M.; ROY, P.S.; PARTHASARATHY, V. Evaluation of LANDSAT and airborne multispectral data and aerial photographs for mapping forest features and phenomena in a part of the Godavari basin. Int. J. Remote Sens. 1985, 6, 419–431. [Google Scholar] [CrossRef]
- Ambastha, K.R.; Jha, C.S. Geospatial analysis of Tamil Nadu Eastern Ghats forest types at landscape level with reference to fragmentation and species diversity. J. Indian Soc. Remote Sens. 2010, 38, 453–463. [Google Scholar] [CrossRef]
- Gairola, S.; Procheş, Ş.; Rocchini, D. High-resolution satellite remote sensing: a new frontier for biodiversity exploration in Indian Himalayan forests. Int. J. Remote Sens. 2013, 34, 2006–2022. [Google Scholar] [CrossRef]
- Jha, C.S.; Fararoda, R.; Singhal, J.; Reddy, C.S.; Rajashekar, G.; Maity, S.; Patnaik, C.; Das, A.K.; Misra, A.; Singh, C.P.; et al. Characterization of Species Diversity and Forest Health using AVIRIS-NG Hyperspectral Remote Sensing Data. Curr. Sci. 2019, 116, 1124. [Google Scholar] [CrossRef]
- Thompson, D.R.; Gao, B.-C.; Green, R.O.; Roberts, D.A.; Dennison, P.E.; Lundeen, S.R. Atmospheric correction for global mapping spectroscopy: ATREM advances for the HyspIRI preparatory campaign. Remote Sens. Environ. 2015, 167, 64–77. [Google Scholar] [CrossRef]
- Soenen, S.A.; Peddle, D.R.; Coburn, C.A. SCS+C: a modified Sun-canopy-sensor topographic correction in forested terrain. IEEE Trans. Geosci. Remote Sens. 2005, 43, 2148–2159. [Google Scholar] [CrossRef]
- Wanner, W.; Li, X.; Strahler, A.H. On the derivation of kernels for kernel-driven models of bidirectional reflectance. J. Geophys. Res. Atmos. 1995, 100, 21077–21089. [Google Scholar] [CrossRef]
- Kindt, R.; Coe, R. Tree Diversity Analysis. A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies; World Agro-forestry Center (ICRAF): Nairobi, Kenya, 2006. [Google Scholar]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cornwell, W.K.; Schwilk, D.W.; Ackerly, D.D. A Trait-Based Test for Habitat Filtering: Convex Hull Volume. Ecology 2006, 87, 1465–1471. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Avolio, M.L.; Forrestel, E.J.; Chang, C.C.; Pierre, K.J.L.; Burghardt, K.T.; Smith, M.D. Demystifying dominant species. New Phytol. 2019, 223, 1106–1126. [Google Scholar] [CrossRef]
- Ricklefs, R.E.; Miller, G.L. Ecology, 4th ed.; W.H. Freeman & Co: New York, NY, USA, 2000; ISBN 978-0-7167-2829-0. [Google Scholar]
- Ricklefs, R.E. Disintegration of the Ecological Community. Am. Nat. 2008, 172, 741–750. [Google Scholar] [CrossRef]
- Scheiter, S.; Langan, L.; Higgins, S.I. Next-generation dynamic global vegetation models: learning from community ecology. New Phytol. 2013, 198, 957–969. [Google Scholar] [CrossRef]
- Engelbrecht, B.M.J.; Comita, L.S.; Condit, R.; Kursar, T.A.; Tyree, M.T.; Turner, B.L.; Hubbell, S.P. Drought sensitivity shapes species distribution patterns in tropical forests. Nature 2007, 447, 80–82. [Google Scholar] [CrossRef]
- Randin, C.F.; Ashcroft, M.B.; Bolliger, J.; Cavender-Bares, J.; Coops, N.C.; Dullinger, S.; Dirnböck, T.; Eckert, S.; Ellis, E.; Fernández, N.; et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 2020, 239, 111626. [Google Scholar] [CrossRef]
- Rocchini, D.; Hortal, J.; Lengyel, S.; Lobo, J.M.; Jiménez-Valverde, A.; Ricotta, C.; Bacaro, G.; Chiarucci, A. Accounting for uncertainty when mapping species distributions: The need for maps of ignorance. Prog. Phys. Geogr. Earth Environ. 2011, 35, 211–226. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Sánchez-Azofeifa, A.; Castro-Esau, K. Intra- and inter-class spectral variability of tropical tree species at La Selva, Costa Rica: Implications for species identification using HYDICE imagery. Remote Sens. Environ. 2006, 105, 129–141. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gamon, J.A. Remote sensing of plant functional types. New Phytol. 2010, 186, 795–816. [Google Scholar] [CrossRef]
- Modzelewska, A.; Fassnacht, F.E.; Stereńczak, K. Tree species identification within an extensive forest area with diverse management regimes using airborne hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101960. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Puletti, N.; Hawthorne, W.; Liesenberg, V.; Corona, P.; Papale, D.; Chen, Q.; Valentini, R. Discrimination of tropical forest types, dominant species, and mapping of functional guilds by hyperspectral and simulated multispectral Sentinel-2 data. Remote Sens. Environ. 2016, 176, 163–176. [Google Scholar] [CrossRef]
- Jetz, W.; Cavender-Bares, J.; Pavlick, R.; Schimel, D.; Davis, F.W.; Asner, G.P.; Guralnick, R.; Kattge, J.; Latimer, A.M.; Moorcroft, P.; et al. Monitoring plant functional diversity from space. Nat. Plants 2016, 2, 1–5. [Google Scholar] [CrossRef]
- Albert, C.H.; Grassein, F.; Schurr, F.M.; Vieilledent, G.; Violle, C. When and how should intraspecific variability be considered in trait-based plant ecology? Perspect. Plant Ecol. Evol. Syst. 2011, 13, 217–225. [Google Scholar] [CrossRef]
- Siefert, A.; Violle, C.; Chalmandrier, L.; Albert, C.H.; Taudiere, A.; Fajardo, A.; Aarssen, L.W.; Baraloto, C.; Carlucci, M.B.; Cianciaruso, M.V.; et al. A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 2015, 18, 1406–1419. [Google Scholar] [CrossRef] [PubMed]
- Poorter, L.; Castilho, C.V.; Schietti, J.; Oliveira, R.S.; Costa, F.R.C. Can traits predict individual growth performance? A test in a hyperdiverse tropical forest. New Phytol. 2018, 219, 109–121. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Amarasekare, P.; Araújo, M.S.; Bürger, R.; Levine, J.M.; Novak, M.; Rudolf, V.H.W.; Schreiber, S.J.; Urban, M.C.; Vasseur, D.A. Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 2011, 26, 183–192. [Google Scholar] [CrossRef]
- Svenning, J.-C.; Kinner, D.A.; Stallard, R.F.; Engelbrecht, B.M.J.; Wright, S.J. Ecological Determinism in Plant Community Structure Across a Tropical Forest Landscape. Ecology 2004, 85, 2526–2538. [Google Scholar] [CrossRef]
- Stavros, E.N.; Schimel, D.; Pavlick, R.; Serbin, S.; Swann, A.; Duncanson, L.; Fisher, J.B.; Fassnacht, F.; Ustin, S.; Dubayah, R.; et al. ISS observations offer insights into plant function. Nat. Ecol. Evol. 2017, 1, 0194. [Google Scholar] [CrossRef]
- van Ewijk, K.Y.; Randin, C.F.; Treitz, P.M.; Scott, N.A. Predicting fine-scale tree species abundance patterns using biotic variables derived from LiDAR and high spatial resolution imagery. Remote Sens. Environ. 2014, 150, 120–131. [Google Scholar] [CrossRef]
- Hoffmann, S.; Schmitt, T.M.; Chiarucci, A.; Irl, S.D.H.; Rocchini, D.; Vetaas, O.R.; Tanase, M.A.; Mermoz, S.; Bouvet, A.; Beierkuhnlein, C. Remote sensing of β-diversity: Evidence from plant communities in a semi-natural system. Appl. Veg. Sci. 2019, 22, 13–26. [Google Scholar] [CrossRef]
| Sites | Location | Area (km2) | Average Elevation (m) | Rainfall (mm) | Temperature (°C) | |
|---|---|---|---|---|---|---|
| Lat (°N) | Lon (°E) | |||||
| SWS | 21.88 | 73.65 | 607.70 | 287 | 1210.66 | 27.00 |
| VNP | 20.82 | 73.44 | 23.99 | 169 | 1557.72 | 26.52 |
| MTR | 11.73 | 76.46 | 688.00 | 1233 | 1731.88 | 24.84 |
| Sites | Flight Elevation (km) | Image Acquisition Date | Image Acquisition Time (GMT +5:30) | Flight Footprint Length (km) | Cloud Cover |
|---|---|---|---|---|---|
| MTR | 4.83 | 05-Jan-2016 | 10:62:47–12:81:39 | 42.22 | Clear to hazy |
| SWS | 4.15 | 08-Feb-2016 | 12:58:43–13:61:58 | 44.85 | Clear |
| VNP | 4.16 | 09-Feb-2016 | 11:02:17–11:57:46 | 13.32 | Clear |
| Sites | Height (m) | Canopy Area (m2) | ||||
|---|---|---|---|---|---|---|
| Mean (± SD) | Min | Max | Mean (± SD) | Min | Max | |
| SWS | 12.48 ± 4.80 | 2.60 | 25.80 | 55.91 ± 65.27 | 2.41 | 613.80 |
| VNP | 18.31 ± 6.30 | 3.50 | 35.00 | 71.89 ± 70.44 | 2.55 | 446.93 |
| MTR | 22.90 ± 7.56 | 6.00 | 39.00 | 75.45 ± 74.66 | 2.41 | 638.20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaurasia, A.N.; Dave, M.G.; Parmar, R.M.; Bhattacharya, B.; Marpu, P.R.; Singh, A.; Krishnayya, N.S.R. Inferring Species Diversity and Variability over Climatic Gradient with Spectral Diversity Metrics. Remote Sens. 2020, 12, 2130. https://doi.org/10.3390/rs12132130
Chaurasia AN, Dave MG, Parmar RM, Bhattacharya B, Marpu PR, Singh A, Krishnayya NSR. Inferring Species Diversity and Variability over Climatic Gradient with Spectral Diversity Metrics. Remote Sensing. 2020; 12(13):2130. https://doi.org/10.3390/rs12132130
Chicago/Turabian StyleChaurasia, Amrita N., Maulik G. Dave, Reshma M. Parmar, Bimal Bhattacharya, Prashanth R. Marpu, Aditya Singh, and N. S. R. Krishnayya. 2020. "Inferring Species Diversity and Variability over Climatic Gradient with Spectral Diversity Metrics" Remote Sensing 12, no. 13: 2130. https://doi.org/10.3390/rs12132130
APA StyleChaurasia, A. N., Dave, M. G., Parmar, R. M., Bhattacharya, B., Marpu, P. R., Singh, A., & Krishnayya, N. S. R. (2020). Inferring Species Diversity and Variability over Climatic Gradient with Spectral Diversity Metrics. Remote Sensing, 12(13), 2130. https://doi.org/10.3390/rs12132130