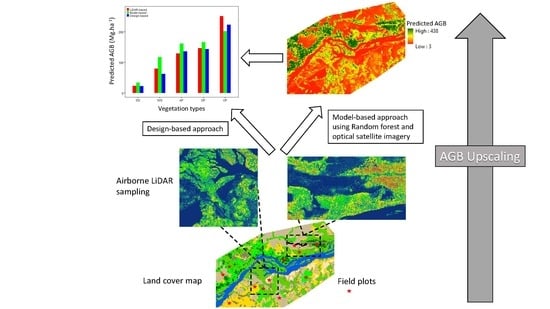
Airborne Lidar Sampling Pivotal for Accurate Regional AGB Predictions from Multispectral Images in Forest-Savanna Landscapes
Abstract
1. Introduction
2. Material and Methods
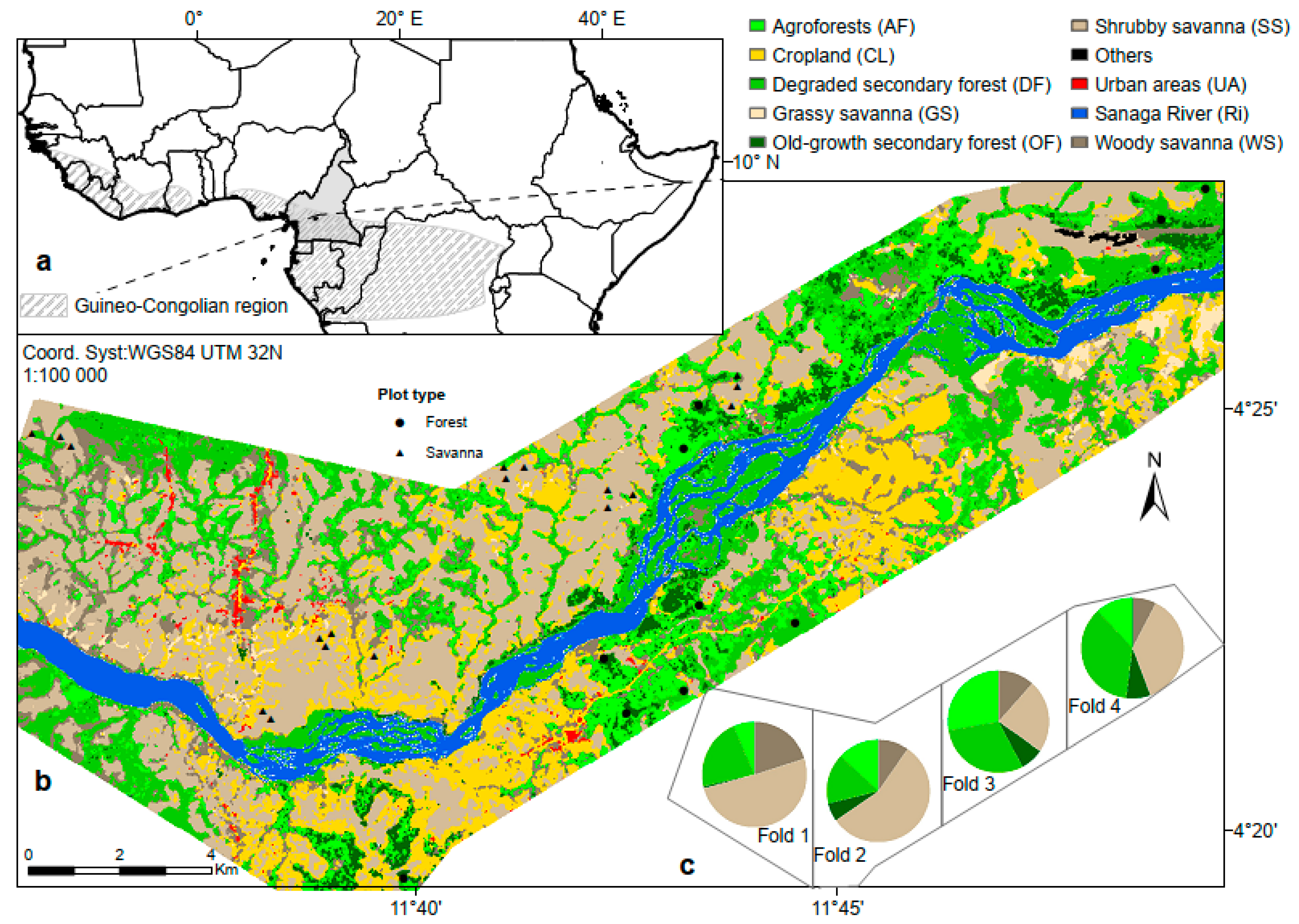
2.1. Study Site
2.2. Data Acquisition and Processing
2.2.1. Field Inventory Data
2.2.2. Airborne LiDAR Data
2.2.3. Spaceborne Data
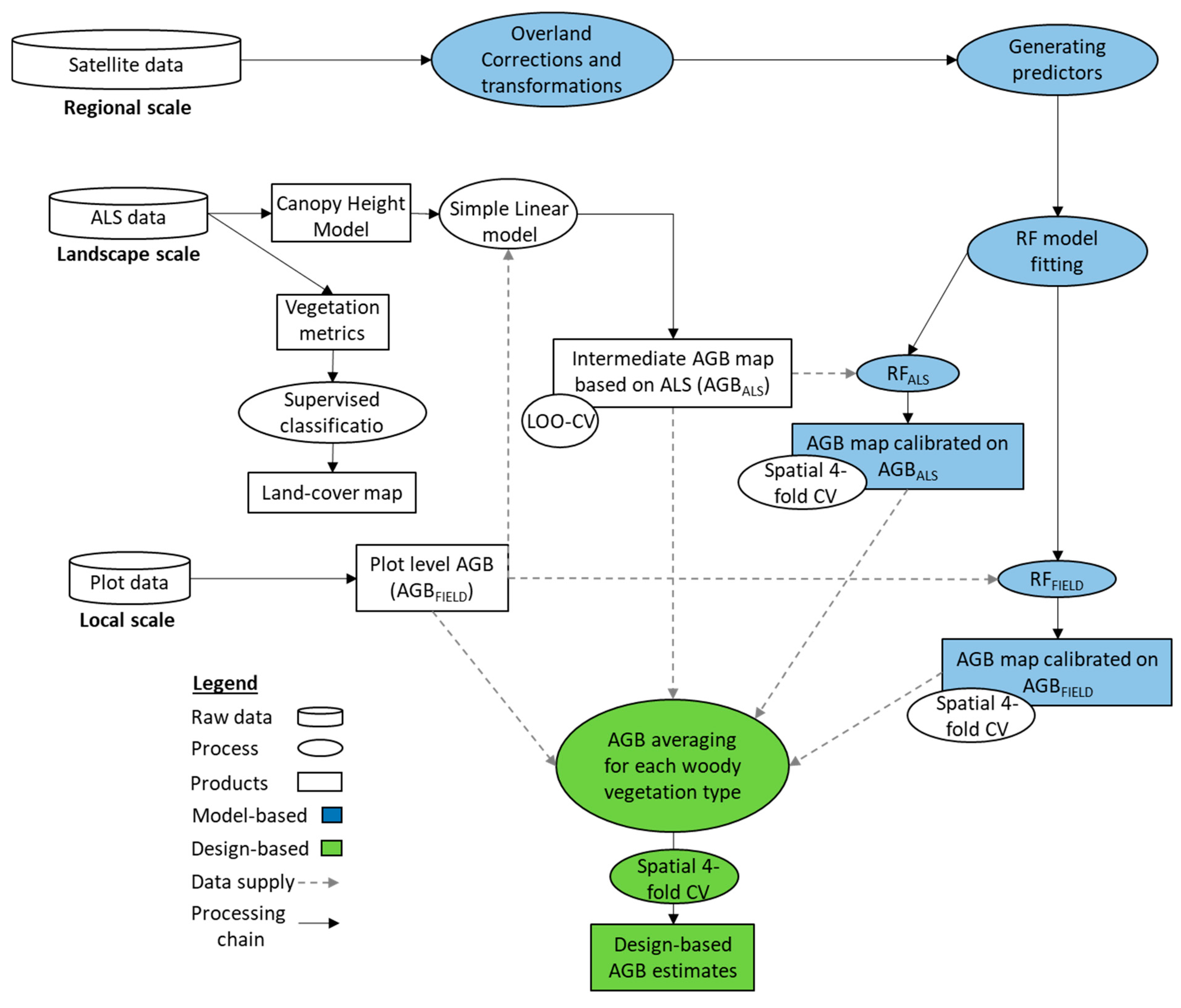
2.3. Upscaling AGB from Field to Spaceborne Measurements
2.3.1. Field-Based AGB Estimates
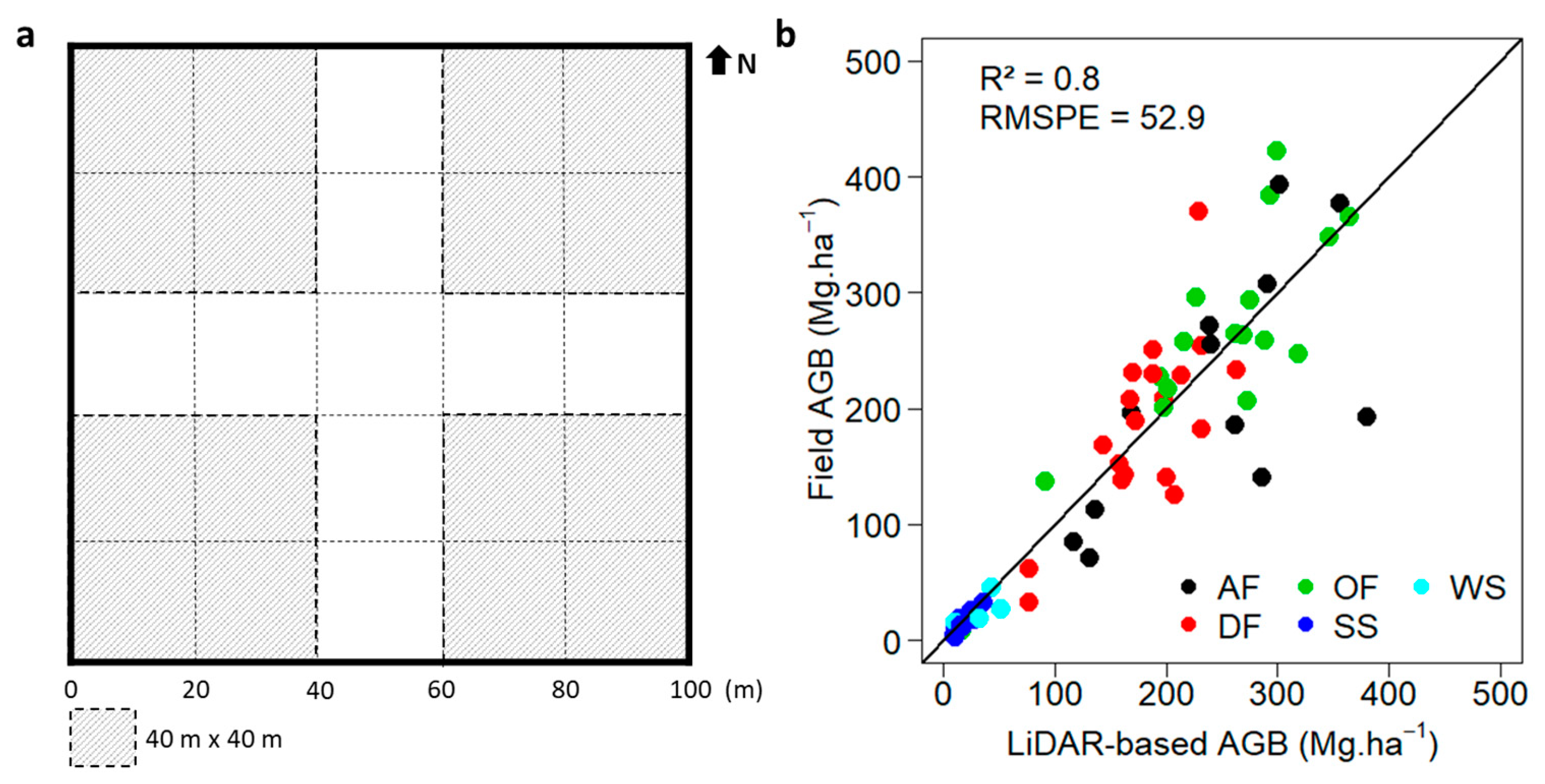
2.3.2. LiDAR-Based Reference AGB Map and Land-Cover Classification
2.3.3. Design-Based AGB Estimates
2.3.4. Model-Based AGB Estimates
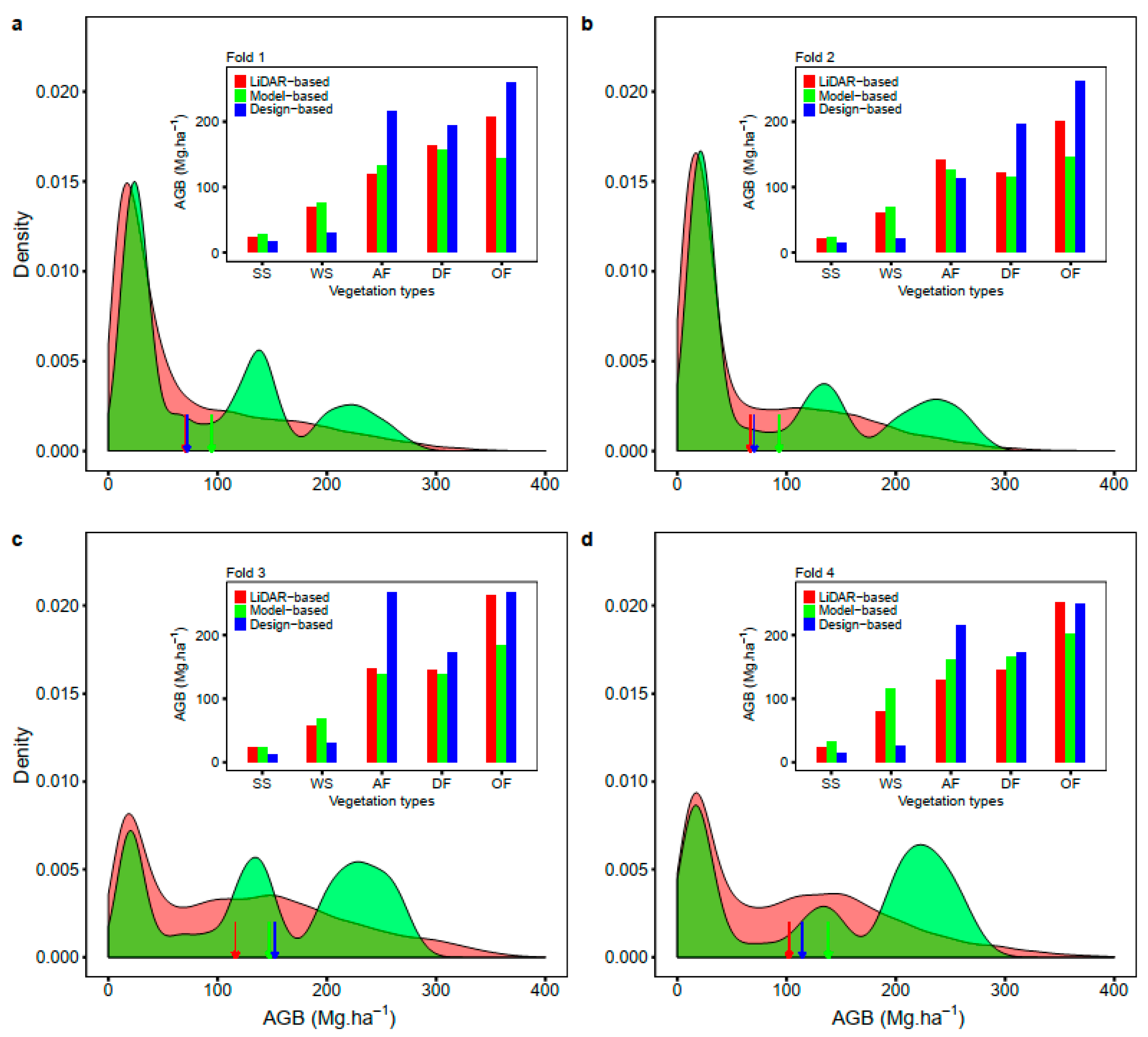
2.3.5. Four-Fold Cross-Validation
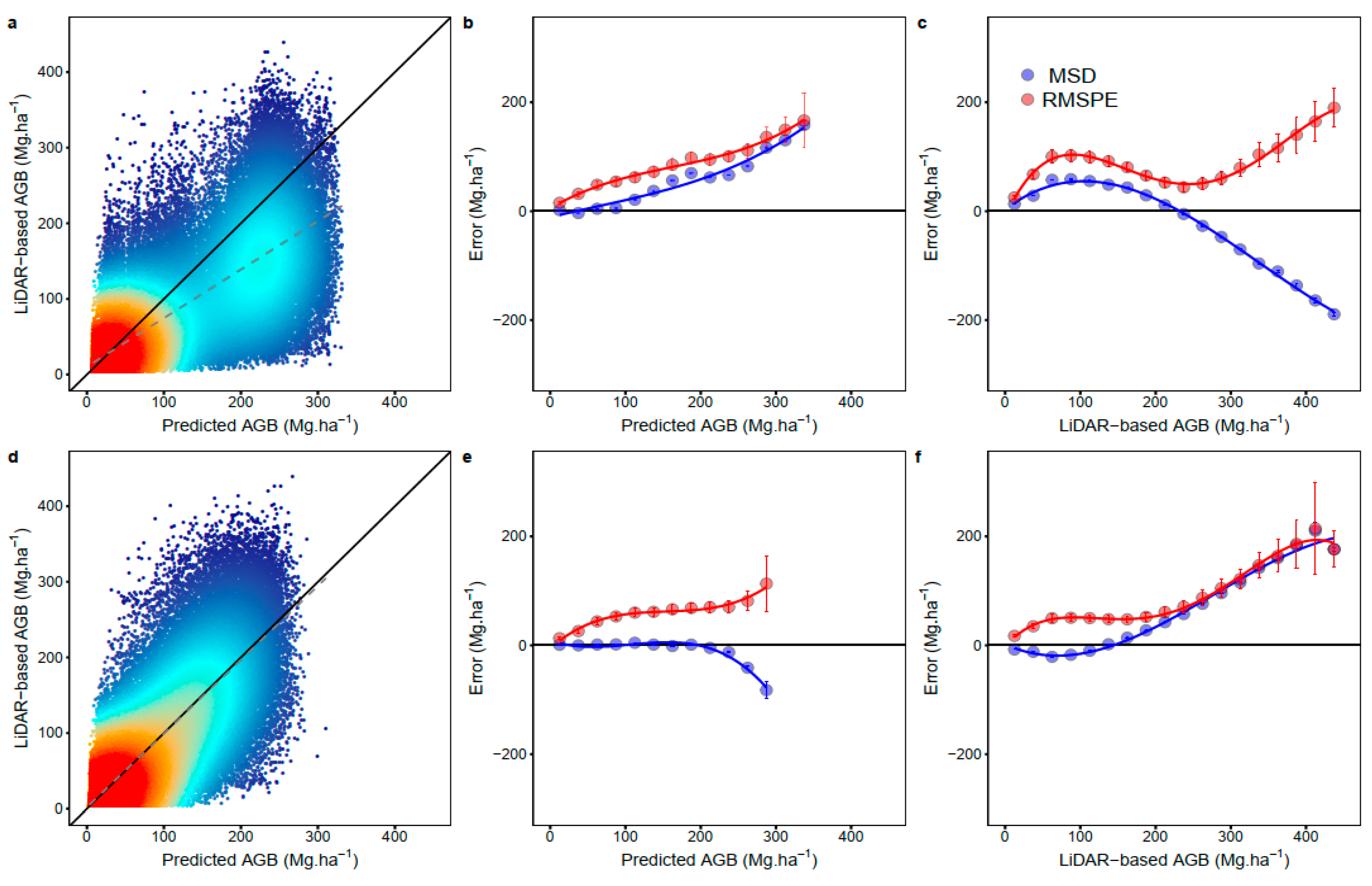
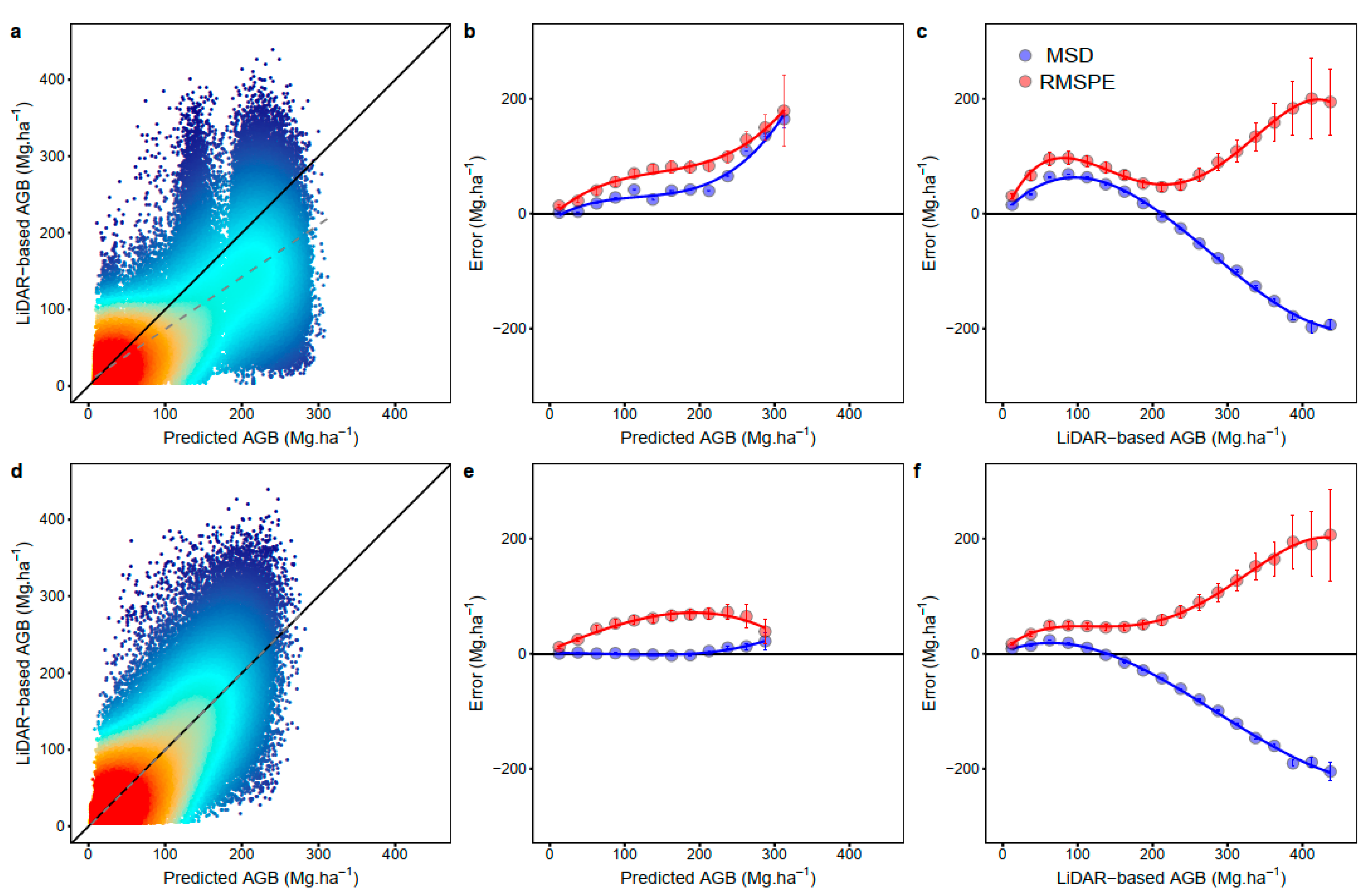
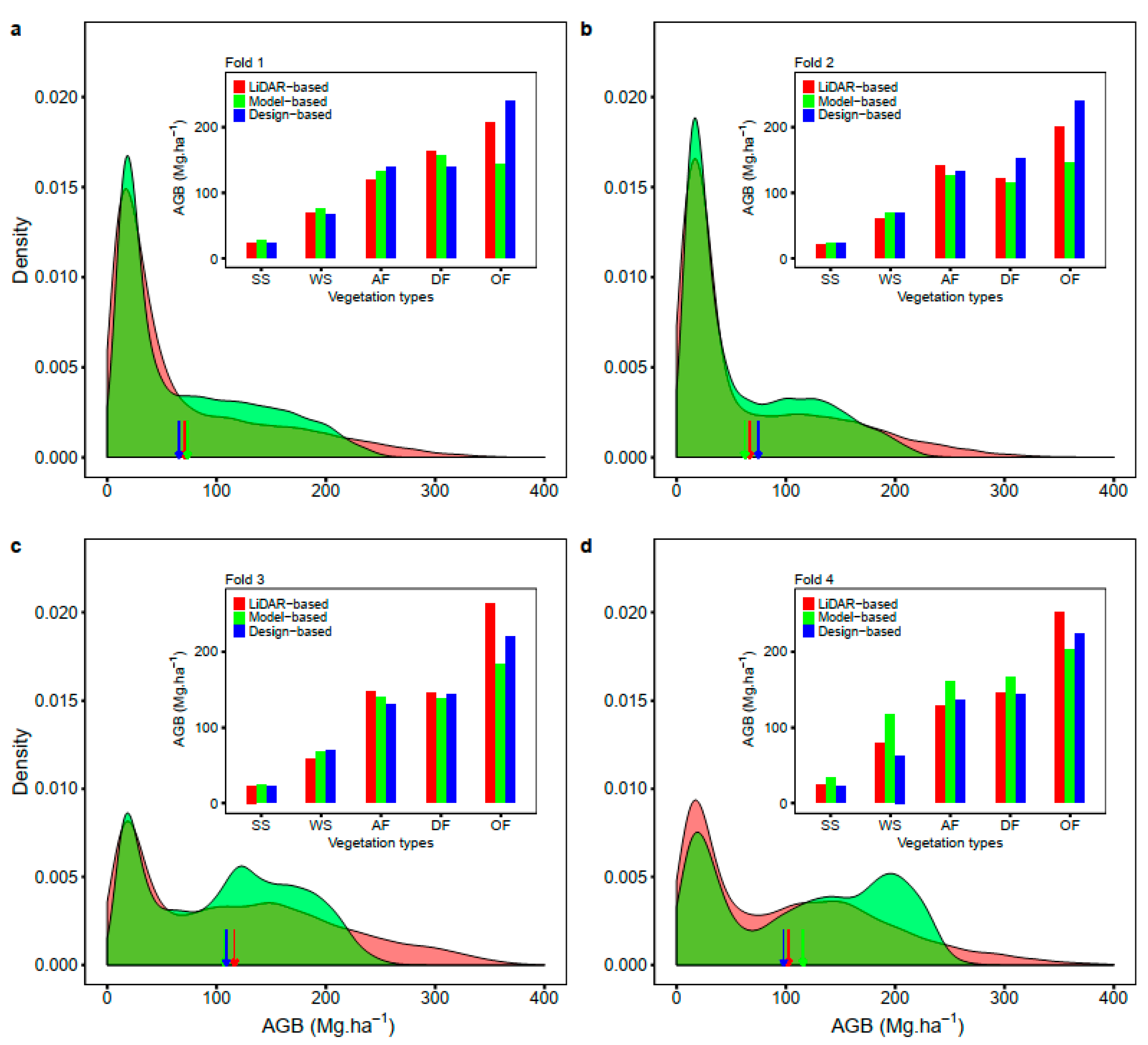
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C

Appendix D
Appendix E
References
- Ciais, P.; Bombelli, A.; Williams, M.; Piao, S.L.; Chave, J.; Ryan, C.M.; Henry, M.; Brender, P.; Valentini, R. The carbon balance of Africa: Synthesis of recent research studies. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 2038–2057. [Google Scholar] [CrossRef]
- Ciais, P.; Piao, S.-L.; Cadule, P.; Friedlingstein, P.; Chedin, A. Variability and recent trends in the African terrestrial carbon balance. Biogeosciences 2009, 6, 1935–1948. [Google Scholar] [CrossRef]
- Valentini, R.; Arneth, A.; Bombelli, A.; Castaldi, S.; Gatti, R.C.; Chevallier, F.; Ciais, P.; Grieco, E.; Hartmann, J.; Henry, M.; et al. A full greenhouse gases budget of Africa: Synthesis, uncertainties, and vulnerabilities. Biogeosciences 2014, 11, 381–407. [Google Scholar] [CrossRef]
- Williams, C.; Hanan, N.; Neff, J.; Scholes, R.J.; A Berry, J.; Denning, A.S.; Baker, D.F. Africa and the global carbon cycle. Carbon Balance Manag. 2007, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Richard, A.; Pekka, E.; Werner, A.; Oliver, L.; Simon, L.; Josep, G.; Robert, B.; Stephen, W.; David, A. A large and persistent carbon sink in the world’s forests. Larg. Persistent Carbon Sink World For. 2011, 333, 988–993. [Google Scholar] [CrossRef]
- Avitabile, V.; Camia, A. An assessment of forest biomass maps in Europe using harmonized national statistics and inventory plots. For. Ecol. Manag. 2018, 409, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Réjou-Méchain, M.; Barbier, N.; Couteron, P.; Ploton, P.; Vincent, G.; Herold, M.; Mermoz, S.; Saatchi, S.; Chave, J.; De Boissieu, F.; et al. Upscaling Forest Biomass from Field to Satellite Measurements: Sources of Errors and Ways to Reduce Them. Surv. Geophys. 2019, 40, 881–911. [Google Scholar] [CrossRef]
- King, M.D.; Platnick, S. Spatial and Temporal Distribution of Tropospheric Clouds observed by MODIS onboard the Terra and Aqua Satellites. Four. Transform. Spectrosc. Hyperspec. Imaging Sound. Env. 2005, 51, 3826–3852. [Google Scholar] [CrossRef]
- Morton, D.C.; Nagol, J.; Carabajal, C.C.; Rosette, J.; Palace, M.; Cook, B.D.; Vermote, E.F.; Harding, D.J.; North, P. Amazon forests maintain consistent canopy structure and greenness during the dry season. Nature 2014, 506, 221–224. [Google Scholar] [CrossRef]
- Song, C.; Woodcock, C.E.; Seto, K.C.; Lenney, M.P.; Macomber, S.A. Classification and Change Detection Using Landsat TM Data. Remote. Sens. Environ. 2001, 75, 230–244. [Google Scholar] [CrossRef]
- Baccini, A.; Asner, G.P. Improving pantropical forest carbon maps with airborne LiDAR sampling. Carbon Manag. 2013, 4, 591–600. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaró, J. Mapping tropical forest carbon: Calibrating plot estimates to a simple LiDAR metric. Remote. Sens. Environ. 2014, 140, 614–624. [Google Scholar] [CrossRef]
- Xu, L.; Saatchi, S.S.; Shapiro, A.; Meyer, V.; Ferraz, A.; Yang, Y.; Bastin, J.-F.; Banks, N.; Boeckx, P.; Verbeeck, H.; et al. Spatial Distribution of Carbon Stored in Forests of the Democratic Republic of Congo. Sci. Rep. 2017, 7, 15030. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P. Tropical forest carbon assessment: Integrating satellite and airborne mapping approaches. Environ. Res. Lett. 2009, 4, 034009. [Google Scholar] [CrossRef]
- Jha, N.; Tripathi, N.K.; Chanthorn, W.; Brockelman, W.; Nathalang, A.; Pélissier, R.; Pimmasarn, S.; Ploton, P.; Sasaki, N.; Virdis, S.G.P.; et al. Forest aboveground biomass stock and resilience in a tropical landscape of Thailand. Biogeosciences 2020, 17, 121–134. [Google Scholar] [CrossRef]
- Réjou-Méchain, M.; Tymen, B.; Blanc, L.; Fauset, S.; Feldpausch, T.; Monteagudo, A.; Phillips, O.L.; Richard, H.; Chave, J. Using repeated small-footprint LiDAR acquisitions to infer spatial and temporal variations of a high-biomass Neotropical forest. Remote. Sens. Environ. 2015, 169, 93–101. [Google Scholar] [CrossRef]
- Adhikari, H.; Heiskanen, J.; Siljander, M.; Maeda, E.; Heikinheimo, V.; Pellikka, P.K. Determinants of Aboveground Biomass across an Afromontane Landscape Mosaic in Kenya. Remote. Sens. 2017, 9, 827. [Google Scholar] [CrossRef]
- Ordway, E.M.; Asner, G.P. Carbon declines along tropical forest edges correspond to heterogeneous effects on canopy structure and function. Proc. Natl. Acad. Sci. USA 2020, 117, 7863–7870. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Liu, J.; Su, Y.; Guo, Q.; Qiu, P.; Wu, X. Estimating aboveground biomass of the mangrove forests on northeast Hainan Island in China using an upscaling method from field plots, Uav-Lidar data and Sentinel-2 imagery. Int. J. Appl. Earth Obs. Geoinform. 2020, 85, 101986. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J.; Anderson, C.B.; E Knapp, D.; E Martin, R.; Kennedy-Bowdoin, T.; Van Breugel, M.; Davies, S.J.; Hall, J.S.; Muller-Landau, H.C.; et al. High-fidelity national carbon mapping for resource management and REDD. Carbon Balance Manag. 2013, 8, 7. [Google Scholar] [CrossRef]
- Hirata, Y.; Furuya, N.; Saito, H.; Pak, C.; Leng, C.; Sokh, H.; Vuthy, M.; Kajisa, T.; Ota, T.; Mizoue, N. Object-Based Mapping of Aboveground Biomass in Tropical Forests Using LiDAR and Very-High-Spatial-Resolution Satellite Data. Remote. Sens. 2018, 10, 438. [Google Scholar] [CrossRef]
- Baccini, A.; Goetz, S.J.; Walker, W.S.; Laporte, N.T.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.S.A.; Dubayah, R.; Friedl, M.A.; et al. Estimated carbon dioxide emissions from tropical deforestation improved by carbon-density maps. Nat. Clim. Chang. 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Csillik, O.; Kumar, P.; Mascaro, J.; O’Shea, T.; Asner, G.P. Monitoring tropical forest carbon stocks and emissions using Planet satellite data. Sci. Rep. 2019, 9, 17831–17912. [Google Scholar] [CrossRef]
- Asner, G.P.; Brodrick, P.G.; Philipson, C.D.; Vaughn, N.R.; Martin, R.E.; E Knapp, D.; Heckler, J.; Evans, L.J.; Jucker, T.; Goossens, B.; et al. Mapped aboveground carbon stocks to advance forest conservation and recovery in Malaysian Borneo. Boil. Conserv. 2018, 217, 289–310. [Google Scholar] [CrossRef]
- Buendia, E.; Tanabe, K.; Kranjc, A.; Baasansuren, J.; Fukuda, M.; Ngarize, S.; Osako, A.; Pyrozhenko, Y.; Shermanau, P.; Federici, S. Refinement To the 2006 Ipcc Guidelines for National Greenhouse Gas Inventories; IPCC: Geneva, Switzerland, 2019. [Google Scholar]
- McRoberts, R.E.; Næsset, E.; Liknes, G.C.; Chen, Q.; Walters, B.; Saatchi, S.; Herold, M. Using a Finer Resolution Biomass Map to Assess the Accuracy of a Regional, Map-Based Estimate of Forest Biomass. Surv. Geophys. 2019, 40, 1001–1015. [Google Scholar] [CrossRef]
- Diziain, R. Word Atlas Of Agriculture; International Association of Agricultural Economists/Committee for the World Atlas of Agriculture: Novara, Italy, 1976. [Google Scholar]
- Djoufack, M.V.; Fonteine, B.; Tsalefac, M. Étude Multi-Echelles des Précipitations et du Couvert Végétal au Cameroun: Analyses Spatiales, Tendances Temporelles, Facteurs Climatiques et Anthropiques de Variabilité du NDVI; Université de Bourgogne et Université de Yaoundé I: Yaounde, Cameroon, 2011. [Google Scholar]
- Food andAgriculture Organization (FAO). Unesco Soil Map of the World 1:5 000 000; Unesco: Paris, France, 1977. [Google Scholar]
- Jagoret, P.; Michel-Dounias, I.; Snoeck, D.; Ngnogué, H.T.; Malézieux, E. Afforestation of savannah with cocoa agroforestry systems: A small-farmer innovation in central Cameroon. Agrofor. Syst. 2012, 86, 493–504. [Google Scholar] [CrossRef]
- Daget, P.; Poissonet, J. Prairies et Pâturages Méthodes d’Etude de Terrain et Interprétations; Cnrs/ Cir; Umr Selment (Systèmes d’Elevage Méditerranéens et Tropicaux): Montpellier, France, 2010; 955p. [Google Scholar]
- Réjou-Méchain, M.; Tanguy, A.; Piponiot, C.; Chave, J.; Hérault, B. Biomass: An package for estimating above-ground biomass and its uncertainty in tropical forests. Methods Ecol. Evol. 2017, 8, 1163–1167. [Google Scholar] [CrossRef]
- Jean-Romain, R.; David, A. Lidr: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. Remote Sens. 2019, 11. [Google Scholar] [CrossRef]
- Defence, A. Geoland2-BioPar Methods Compendium of MERIS FR Biophysical Products. GISci. Remote Sens. 2014, 52. [Google Scholar]
- Kneizys, F.X.; Shettle, E.P.; Gallery, W.O.; Chetwynd, J.H. Atmospheric Transmittance/Radiance: Computer Code Lowtran 5. Atmos. Trans. Rad. Com. Code Low. 1980. [Google Scholar]
- Jacquemoud, S.; Baret, F. Prpspect: A model of leaf optical properties spectra. Remote. Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflectance modeling: The Sail model. Remote. Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef]
- Chave, J.; Coomes, D.; Jansen, S.; Lewis, S.L.; Swenson, N.G.; Zanne, A.E. Towards a worldwide wood economics spectrum. Ecol. Lett. 2009, 12, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Zanne, A.E.; Lopez-Gonzalez, G.; Coomes, D.A.; Ilic, J.; Jansen, S.; Lewis, S.L.; Miller, R.B.; Swenson, N.G.; Wiemann, M.C.; Chave, J.; et al. Global Wood Density Database. 2009. Available online: http://hdl.handle.net/10255/dryad.235 (accessed on 16 March 2020).
- Chave, J.; Réjou-Méchain, M.; Burquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Boil. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Colgan, M.S.; Asner, G.P.; Swemmer, T.; Swemmer, A. Harvesting tree biomass at the stand level to assess the accuracy of field and airborne biomass estimation in savannas. Ecol. Appl. 2013, 23, 1170–1184. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote. Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.; Gao, X.; Ferreira, L. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote. Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote. Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Kross, A.; Seaquist, J.W.; Roulet, N.T.; Fernandes, R.; Sonnentag, O. Estimating carbon dioxide exchange rates at contrasting northern peatlands using MODIS satellite data. Remote. Sens. Environ. 2013, 137, 234–243. [Google Scholar] [CrossRef]
- Sharma, R.; Chakraborty, A.; Joshi, P.K. Geospatial quantification and analysis of environmental changes in urbanizing city of Kolkata. Environ. Monit. Assess. 2014, 187, 187. [Google Scholar] [CrossRef]
- Särndal, C.-E.; Thomsen, I.; Hoem, J.M.; Lindley, D.V.; Barndorff-Nielsen, O.; Dalenius, T. Design-Based and Model-Based Inference in Survey Sampling [with Discussion and Reply]. Scand. J. Stat. 1978, 5, 27–52. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Andy, L.; Matthew, W. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Mariana, B.; Lucian, D. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of spatial predictor variable selection in machine learning applications–Moving from data reproduction to spatial prediction. Ecol. Modell. 2019, 411. [Google Scholar] [CrossRef]
- Bouvet, A.; Mermoz, S.; Le Toan, T.; Villard, L.; Mathieu, R.; Naidoo, L.; Asner, G.P. An above-ground biomass map of African savannahs and woodlands at 25 m resolution derived from Alos Palsar. Remote. Sens. Environ. 2018, 206, 156–173. [Google Scholar] [CrossRef]
- Timothy, D.; Onisimo, M.; Cletah, S.; Adelabu, S.; Tsitsi, B. Remote sensing of aboveground forest biomass: A review. Trop. Ecol. 2016, 57, 125–132. [Google Scholar]
- Zhang, L.; Xu, M.; Qiu, S.; Li, R.; Zhao, H.; Shang, H.; Lai, C.; Zhang, W. Improving the estimate of forest biomass carbon storage by combining two forest inventory systems. Scand. J. For. Res. 2016, 32, 1–9. [Google Scholar] [CrossRef]
- Quegan, S.; Le Toan, T.; Chave, J.; Dall, J.; Exbrayat, J.-F.; Minh, D.H.T.; Lomas, M.; D’Alessandro, M.M.; Paillou, P.; Papathanassiou, K.; et al. The European Space Agency Biomass mission: Measuring forest above-ground biomass from space. Remote. Sens. Environ. 2019, 227, 44–60. [Google Scholar] [CrossRef]
| Country | Multispectral | Resolution (m) | Model Type | Model Validation | R2 | RMSPE (Mg.ha−1) | Ref. |
|---|---|---|---|---|---|---|---|
| Colombia & Peru | MODIS | 500 | Random Forest | strat 1 (10%) | 0.86 | 31.4–35.2 | [11] |
| China | Sentinel 2 | 10 | Random Forest | strat 2 | 0.62 | 50.36 | [19] |
| Panama | Landsat 5,7 | 100 | Random Forest | strat 1 (30%) | 0.62 | 45 | [20] |
| Cambodia | QuickBird | 1.5 | Multiple regression | - | 0.73 | 42.8 | [21] |
| Democratic Republic of Congo | Landsat 8 | 100 | Maximum Entropy | strat 3 | 0.76 | 61.29 | [13] |
| strat 4 | 0.65 | 62.16 | |||||
| Pantropical | MODIS | 500 | Random Forest | strat 1 (10%) | 0.71–0.83 | 38–50 | [22] |
| Peru | Planet Dove | 100 | Random Forest | strat 1 (20%) | 0.7 | 50.76 | [23] |
| Malaysian Borneo | Landsat 8 | 30 | Deep learning | strat 2 | 0.7 | 83.2 | [24] |
| Sites | No. Stem (ind.ha−1) | Lorey’s Height (m) | Basal Area (m2.ha−1) | Woody Biomass (Mg.ha−1) |
|---|---|---|---|---|
| Forest | 392 (216–538) | 27 (24–33) | 28 (19–35) | 234 (80–422) |
| Savanna | 239 (50–550) | 7 (5–10) | 18 (12–28) | 21 (1–133) |
| Designation | Spectral band | Candidate predictors | ||||
|---|---|---|---|---|---|---|
| S. 6/7 | L. 8 | S. 2 | S. 6/7 | L. 8 | S. 2 | |
| Blue | - | B2 | B2 | |||
| Green | B2 | B3 | B3 | x | x | x |
| Red | B3 | B4 | B4 | x | x | x |
| Red1 | - | - | B5 | x | ||
| Red2 | - | - | B6 | x | ||
| Red3 | - | - | B7 | x | ||
| NIR | B4 | B5 | B8 | x | x | x |
| Red4 | - | - | B8a | x | ||
| SWIR1 | - | B6 | B11 | x | x | |
| SWIR2 | - | B7 | B12 | x | x | |
| Vegetation indices | ||||||
| Equations | References | |||||
| RGR | (Red/Green) | [42] | x | x | x | |
| NIRGR | (NIR/Green) | x | x | x | ||
| NDVI | (NIR - Red)/(NIR + Red) | [43] | x | x | x | |
| EVI | 2.5*[(NIR - Red)/(1 + NIR + 6*Red -7.5*Blue)] | [44] | x | x | x | |
| SR | (NIR/RED) | [45] | x | x | x | |
| SAVI | (NIR - Red)/(NIR + Red + L)*1.5 with L = 0.5 | [44] | x | |||
| IRECI | (NIR - Red)/(Red1/Red2) | [46] | x | |||
| S2REP | [705 + 35*(0.5*(Red3 + Red)/2) - NIR)/(Red2 - NIR)] | [46] | x | |||
| NDVI1 | (NIR - Red1)/(NIR + Red1) | [47] | x | |||
| NDVI2 | (NIR - Red2)/(NIR + Red2) | [47] | x | |||
| NDVI3 | (NIR - Red3)/(NIR + Red3) | [47] | x | |||
| NDVI4 | (NIR - Red4)/(NIR + Red4) | [48] | x | |||
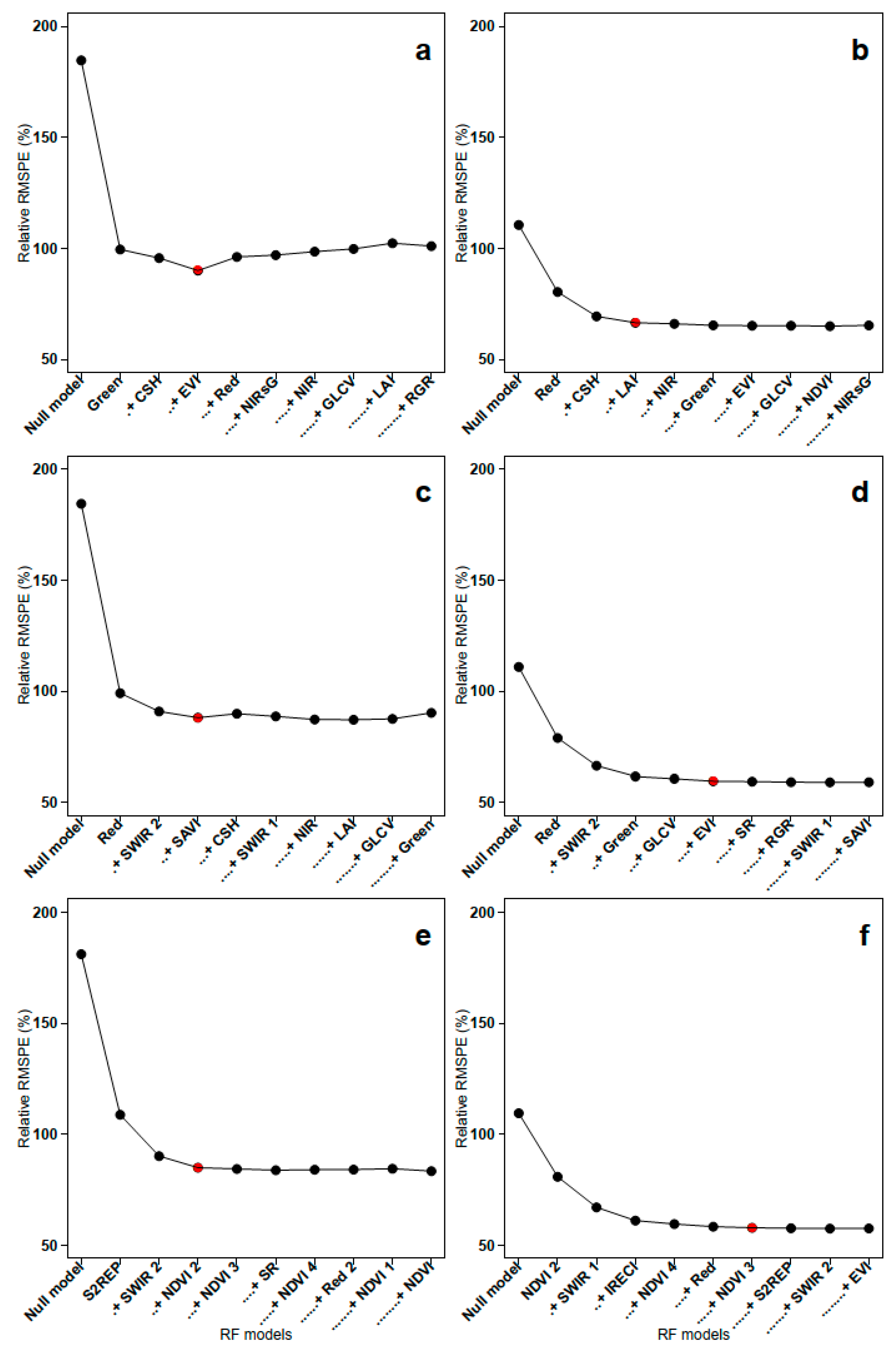
| Sensor | RFFIELD Models | R2 | RMSPE | Relative RMSPE | RFALS Models | R2 | RMSPE | Relative RMSPE |
|---|---|---|---|---|---|---|---|---|
| S. 6/7 | Green + CSF + EVI | 0.58 | 65 | 90 | Red + CSF + LAI | 0.62 | 48.3 | 66.5 |
| L. 8 | Red + SWIR 2 + SAVI | 0.61 | 64.8 | 88 | Red + SWIR 2 + Green + fCover + EVI | 0.7 | 43.1 | 60 |
| S. 2 | S2REP + SWIR 2 + NDVI 2 | 0.58 | 63.2 | 85 | NDVI 2 + SWIR 2 + IRECI + NDVI 4 + Red + NDVI 3 | 0.7 | 42.8 | 58 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagang, L.B.T.; Ploton, P.; Sonké, B.; Poilvé, H.; Couteron, P.; Barbier, N. Airborne Lidar Sampling Pivotal for Accurate Regional AGB Predictions from Multispectral Images in Forest-Savanna Landscapes. Remote Sens. 2020, 12, 1637. https://doi.org/10.3390/rs12101637
Sagang LBT, Ploton P, Sonké B, Poilvé H, Couteron P, Barbier N. Airborne Lidar Sampling Pivotal for Accurate Regional AGB Predictions from Multispectral Images in Forest-Savanna Landscapes. Remote Sensing. 2020; 12(10):1637. https://doi.org/10.3390/rs12101637
Chicago/Turabian StyleSagang, Le Bienfaiteur T., Pierre Ploton, Bonaventure Sonké, Hervé Poilvé, Pierre Couteron, and Nicolas Barbier. 2020. "Airborne Lidar Sampling Pivotal for Accurate Regional AGB Predictions from Multispectral Images in Forest-Savanna Landscapes" Remote Sensing 12, no. 10: 1637. https://doi.org/10.3390/rs12101637
APA StyleSagang, L. B. T., Ploton, P., Sonké, B., Poilvé, H., Couteron, P., & Barbier, N. (2020). Airborne Lidar Sampling Pivotal for Accurate Regional AGB Predictions from Multispectral Images in Forest-Savanna Landscapes. Remote Sensing, 12(10), 1637. https://doi.org/10.3390/rs12101637