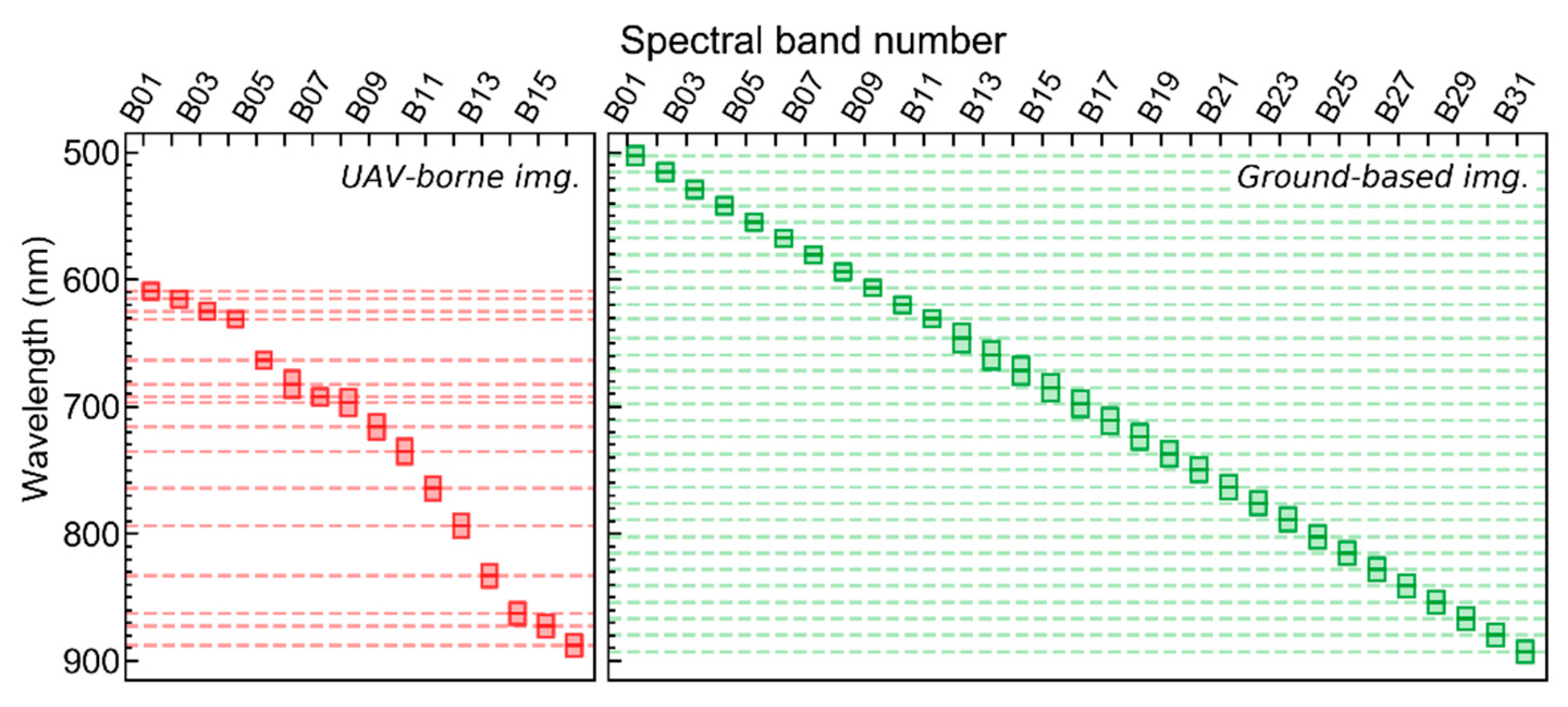
1. Introduction
Site-specific crop management and characterization of different cultivars in breeding trials (i.e., phenotyping) are examples of tasks demanding the description of vegetation biochemical and biophysical properties with high spatial and temporal resolution [
1,
2]. Recent advances in sensing solutions and subsequent data analysis targeting these applications can provide alternatives to increase feasibility of the detailed description of crop traits and plant response to stress [
3,
4,
5]. In this context, proximal or remote radiometric measurements of the vegetation canopy spectral response on discrete wavelength intervals in the visible (Vis), near infrared (NIR), and shortwave infrared (SWIR) have physically based relationships with leaf and canopy properties, and therefore have potential to be used for spatially explicit estimation of crop traits.
More specifically concerning biotic stress monitoring, assessment of disease incidence and severity frequently relies on visual rating, which is a time consuming activity susceptible to errors related to several factors, such as the complexity of the disease symptoms presented by plants and the level of experience of the professional performing the evaluation [
6]. Alternative solutions for identifying effects of pathogen development on crop traits based on radiometric measurements in the optical domain have been introduced for spectra acquired at leaf and canopy levels [
7,
8,
9,
10,
11,
12,
13,
14]. Although methods focusing on data acquired at leaf level demonstrate, in general, that a strong relationship between spectral changes and disease development exists, the same is not always observed in studies based on measurements acquired at canopy level. This fact is particularly true if discrimination between healthy and diseased vegetation is intended to be made during early stages of the pathogen development or under low disease severity levels [
15]. As described by Behmann et al. [
16], some aspects adding complexity and decreasing accuracy of early assessment of disease effects on crop traits based on spectral properties are: multiple factors simultaneously affecting the crop spectral response, besides disease-related changes (e.g., effects of nutrient and water availability or natural plant senescence); variability of canopy structure (e.g., leaf inclination), which together with changes in view-geometry and illumination conditions may have considerable impact on canopy reflectance measurements, in particular for data with very high spatial resolution; low signal-to-noise ratio for the spectra acquired; and the fact that changes occurring due to early disease development are subtle (pre-visual), which makes it difficult to obtain reference data (labels) at a more detailed scale than the plant level or without being mixed with information corresponding to healthy tissue and background.
Despite these limitations, several authors have reported successful discrimination between healthy and diseased crop patches and plants based on high resolution imagery acquired at canopy level by sensors mounted on Unmanned Aerial Vehicles (UAVs) or other airborne platforms. Many of the studies performed dedicate attention to the discrimination of diseased vegetation in perennial crops (e.g., Huanglongbing in citrus, leafroll disease and
Flavescence dorée in grapevine, verticilum wilt and
Xylella fastidiosa on olive trees, and red leaf blotch on almond orchards), using UAV-acquired multi- or hyperspectral data in the Vis-NIR, frequently coupled with thermal imagery and measurements of sun-induced fluorescence [
13,
14,
17,
18,
19,
20,
21,
22]. For annual crops, studies have also been conducted, for example, on yellow rust and powdery mildew in wheat [
23,
24] and on downy mildew in opium poppy plants [
25] based on airborne or UAV multi- or hyperspectral imagery in the Vis-NIR-SWIR and thermal infrared domains. In these studies, multiple features derived from the spectral information acquired have been tested to assess the impacts of disease incidence on crop traits, such as reflectance in single spectral bands, calculation of spectral distance metrics, derivation of vegetation indices, and estimation of crop traits from spectral measurements based on radiative transfer model (RTM) inversion. Usually, the features derived are subsequently used in parametric statistical analysis (e.g., analysis of variance and groups means test) or in parametric or non-parametric modelling frameworks for assessment of disease incidence or severity using classification or quantification methods (e.g., linear and quadratic discriminant analysis, support vector machines, classification and regression trees, etc.). With the variety of methods and features used, variable performance has also been reported for the discrimination between healthy and diseased plants and for the quantification of disease severity in the targeted areas. However, in most cases authors indicate that methods relying on optical imagery acquired at canopy level are sensitive enough to allow timely detection of disease incidence or accurate quantification of its severity.
Besides UAVs or other airborne platforms, ground-based imaging systems have been evaluated in studies concerning disease assessment based on optical data [
26,
27,
28,
29,
30]. In this case, the authors also focused on pathogens affecting different perennial and annual crops (e.g., Huanglongbing in citrus, cercospora leaf spot in sugarbeet, tulip breaking virus, yellow rust and fusarium head blight in wheat and barley) and tested several methods for discriminating diseased and healthy plants, or to quantify disease severity, achieving variable discriminative potential and accuracy. In addition to airborne or ground-based imaging system, a considerable number of studies have employed point-based spectral readings (mostly hemispherical-conical reflectance measurements [
31]) at canopy level to evaluate the development of different pathogens, and similarly to other sensing approaches, reported variable degrees of accuracy or discriminative potential for the information acquired [
32,
33,
34,
35,
36].
Consequently, considerable evidence exists that canopy based spectral data may provide useful information for discriminating between diseased and healthy plants or for assessing disease severity due to the direct impact of pathogen development on biochemical and biophysical properties of vegetation at leaf and canopy scales [
22]. However, disease symptoms, resulting from pathogen development and from plant response to infection, are to certain extent specific to each crop and disease considered [
9,
10]. Therefore, not all results obtained in a specific context can be generalized to others. Also, many studies performing data acquisition at canopy level do not include a detailed description of the relationship between disease development and changes in crop spectral response, or do not discuss the implications of these changes in the results obtained during discrimination of healthy and diseased areas, or during modeling of disease severity. This fact may be attributed to the lack of detail in the available datasets, mainly regarding spatial and spectral resolution or timely assessment of disease development. For example, some studies [
14,
18,
20,
24] adopt plants or canopy patches with up to 5–10% disease severity to characterize low pathogen incidence, which may be a relatively high value for other crops or pathogens, depending on the management practice to be implemented and on how early the detection need to be made.
Regarding late blight (
Phytophthora infestans) incidence in potato (
Solanum tuberosum), only a few studies are available relating disease development and crop heathy monitoring based on UAV imagery or other spectral datasets acquired at canopy level. Considering the importance of late blight assessment for potato management, as emphasized by Cooke et al. [
37], this topic is certainly of interest. Sugiura et al. [
38] presented an approach for assessing late blight severity using UAV optical imagery. This method involves RGB image color transformation and pixel-wise classification based on a threshold optimization procedure. Results obtained by these authors are relatively accurate, with reported R
2 between the area under the disease progress curve estimated visually and by the image-based approach, varying between 0.73 and 0.77. Duarte-Carvajalino et al. [
39] performed machine learning-based estimation of late blight severity using very high resolution imagery acquired over the growing season, with a modified camera registering blue, red, and NIR bands. Despite using considerably different prediction approaches in comparison with that described by Sugiura et al. [
38], the performance reported was similar in both studies. Other authors only described qualitative evaluation of late blight incidence using UAV multispectral imagery [
40,
41] or only assessed effects of advanced stages of the pathogen development on potato traits and crop spectral response using data acquired by an hyperspectral imaging system [
42,
43]. However, studies focusing on UAV or other sources of very high resolution imagery (with sub-decimeter resolution), including validation by means of ground truth data (e.g., measurement of crop traits and assessment of disease severity, etc.), and aiming a detailed description of spectral changes related to early pathogen incidence have not been made so far for potato infection with late blight.
Therefore, the objectives of the present study can be summarized as follows: (i) identify changes on the potato canopy reflectance in the optical domain related to late blight development, in different organic cropping systems (i.e., cultivation with a single cultivar in contrast to a mixture of different cultivars); (ii) compare alterations in the spectra observed using ground-based imagery (pixel size between 0.1 and 0.2 cm) with those detected through UAV data (pixel size between 4 and 5 cm); and (iii) evaluate the potential of UAV imagery for early discrimination of different late blight severity levels in potato, in particular identifying possible detectable changes in the canopy reflectance due to early disease development using sub-decimeter imagery. For that, ground-based and UAV images were analyzed using an up-to-date analytical framework, involving Simplex Volume Maximization (SiVM) and pixel-wise log-likelihood ratio (LLR) calculation, as similarly performed by other authors at leaf and canopy levels [
9,
44], in order to provide a sound basis for conclusions regarding the objectives of this research.
4. Discussion
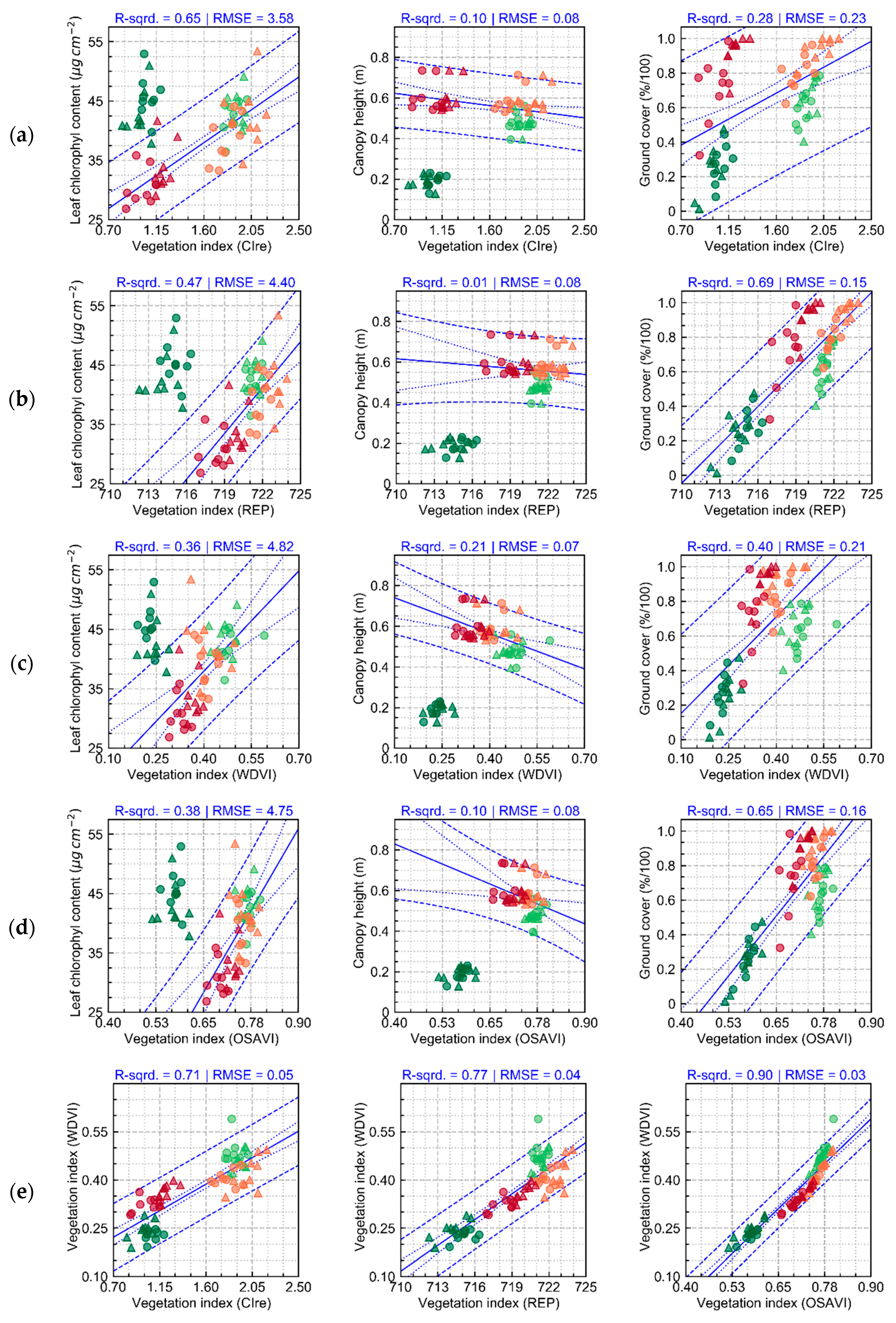
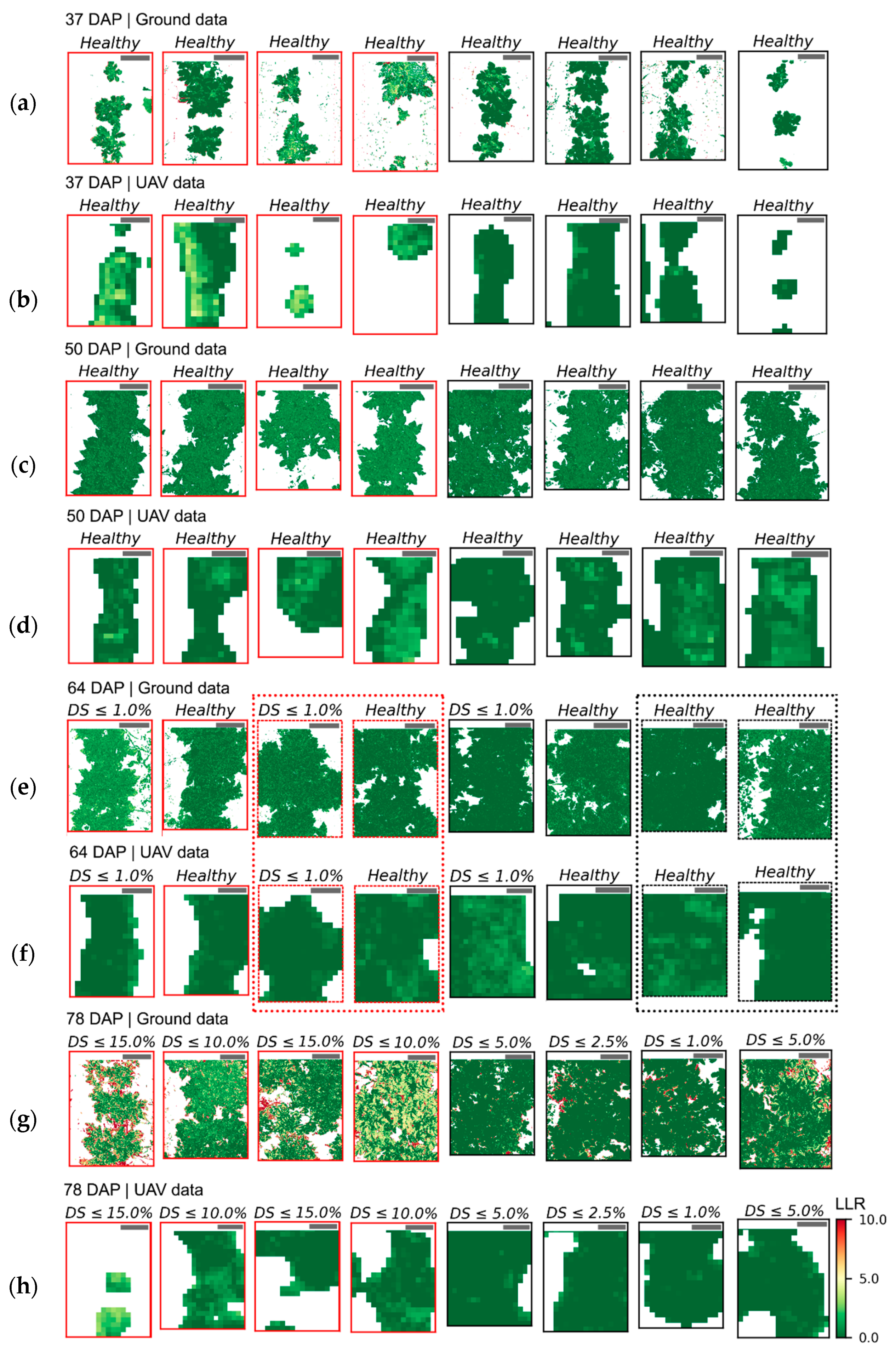
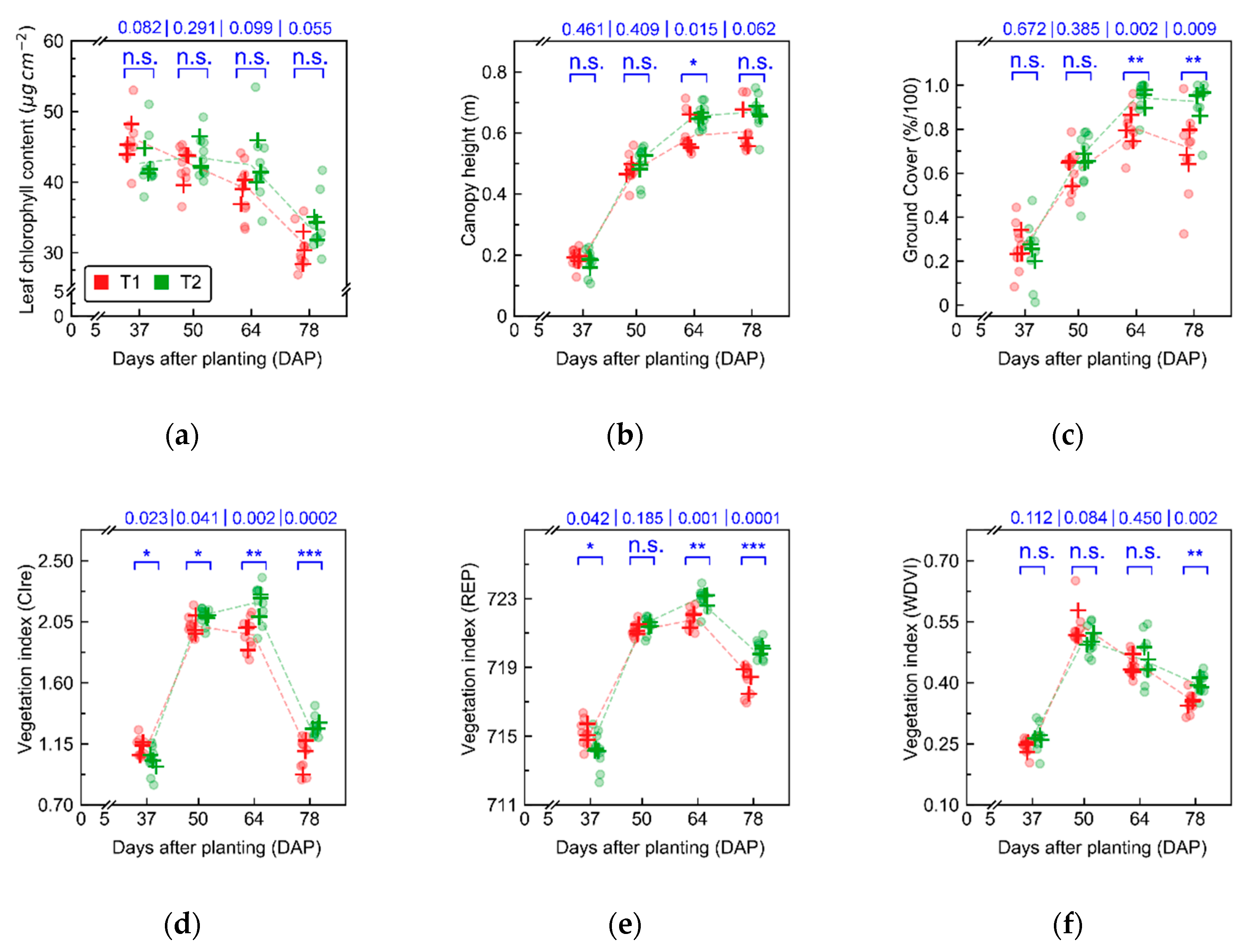
Measurements of crop traits, described in
Section 3.1 (
Figure 3 and
Figure A3), indicate that the first three data acquisitions (from 37 to 64 days after planting—DAP) were performed while crop growth progressed towards full canopy development, which occurred between 50 and 64 DAP. During this period, the main changes observed in crop traits were related to increase in leaf area index for plants cultivated in both treatments. Differences between treatments that were observed in early stages (between 37 and 50 DAP) can be attributed mainly to cultivar intrinsic characteristics. At 64 DAP, more substantial differences between treatments, concerning leaf chlorophyll content and canopy structural traits (e.g., ground cover), were observed, which may be related to initial stages of late blight development (
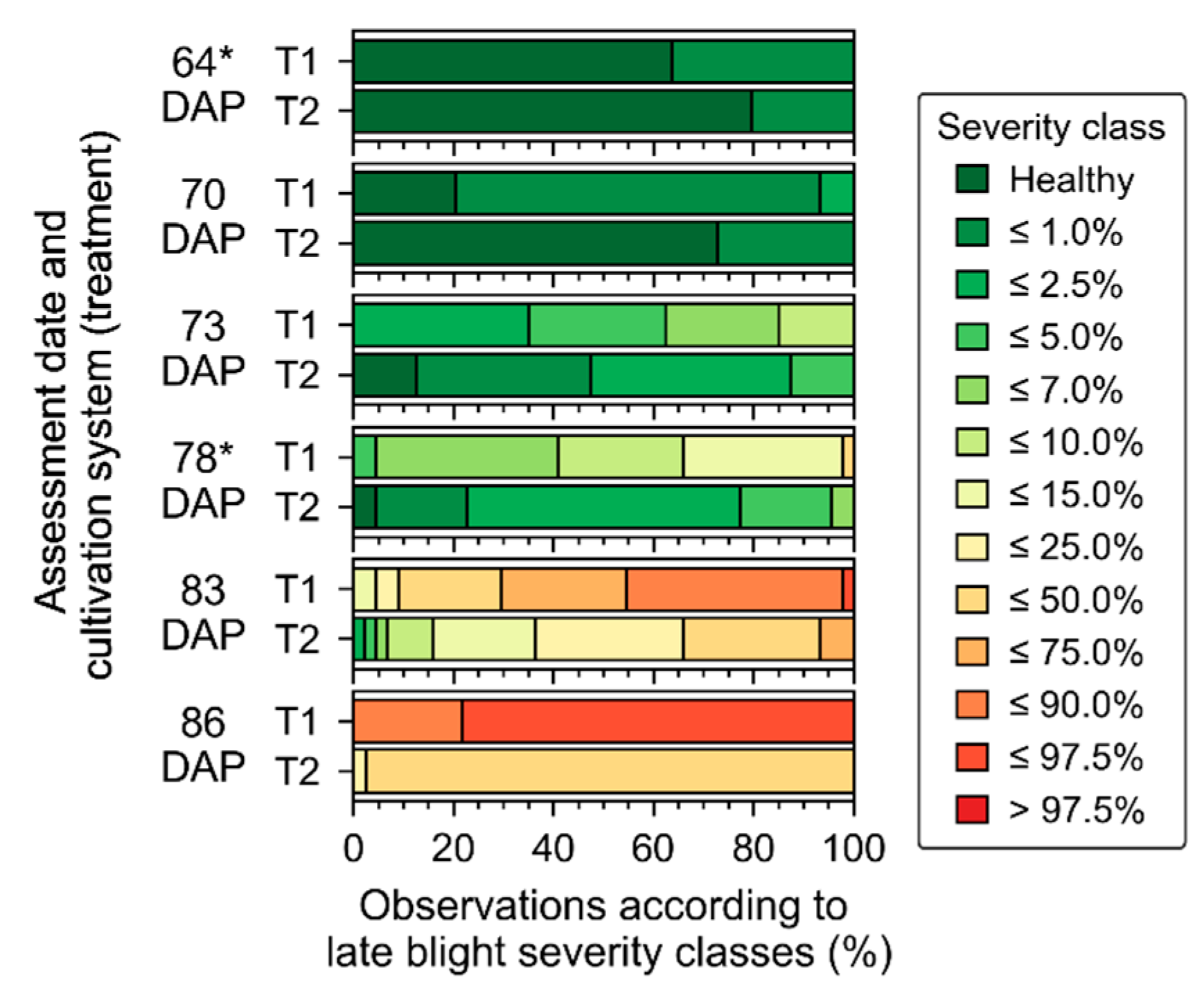
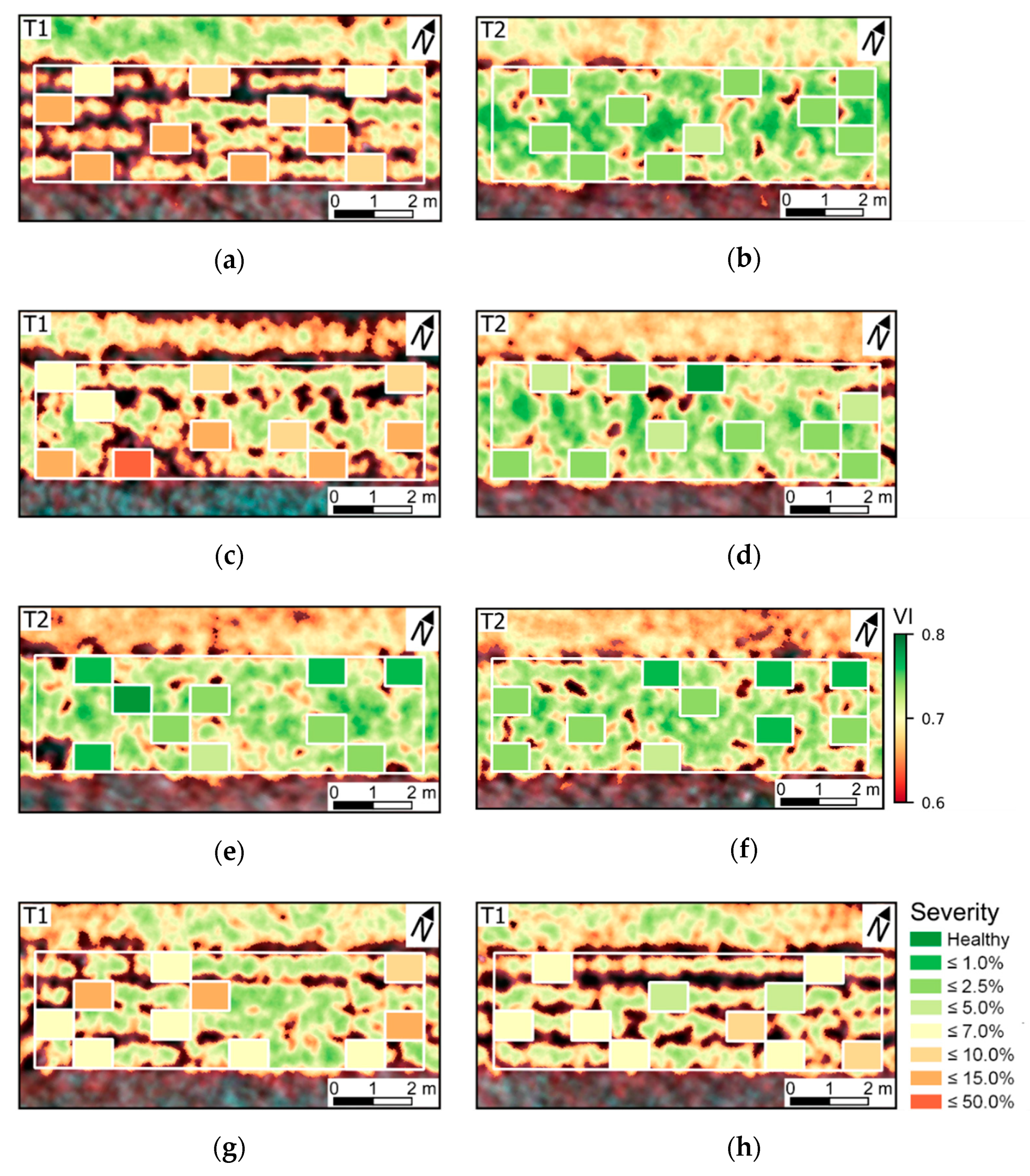
Figure 4). In the last data acquisition (78 DAP), differences between treatments increased following the increase in disease severity in particular for T1 (i.e., “non-mixed” system), which confirms trends observed 64 DAP.
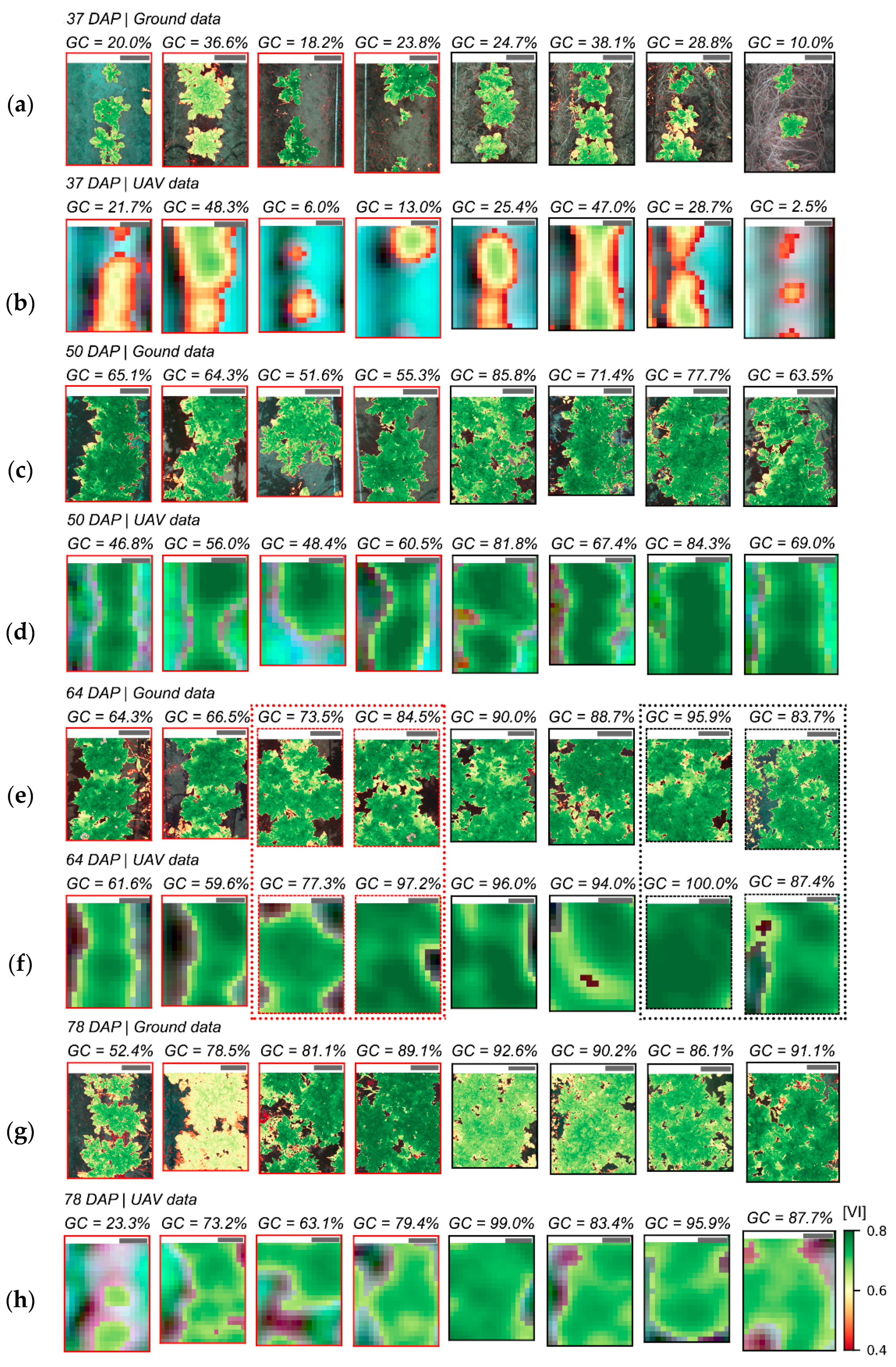
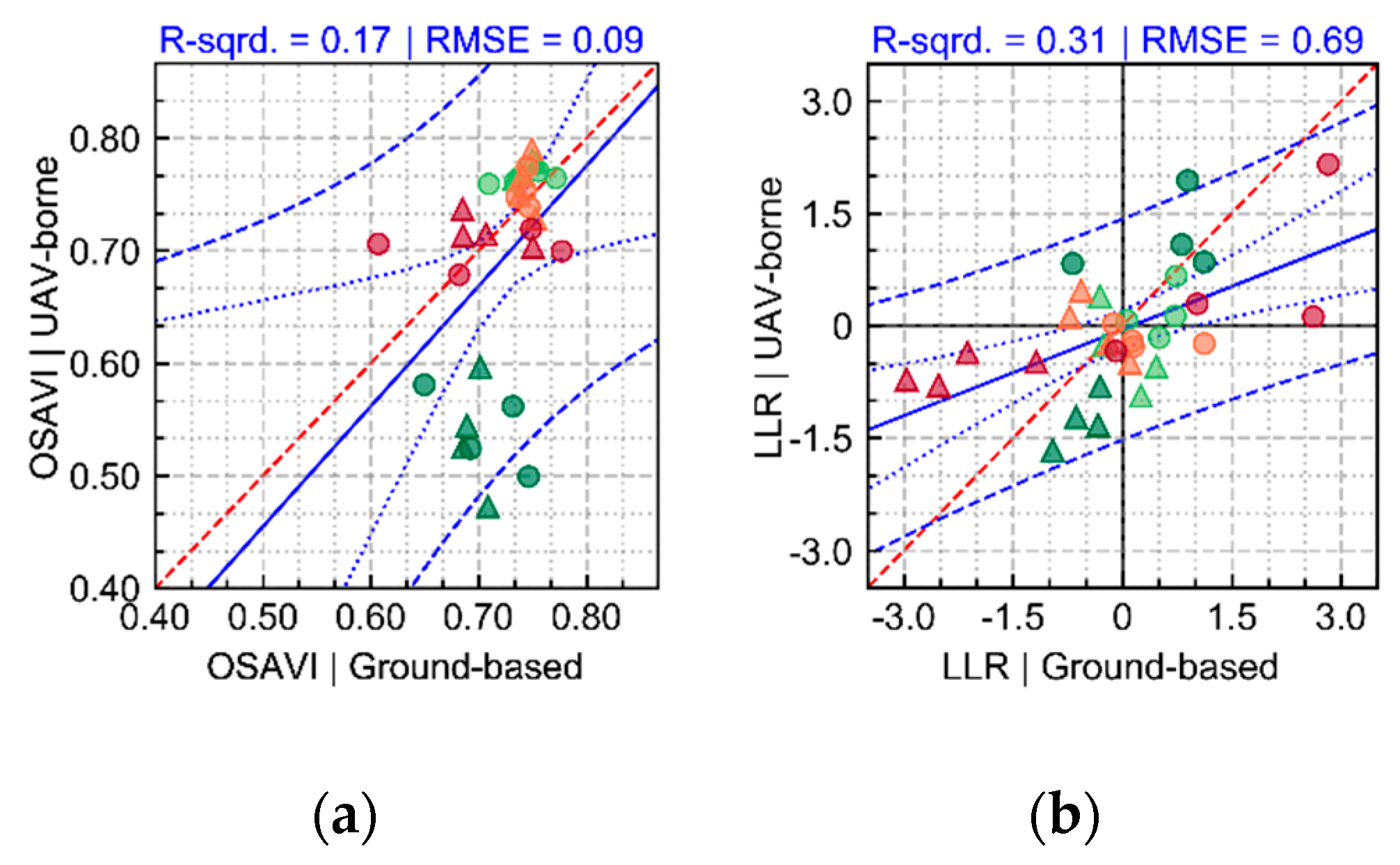
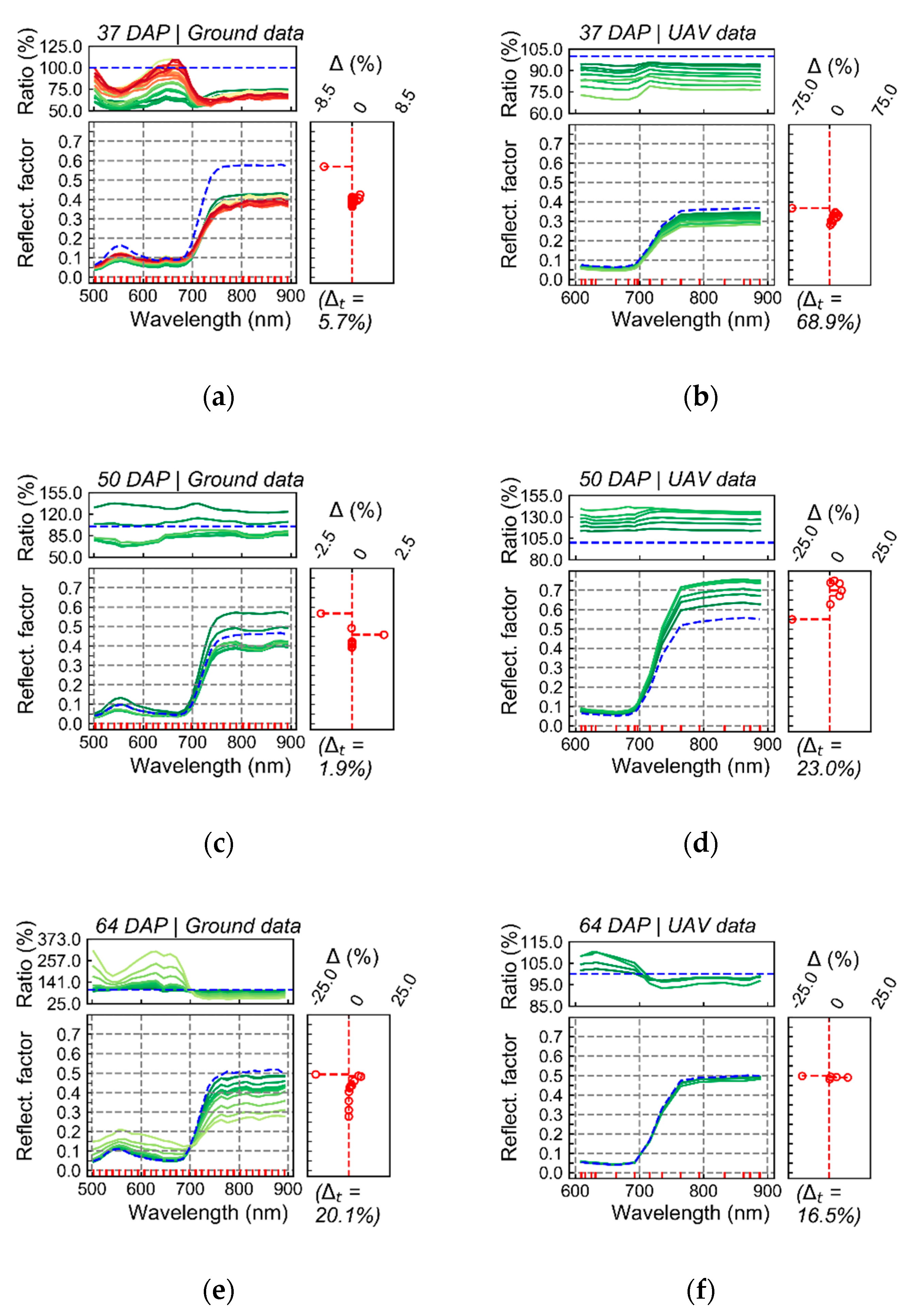
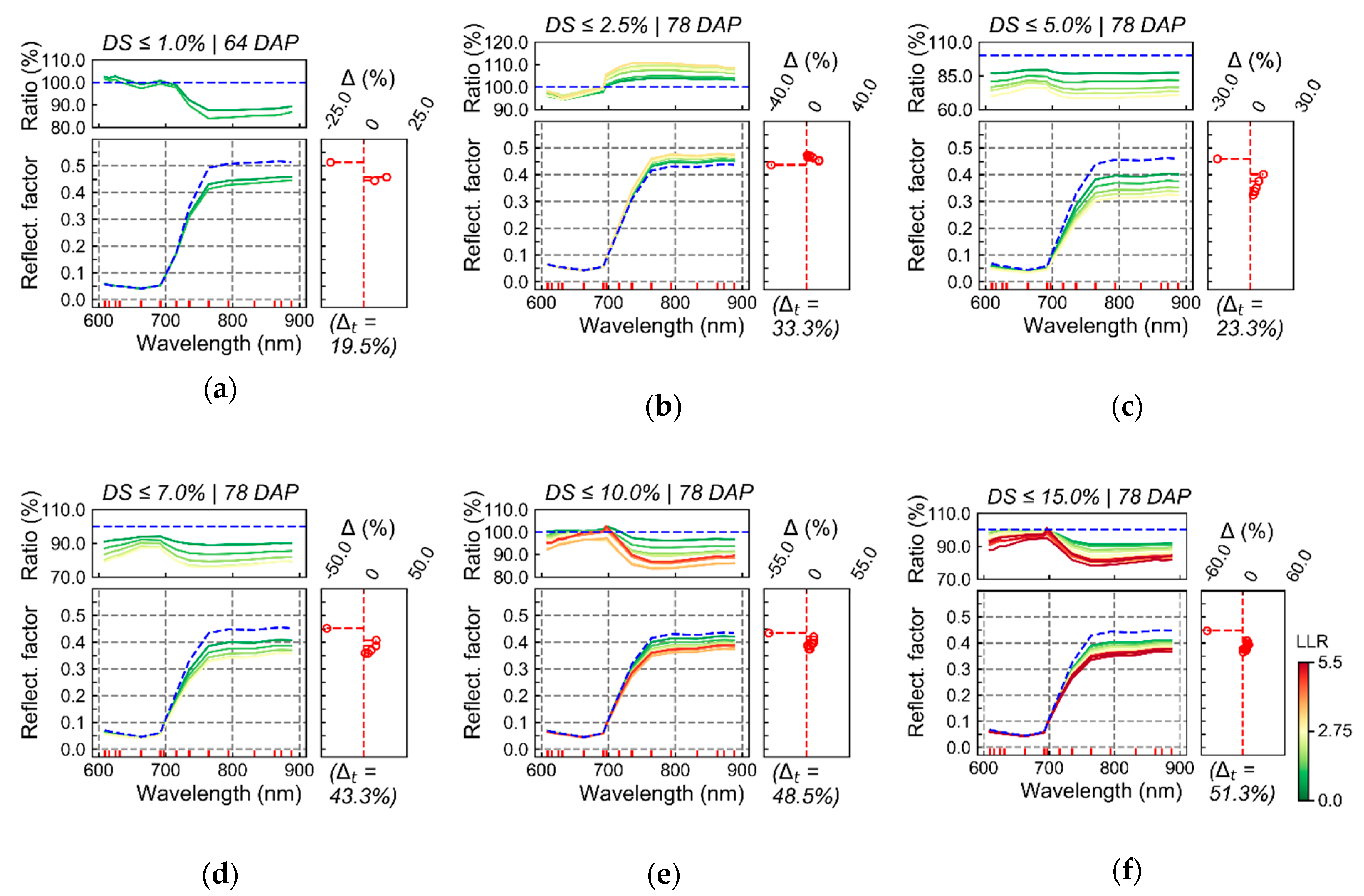
The variability in crop development observed during the growing season, related to disease development or not, could be detected through optical imagery acquired using ground-based or UAV imaging setups. For most of the acquisition dates, patterns observed for ground-based data are comparable to those derived from UAV (
Figure 5 and
Figure A6). However, in some cases disagreement was observed, in particular in the second and third data collection (50 and 64 DAP). Differences between analysis outputs resulting from ground-based and UAV data are mainly related to the higher spatial resolution of the ground-based imagery. The increased resolution allowed a better description of the variability present within the crop canopy, together with a better retention of this variability after background removal due to the potential lower degree of spectral mixing for this dataset.
In addition, spectral variability related to disease incidence could be well described by ground-based imagery (
Figure 5 and
Figure A6a,c,e,g). On the other hand, UAV data indicated trends similar to those observed in the ground-based images, in particular for relatively high disease severity levels, but these trends were attenuated on this data source (
Figure 5 and
Figure A6b,d,f,h). A potential limitation in sensitivity for data acquired at canopy level, and even at leaf level, if spatial resolution is reduced has been indicated by other authors [
15,
78]. However, in the present study it was observed that even at low levels of disease severity (between 2.5 and 5% severity), spectral information related to the disease incidence could be derived from radiometric measurements made by sensors on-board of a UAV platform (
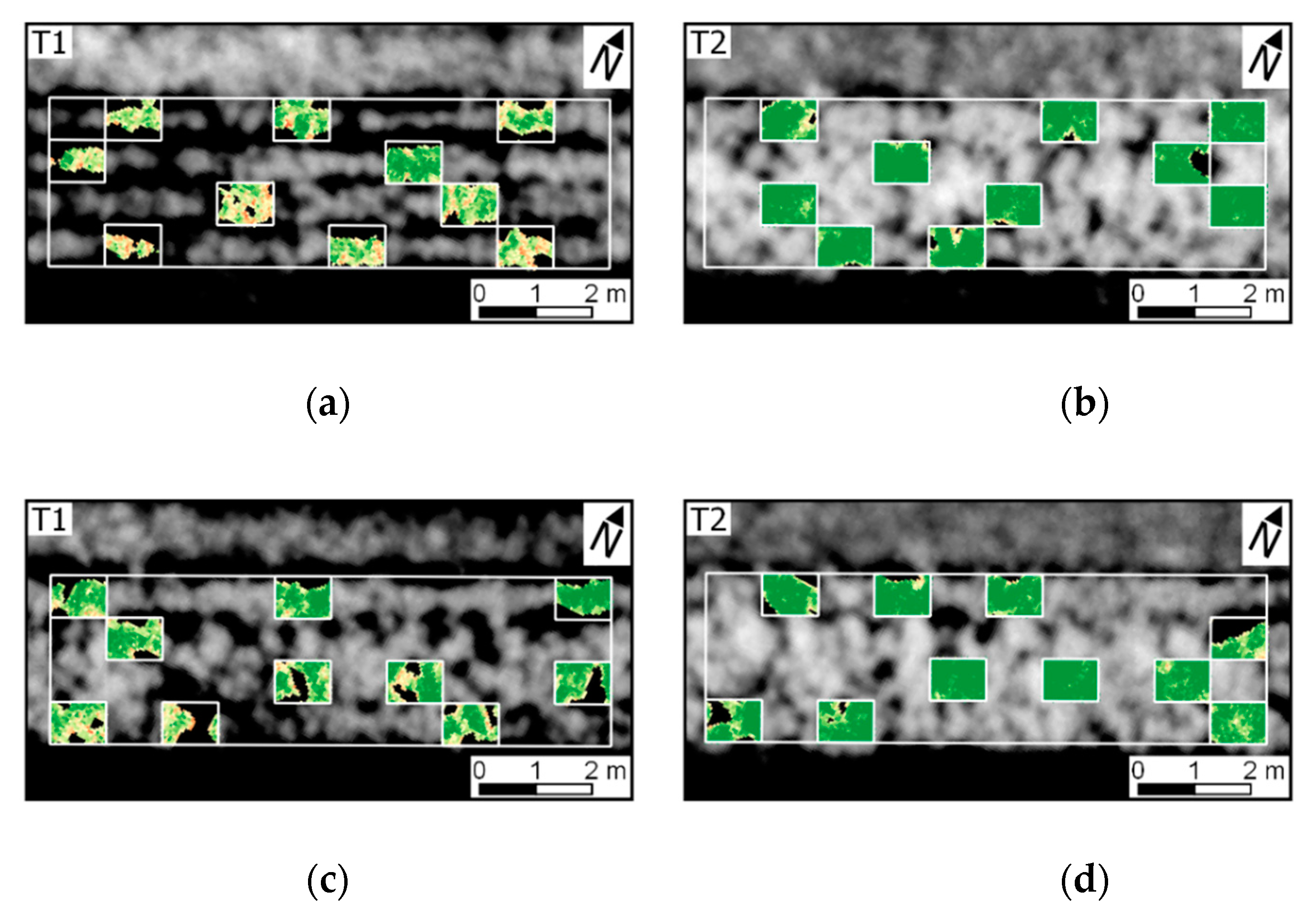
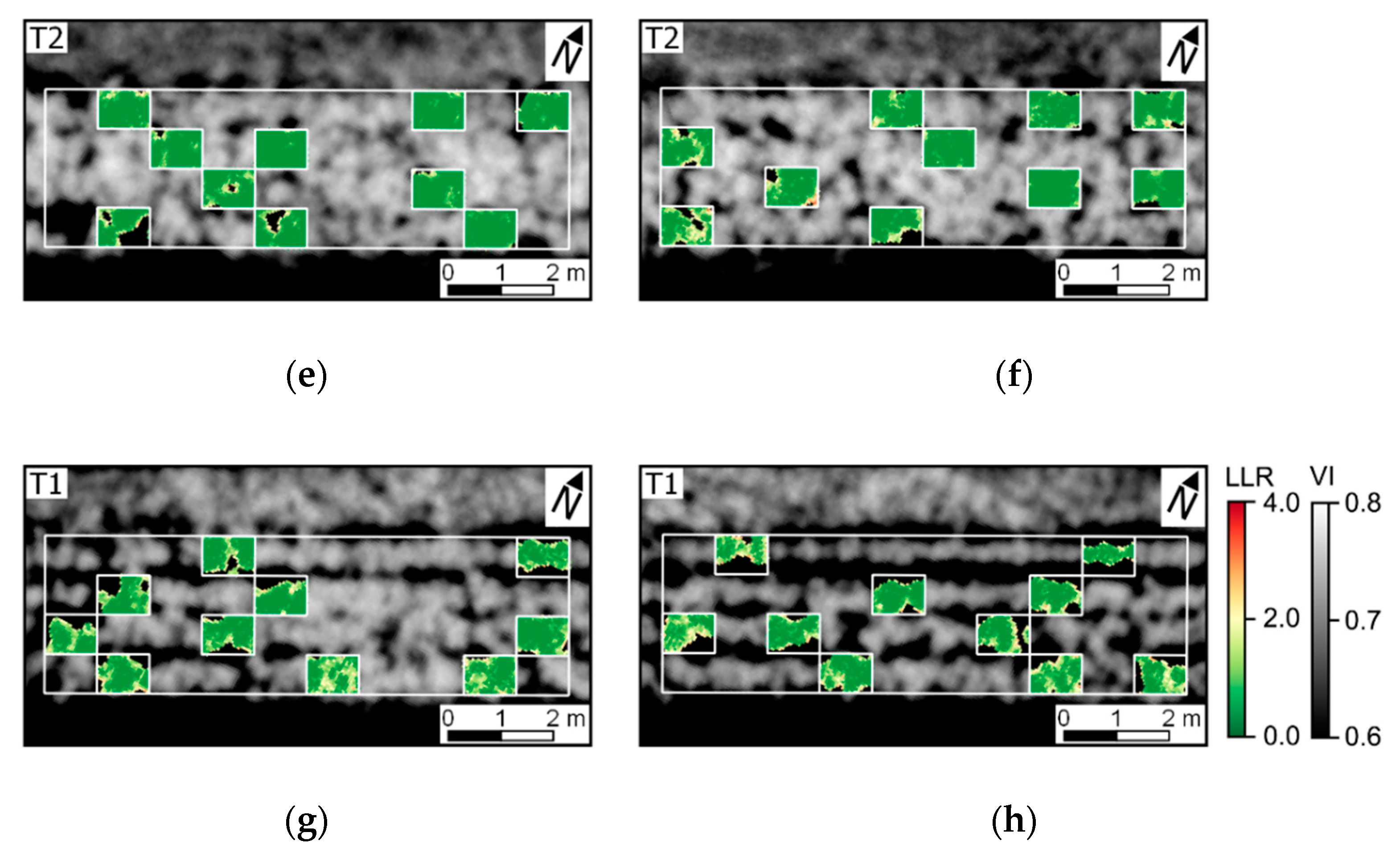
Figure 5,
Figure 6,
Figure 7 and
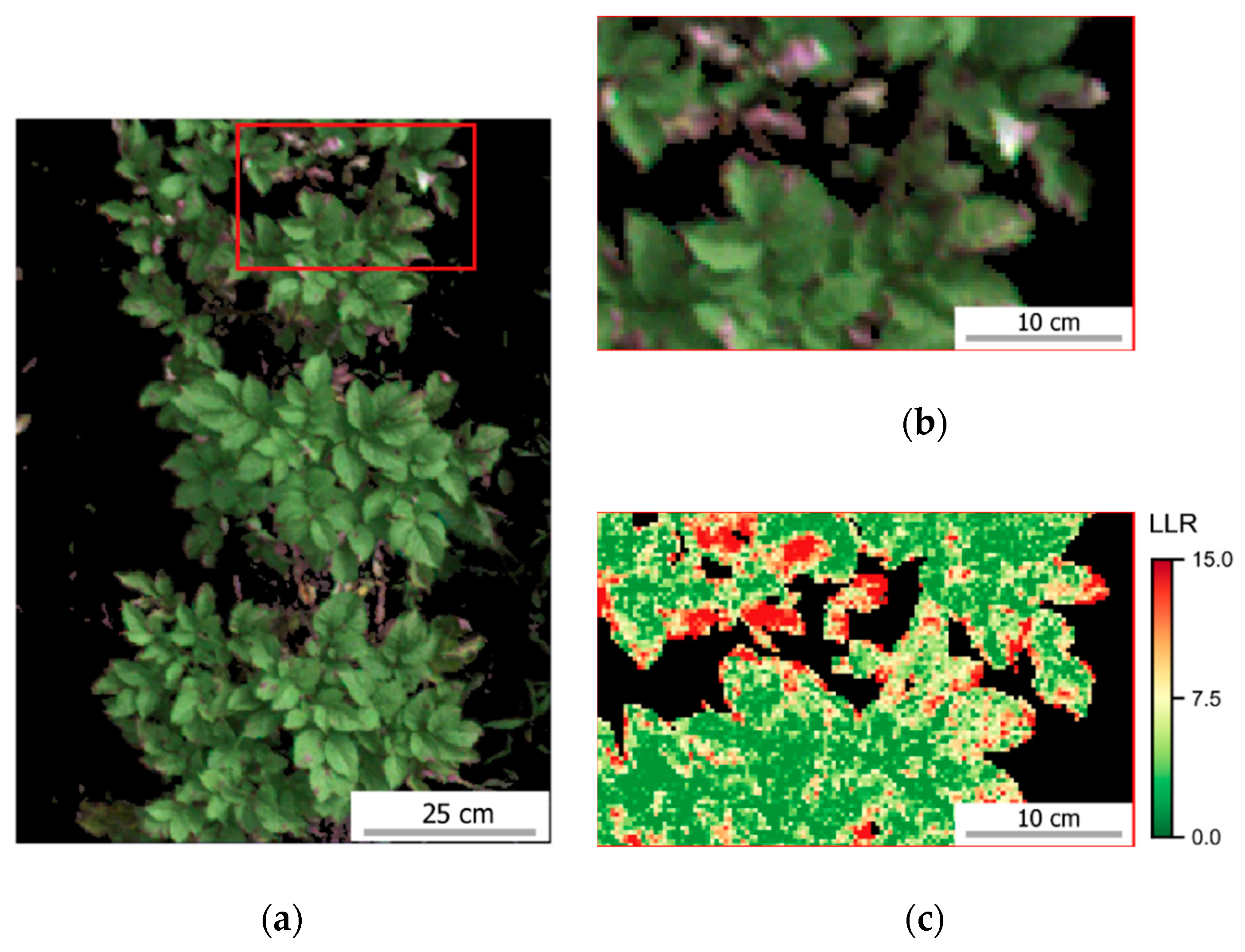
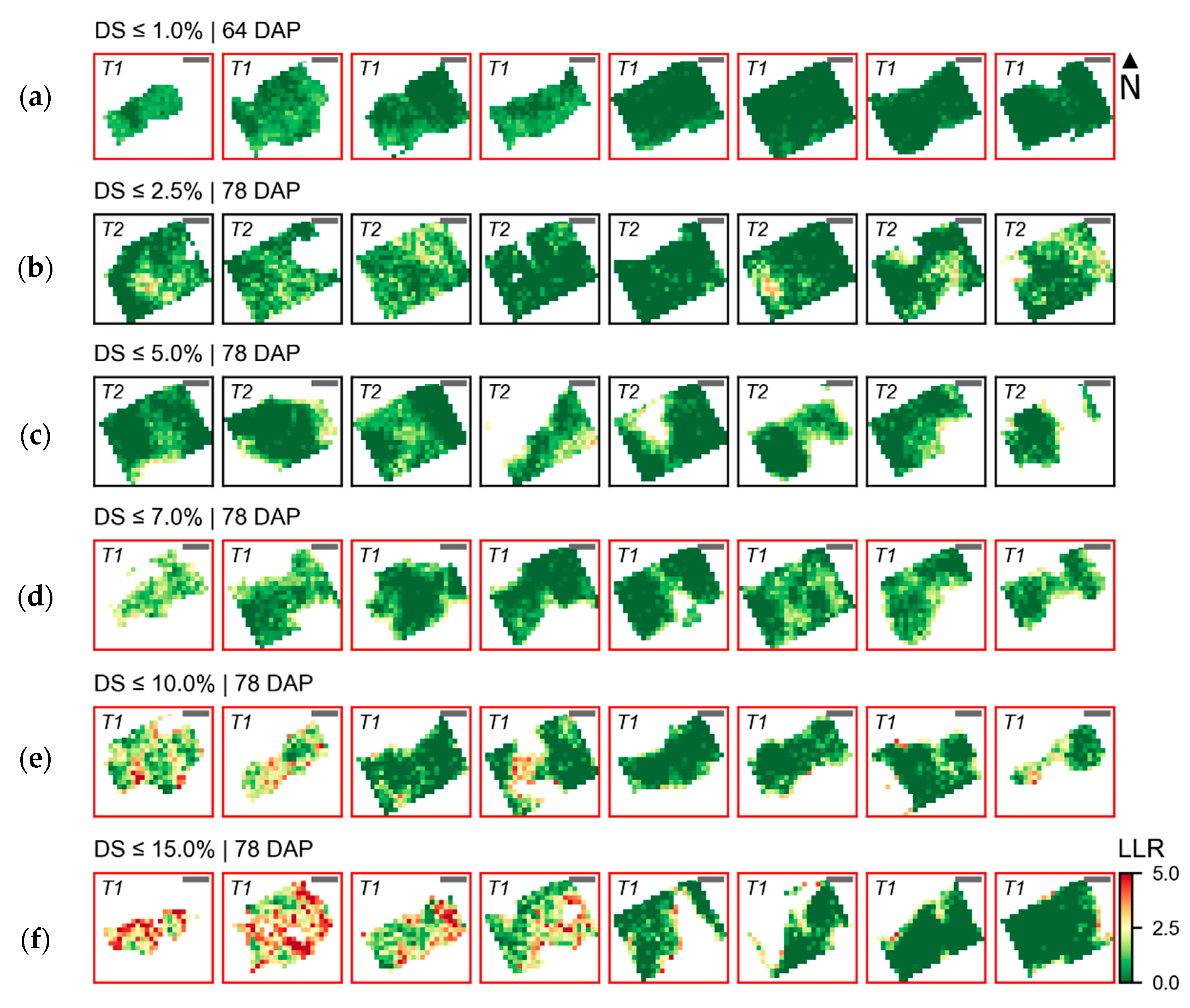
Figure A6), with relatively low spatial resolution (approximately 4–5 cm of ground sampling distance) in comparison with ground-based information (with 0.1–0.2 cm of spatial resolution). This indicates that spectral data acquired at canopy level with sub-decimeter resolution has potential to describe spectral changes related to disease incidence, in particular if analysis targeting the most related spectral information with diseased patches is used (
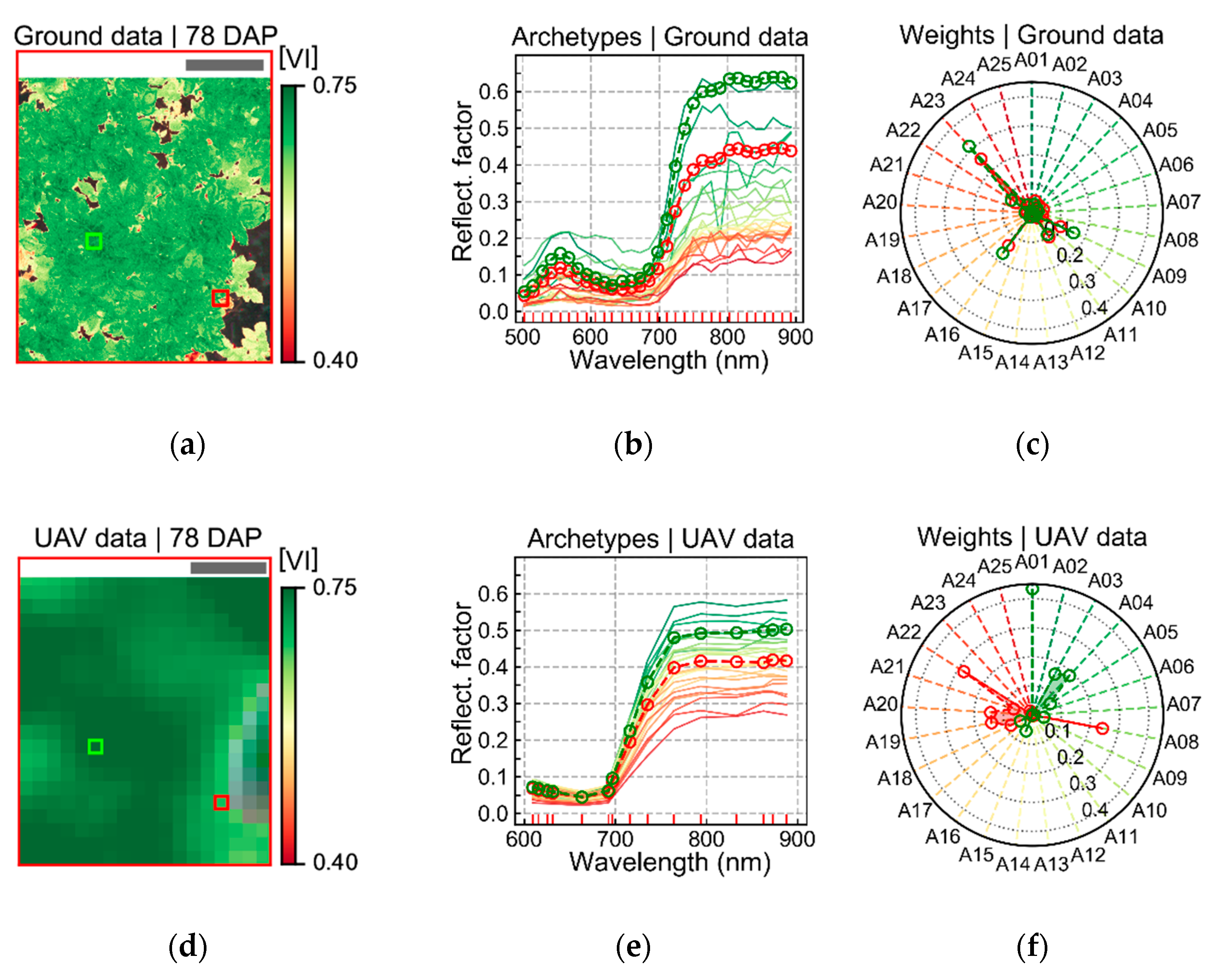
Figure 8). Similar results have been reported in other studies regarding the use of UAV optical imagery to assess disease incidence in other crops [
18,
14,
22]. Generally, in these studies, parametric statistical frameworks (e.g., analysis of variance and groups means test) are used to evaluate the discriminative potential of spectral information regarding disease incidence. This was performed here to compare treatments (
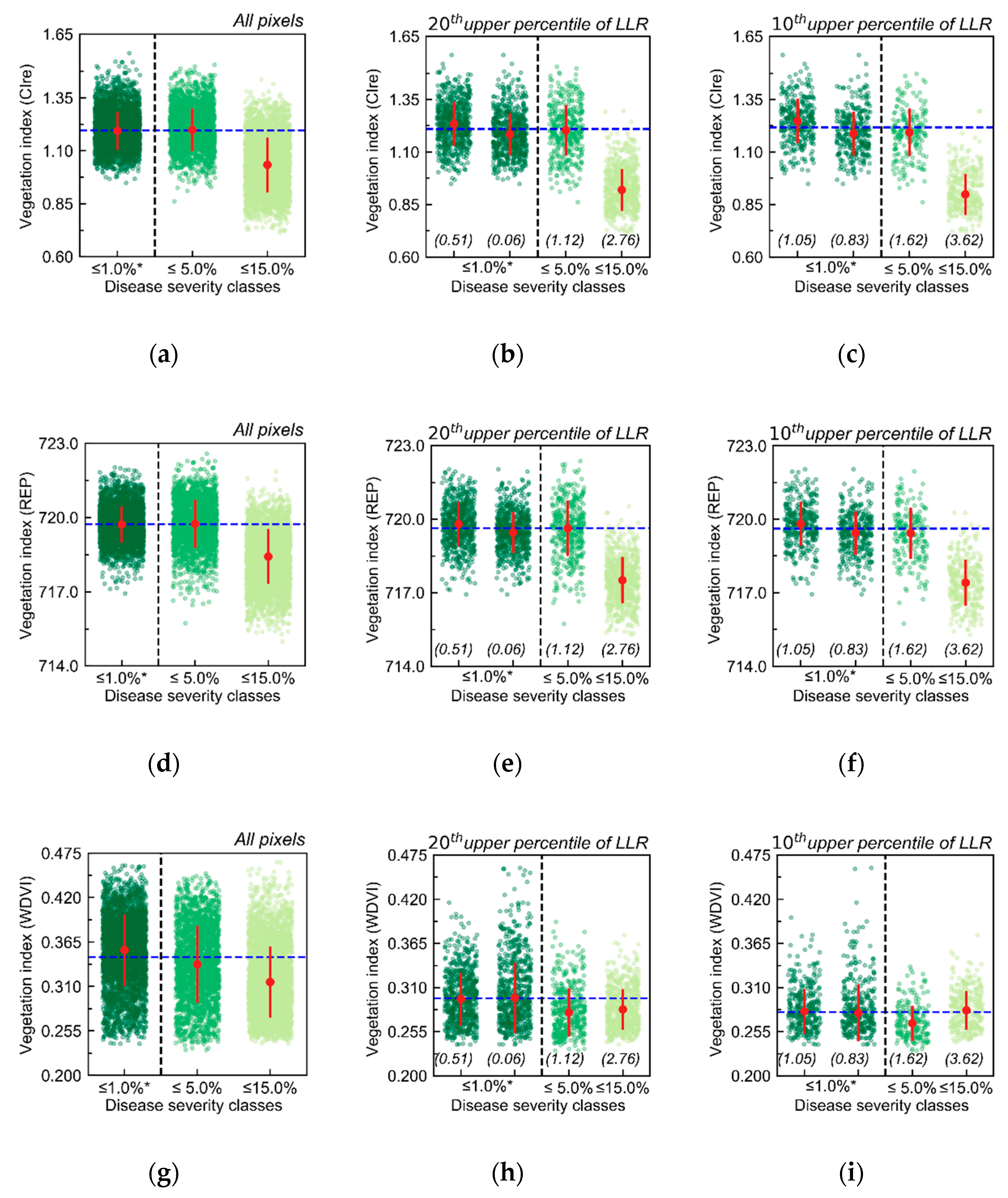
Figure 3) but not to evaluate specific changes related to different disease severity levels. Implementing such analysis for evaluating the impact of different disease severity classes on the canopy reflectance was not possible in this research since the distribution of disease incidence classes differed between treatments and experimental blocks, which could lead to a biased evaluation in this case. On the other hand, the evaluation reported in
Figure 3 and
Figure 8 indicates that discrimination between treatments and different disease severity levels based on aggregate information at sampling unit level (i.e., distribution of vegetation indices for the imaged patches), as frequently performed in other studies, is possible for relatively higher disease severity levels, although focusing on the identification of specific spectral information related to diseased areas improved the characterization of lower levels of disease severity through the spectral information gathered.
An interesting method for late blight monitoring in potato based on optical imagery with very high resolution has been presented by Sugiura et al. [
38]. The solution introduced by these authors provided accurate disease severity prediction based on RGB color transformation and pixel-wise classification through threshold optimization procedure. However, these authors relied on color features rather than on reflectance measurements, which may reduce the flexibility of the approach proposed regarding its application under diverse data acquisition conditions (i.e., with changes in illumination and field of view, etc.), and when disease incidence occurs simultaneously with other abiotic or biotic stress factors. In this sense, optimization for different datasets acquired would be required. Therefore, using reflectance information rather than color-based features could allow mitigation of some of these limitations, in particular if coupled with methods proposed to compensate for illumination changes and to perform BRDF effects correction, as those described by Honkavaara et al. [
47]. Also, using multi- or hyperspectral datasets may allow improvement of discrimination potential concerning the classification of healthy and diseased areas due to increased availability of features potentially related to the effects of disease incidence on the crop canopy traits. More recently, Duarte-Carvajalino et al. [
39] performed machine learning-based retrieval of late blight severity in potato, using a very high resolution camera, similarly to Sugiura et al. [
38] but using a modified set-up (acquiring images in blue, green, and NIR wavelengths instead of conventional RGB). They performed radiometric calibration for the acquired imagery, despite the limitations of the sensor system used, and included in their analysis datasets corresponding to different dates over the growing season. Performance comparable to that obtained by Sugiura et al. [
38] was reported, in particular for models derived using Convolutional Neural Networks, indicating potential for similar applications in this context.
In the present research, in contrast to results previously reported in the literature, an attempt is made to effectively relate disease development over time with spectral changes in a dataset composed of sub-decimeter resolution UAV optical imagery. From the spectral differences observed between treatments and disease severity classes (
Figure 5,
Figure 6,
Figure 7 and
Figure A6), it is possible to conclude that disease incidence, even at relatively low levels, has direct effects on the canopy spectral response measured by sensors similar to that used in the study. On the other hand, intrinsic characteristics of different potato cultivars may potentially affect the spectral response observed and lead to spurious correlation between spectral data and disease severity observations, in particular for low late blight severity levels. This can be an important aspect to consider during the development of future modelling approaches, especially if based on data acquired in agronomic experiments with multiple cultivars.
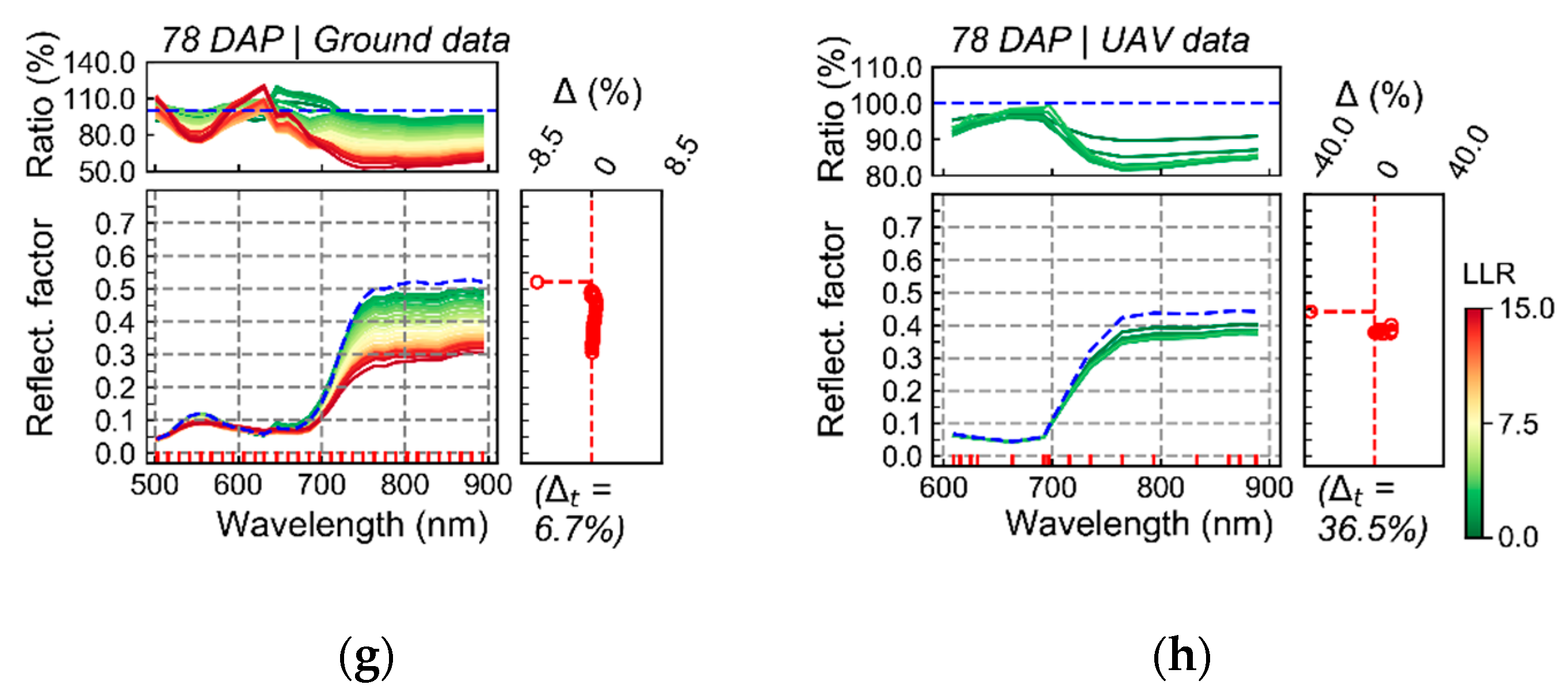
Spectral changes that could be associated with late blight development, mainly based on measurements realized on the last data acquisition (78 DAP), were characterized by reduced reflectance on all spectral bands measured using the UAV sensor (
Figure 6). This indicates that changes detected were strongly related to alterations in the canopy and leaf structure [
19]. In general, as the relationship between the spectral information and the disease severity levels became stronger (i.e., as LLR values increased), the reflectance decreased in all spectral bands for disease classes above 2.5% severity. Expected spectral changes related to pigment content at leaf and canopy levels could mainly be identified in the red-edge region, which is also associated with canopy and leaf structural traits. Deviations in the red region, more directly related to changes in chlorophyll content (i.e., increase in reflectance for diseased vegetation due to lower chlorophyll content), were less evident even for higher levels of disease severity (i.e., above 2.5% severity). These facts indicate that the main areas that could be related to disease development were those with reduced canopy (i.e., LAI) and leaf (i.e., number of layers specifying air/wall interfaces within the leaf mesophyll) structure. Changes related to pigment content at leaf and canopy levels were less pronounced than changes related to canopy and leaf structural alterations.
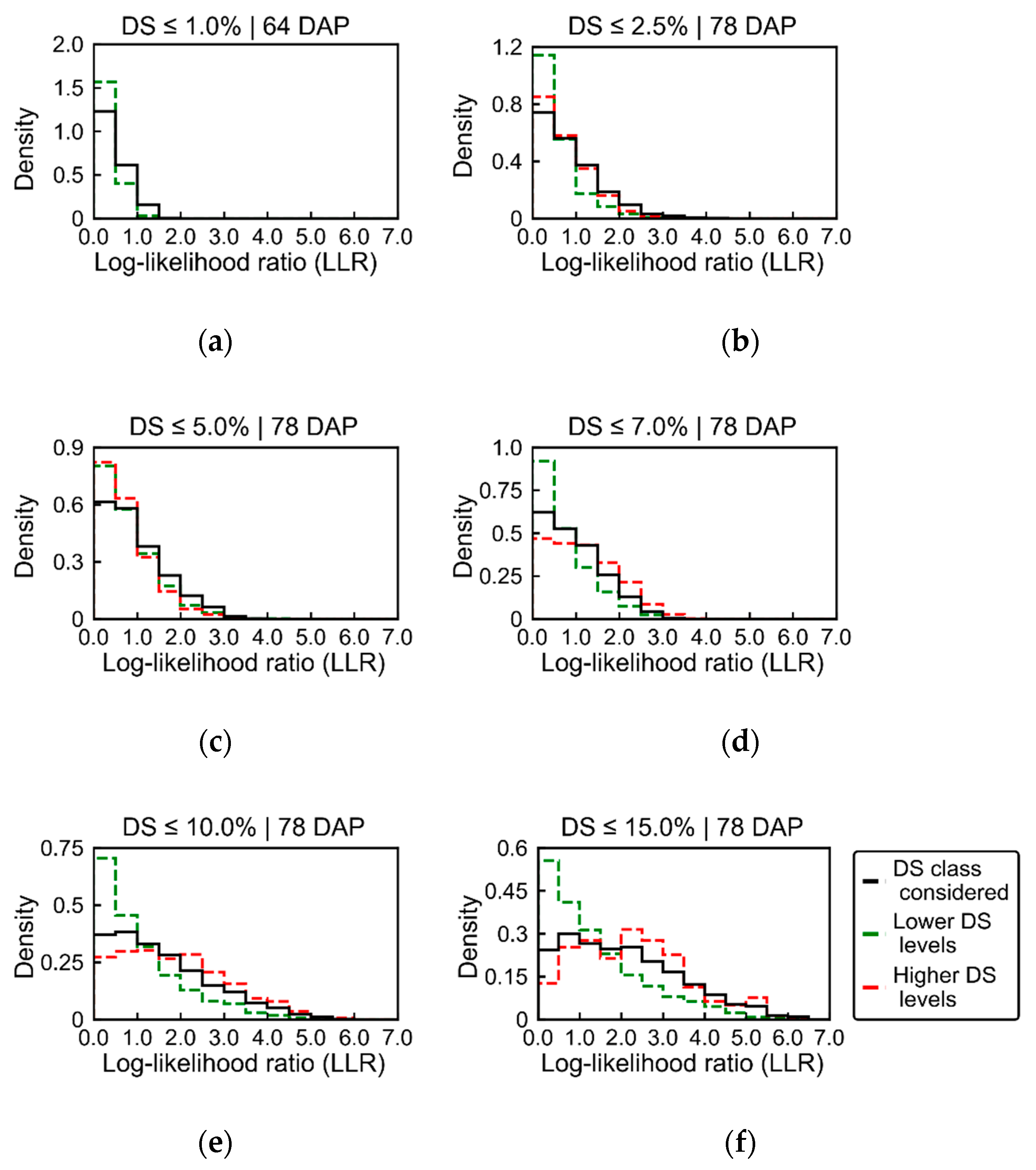
Figure 7,
Figure 8,
Figure 9,
Figure 10 and
Figure 11 confirm these observations and indicate that extremely early alterations related to pigment content degradation in the infected tissues may be more difficult to detect using UAV imagery with the same characteristics as those used in this study. Conversely, alterations in the red and green regions could be observed on ground-based spectral measurements 64 and 78 DAP while, as already described, only small changes could be detected using the UAV imagery for very low disease severity levels on these dates (
Figure 5 and
Figure 6). It is worth noting that for very early infection stage (≤1.0% disease severity) alterations in the visible part of the spectrum were observed in the UAV data (
Figure 6a), but LLR values for the changes observed were very small, indicating that these alterations would probably be difficult to detect in more general applications. Also, changes observed for disease severity between 1.0 and 2.5% (
Figure 6b) followed the opposite trend of that expected, with overall increased reflectance in the Vis-NIR region for spectra related to the disease incidence (i.e., higher LLR values). This is probably due to the association between traits of specific susceptible cultivars(s) to disease incidence, which is indicated by higher values of LLR concentrated in specific spots within the crop canopy (
Figure 7b), explaining the inverse trend observed.
An important final aspect to consider is the relationship between the type of spectral information derived from the UAV imagery acquired and the outputs of the analysis relating spectral information with disease incidence and severity. The UAV data used as input for analysis, with results described in
Section 3, combines all data collected in a given location in the field during the UAV flight. This combined information was derived taking the pixel-wise average for the complete dataset acquire, i.e., considering all scenes obtained over each imaged area. This “average spectral data product”, as thoroughly discussed by Aasen and Bolten [
3], is characterized by reduced influence of angular properties on the reflectance representing a given crop surface. While this is a desirable feature for spectral datasets used to detect very subtle changes in the canopy reflectance, as those related to early disease detection, some sensitivity may be lost regarding the characterization of lower parts of the crop canopy. This can potentially be a reason for the relatively low association between spectral information and disease incidence (i.e., low LLR values) for some image patches with relatively high disease severity (
Figure 7).
Studies focusing on the relationship between reflectance angular properties and crop trait estimation, as those performed by Roosjen et al. [
5] using UAV imagery and Kong et al. [
79] using point-based multi-angular spectral measurements, indicate that considering multiple view-angles may increase the potential for crop trait characterization, and that estimation of properties at lower parts of the crop canopy are better performed based on slightly off-nadir reflectance measurements. The latter is attributed to the fact that off-nadir measurements may have greater probability to correspond to reflected light having interacted with lower parts of the canopy, especially for wavelengths in the visible part of the spectrum. This fact may be particularly relevant for early late blight assessment, since the disease onset generally occurs in lower parts of the canopy, and therefore nadir oriented measurements, may miss the local changes in pigment content occurring in these areas.
5. Conclusions
In this study, the potential of radiometric readings in the optical domain to describe visual ratings regarding the development of potato late blight was evaluated from a perspective of the sensitivity of the spectral information to describe early changes occurring in the infected canopy areas. It was verified that optical data acquired at canopy level with sub-decimeter resolution has potential to provide useful information for detecting late blight incidence and assessing its severity in early stages of disease development (i.e., between 2.5 and 5.0% disease severity). Despite these positive outputs, the main changes detected were related to crop canopy structural traits, and to a lesser extent, to pigment content.
The evaluation performed here focused on post-visual disease symptoms and its relationship with changes in spectral response at canopy level. It was observed that although aggregated information at sampling unit level (i.e., distribution of vegetation indices values) allowed, to a certain extent, to differentiate contrasting treatments and disease severity levels, early detection of late blight might be difficult based on frameworks involving similar approaches. Conversely, better descriptive potential was observed when specific spectral information regarding a given treatment or disease severity level was identified through SiVM and LLR calculation. Based on these last methods, it was possible to identify patterns of spectral changes and their spatial arrangement in the imaged patches. These patterns were related to disease symptoms and their spatial distribution observed in ground-based, very-high resolution imagery. In this regard, the main detectable changes observed by UAV imagery concerned canopy and leaf structural traits, and to a minor degree, pigment content also at leaf and canopy levels. These facts indicate that late blight detection and severity assessment based on UAV imagery in the optical domain with sub-decimeter resolution may rely in particular on the identification of affected areas characterized by reduction in leaf and canopy structure. Relationship with leaf and canopy pigment content was less perceptible than changes related to structural traits. These observations are of interest if one intends to develop a specific framework for late blight detection and severity assessment based on optical imagery acquired at canopy level. In this case, including off-nadir spectral data and reflectance measurements in wavelengths in other spectral regions (i.e., green region), besides red and near-infrared, may increase sensitivity of the approach used, in particular concerning detection of changes in pigment content, considering the saturation effect normally observed in reflectance measurements in the red region under relatively high chlorophyll content levels.