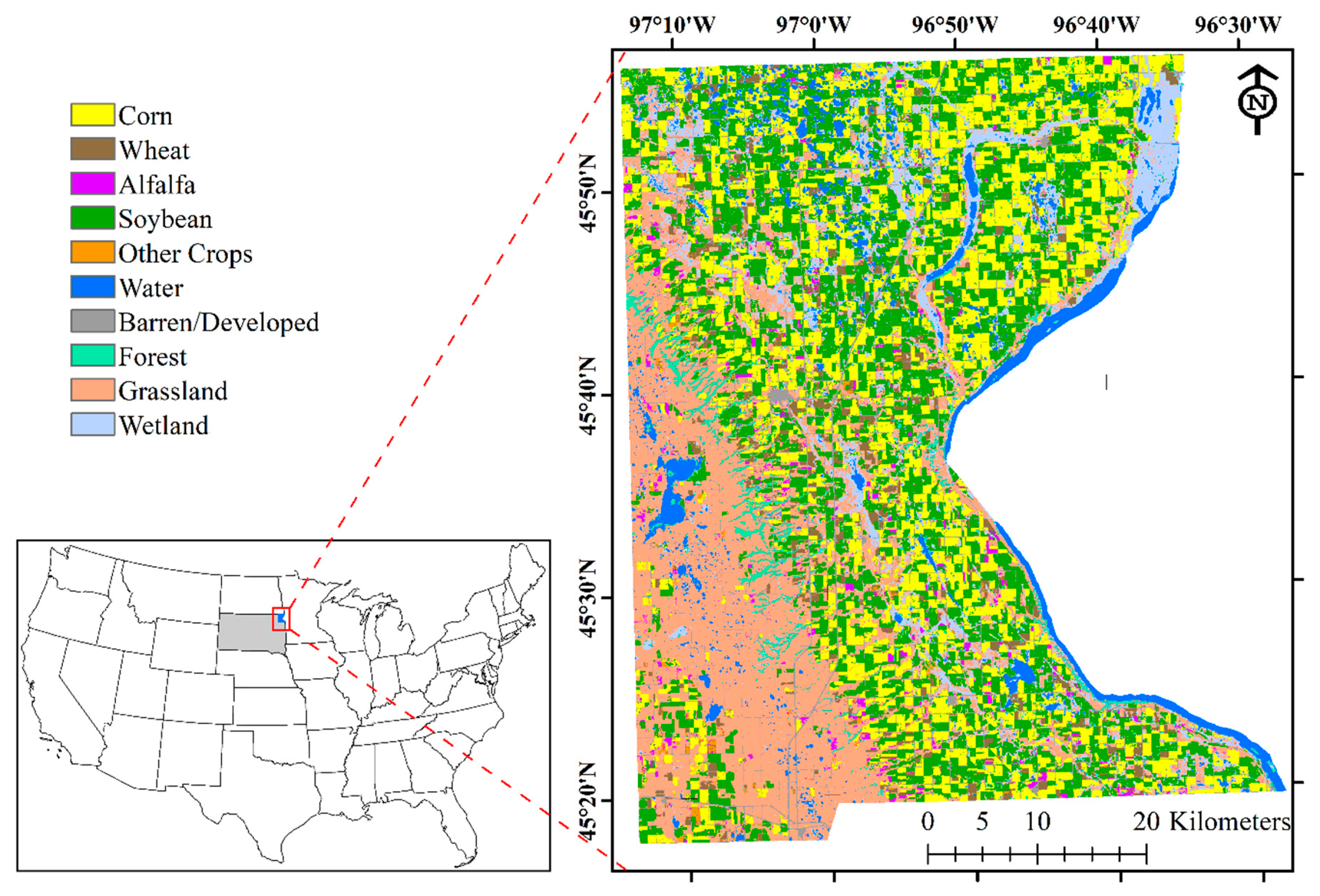
1. Introduction
Knowledge about land use/land cover (LULC) is fundamental for natural resource management, agricultural policy making, and regional and urban planning. Most reliable data sources for LULC information are periodic surveys from governmental agencies, e.g., the National Resource Inventory and the National Agricultural Statistics Service (NASS), both in the United States Department of Agriculture (USDA) [
1,
2]. However, those datasets often lack spatial and temporal details, which prevents a comprehensive analysis of land change. Remote sensing technology can complement field observations and surveys. Conventional classification approaches, such as those applied in the National Land Cover Dataset (NLCD) [
3,
4,
5] or the Cropland Data Layer (CDL) [
6], were developed in an era of data scarcity and limited computational power and data storage. Thus, they have focused on mapping annual land cover from multispectral data from one or just a few image dates. However, in areas with frequent morning cloud cover, collecting even a few cloud-free scenes over a year can be challenging. The recent rapid increase of accessible Earth observation data coupled with improved computing and storage capabilities is leading to the emergence of methods for mapping land cover using multi-date imagery and dense image time series [
7]. Compared to the traditional approach, the use of image time series often improves classification accuracy by incorporating both spectral and temporal profiles [
8,
9,
10].
Land surface phenology (LSP) has been a useful approach to characterize seasonal vegetation dynamics on vegetation index time series [
11]. Over the past few years, several efforts have been made to map LULC using phenological metrics derived from satellite image time series with promising outcomes [
12,
13,
14,
15,
16,
17,
18]. Due to the relatively low return interval of orbital sensors with spatial resolutions finer than 50 m, many studies—with notable exceptions [
13,
16,
18]—have relied on MODIS time series to capture phenological characteristics of land surfaces, thus often producing cover maps at spatial resolutions (e.g., 250–1000 m) that are coarse relative to human land uses, such as agriculture and settlements. To overcome limited temporal coverage of Landsat-like data and map land covers at finer spatial resolutions, Jia et al. [
13] and Kong et al. [
16] fused the MODIS Normalized Different Vegetation Index (NDVI) [
19] with Landsat and Gaofen-1 NDVI time series, respectively. Although each produced land cover maps at finer spatial resolution (30 m for Landsat and 16 m for GF-1), neither Jia et al. [
13] nor Kong et al. [
16] were able to map more than Level-1 NLCD Land Cover Classification System, except for coniferous and broadleaf forest in Kong et al. [
16] (Level-2 NLCD).
In 2016, the United States Geological Survey reorganized the Landsat archive into a tiered collection, namely the Landsat Collections, to facilitate time series analysis and data stacking [
20]. Taking advantage of the Landsat Collections data, Nguyen et al. [
18] performed a phenometrically-based classification for sample areas in South Dakota using all available Tier-1 (highest quality) images from Landsat 5 Thematic Mapper (TM), Landsat 7 Enhanced Thematic Mapper Plus (ETM+), and Landsat 8 Operational Land Imager (OLI). At each pixel, an Enhanced Vegetation Index (EVI) time series calculated from Landsat Collections data was simulated as a convex quadratic function of accumulated growing degree-days (AGDD), i.e., a measure of accumulated heat from January 1 onward whenever the average temperature exceeded 0° Celsius. Results showed that classification using only phenometrics generated from the fitted model could accurately map broad thematic land cover classes (water, developed, grassland) as well as commodity crops (corn/maize, soybean, wheat) in Codington and Roberts counties in South Dakota for two years (2012 and 2014). However, they also pointed out some challenges of the phenometrically-based classification. First, the classification accuracy varied, since the form of the chosen land surface phenology (LSP) model might be more suitable for some certain vegetation types than others. Second, the phenometrically-based classification performed well only for vegetated classes, particularly crops. Third, many cloud/snow/shadow-free observations were needed at each pixel over a year to fit the LSP model well and to avoid data gaps in the predicted land cover map. Regarding the last point, they [
18] also showed that an adequate number of observations could be gathered by combining data from multiple comparable sensors, especially in sidelap zones of Landsat swaths. Finally, in addition to pointing out the challenges of classification based on phenometrics, Nguyen et al. [
18] also discussed the potential opportunity to improve classification accuracy by incorporating both phenological and spectral information.
Here, we explored the challenges of the phenometrically-based classification and a potential way to improve classification accuracy, as demonstrated in [
18]. This study focused on evaluating the performance of alternative land cover classifications using either (1) only phenological metrics derived from either of different land surface phenology (LSP) models: The Convex Quadratic Model, in which EVI2 =
f(AGDD) [
11,
21] and the Hybrid Piecewise Logistic Model, in which EVI2 =
f(day of year) [
22], or (2) a suite of spectral band percentiles and normalized ratios (spectral variables), or (3) both phenological metrics and spectral variables. In our evaluation, we addressed three research questions. The first question was whether the maps from the phenometrics were more accurate than maps from spectral variables alone. As land surface phenology has been a useful tool to characterize the dynamics of the vegetated land surface [
11], we hypothesized that land cover classifications using only phenometrics could be more accurate for vegetated land covers, especially for commodity crops, than those using only spectral variables. The second question asked which set of phenometrics—derived either from the Convex Quadratic Model (CxQ) or from the Hybrid Piecewise Logistic Model (HPLM)—performed better. In the temperate ecosystem, plant development is sensitive to variation in temperature. We hypothesized, therefore, that the Convex Quadratic Model, which links vegetation growth with the progression of thermal time, would be better suited to land cover classification of our study area in northeastern South Dakota. The third question asked whether combining the phenometrics and spectral variables would result in superior performance. Studies have indicated that classification accuracies were improved by incorporating phenological features [
13,
16]. Thus, we hypothesized that classification using a combination of spectral variables and phenometrics would be consistently more accurate than using only phenometrics or spectral variables. To build a more complete picture of classification performance, we ran Random Forest Classifiers (RFC) with different sampling scenarios and sets of input variables.
First, three annual time series of remotely sensed data were constructed, including accumulated growing degree-days from the MODIS 8-day composites of land surface temperatures and 2-band Enhanced Vegetation Index (EVI2) [
23] and spectral variables from surface reflectance products from (1) Landsat Analysis Ready Data (ARD) and (2) Harmonized Landsat Sentinel-2 (HLS) data, separately. At each pixel, EVI2 time series were then fitted to the LSP models, CxQ or HPLM. Phenometrics derived from the fitted LSP models as well as spectral variables were submitted individually and in combination to RFC to map land use/land cover of the study area. Accuracy assessments for both RFC models and predicted land cover maps were reported using both conventional accuracy metrics (overall, producer’s, and user’s accuracies) [
24] and alternatives for kappa [
25].
Our assessment of classification performance is two-fold. First, RFC model performance was evaluated by submitting different input datasets randomly generated from the CDL. Accuracy comparisons between classification scenarios were tested by both Mann–Whitney U [
26] and equivalence tests [
27,
28]. These two tests are based on opposite but complementary evaluation perspectives. The nonparametric U test indicates whether the two sets of accuracy metrics are statistically different, regardless how difference magnitude. The equivalence test, on the other hand, examines whether differences fall within a certain user-defined threshold and, thus, deemed equivalent or are large enough to be deemed not equivalent. The second step was to compare the predicted land cover maps with the CDL.
6. Conclusions
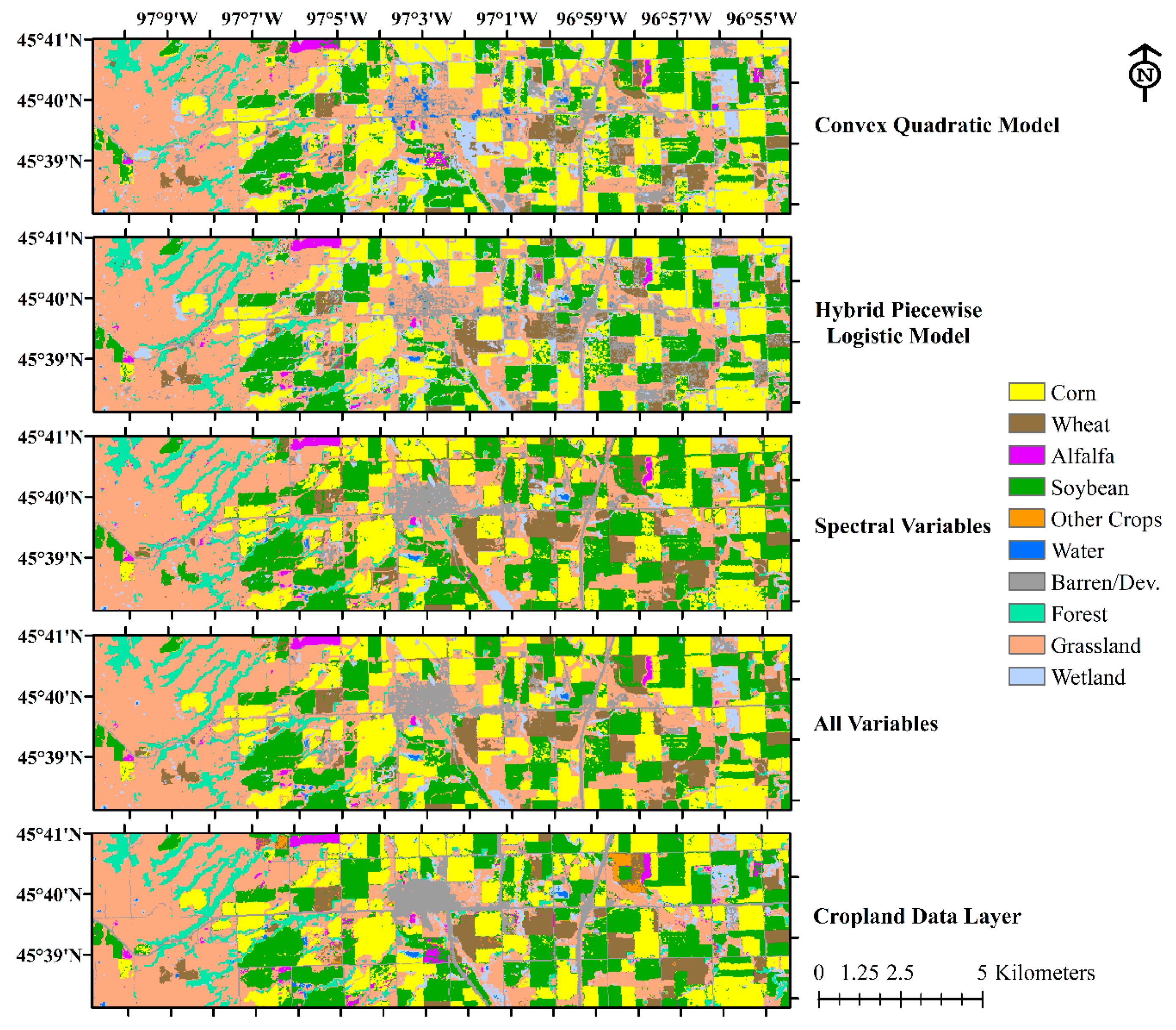
The focus of this study was to evaluate classification accuracy using different sets of input variables derived from either Landsat ARD or HLS time series, including phenometrics generated from two land surface phenology models (CxQ and HPLM), spectral variables, and the combined set of phenometrics and spectral variables. Between the two phenometrically-based classifications, HPLM RFC models exhibited slightly better accuracy but absolute differences in OA were minor (<1%), mostly due to more precise pixel allocation of land cover. Compared to the phenometrically-based RFC models, the spectrally-based RFC models yielded more accurate land cover maps, especially for non-crop cover types. However, the spectrally-based RFC models could not classify wheat accurately. As hypothesized, the most accurate RFC models were retrieved when using both phenometrics and spectral variables as inputs. The combined-variable RFC models overcame weaknesses of both phenometrically-based classifications (low accuracies for non-vegetated covers) and spectrally-based classifications (low accuracies for wheat). The analysis of important variables indicated that classifications of the study area were strongly driven by variables related to the initial green-up phase of seasonal growth and highest EVI2 over the growing season.
We explored land use/land cover classification under different sample pool and sample size scenarios. First, to improve the land cover accuracy of the sample data, both spatial and temporal filters were applied to compensate classification errors of the CDL and offsets between input datasets. The results indicated that a sample pool with a minimum correction of land cover information yielded the most accurate predicted map. Next, land cover classification was also tested with different sample sizes. Although previous findings suggested that a sample size should cover at least 0.25% of the study area to achieve an accurate (OA ≥ 0.90) land cover map, smaller datasets would be acceptable for classification, but should not smaller than 0.05% of the study area, since classification accuracy would decrease rapidly below that threshold.
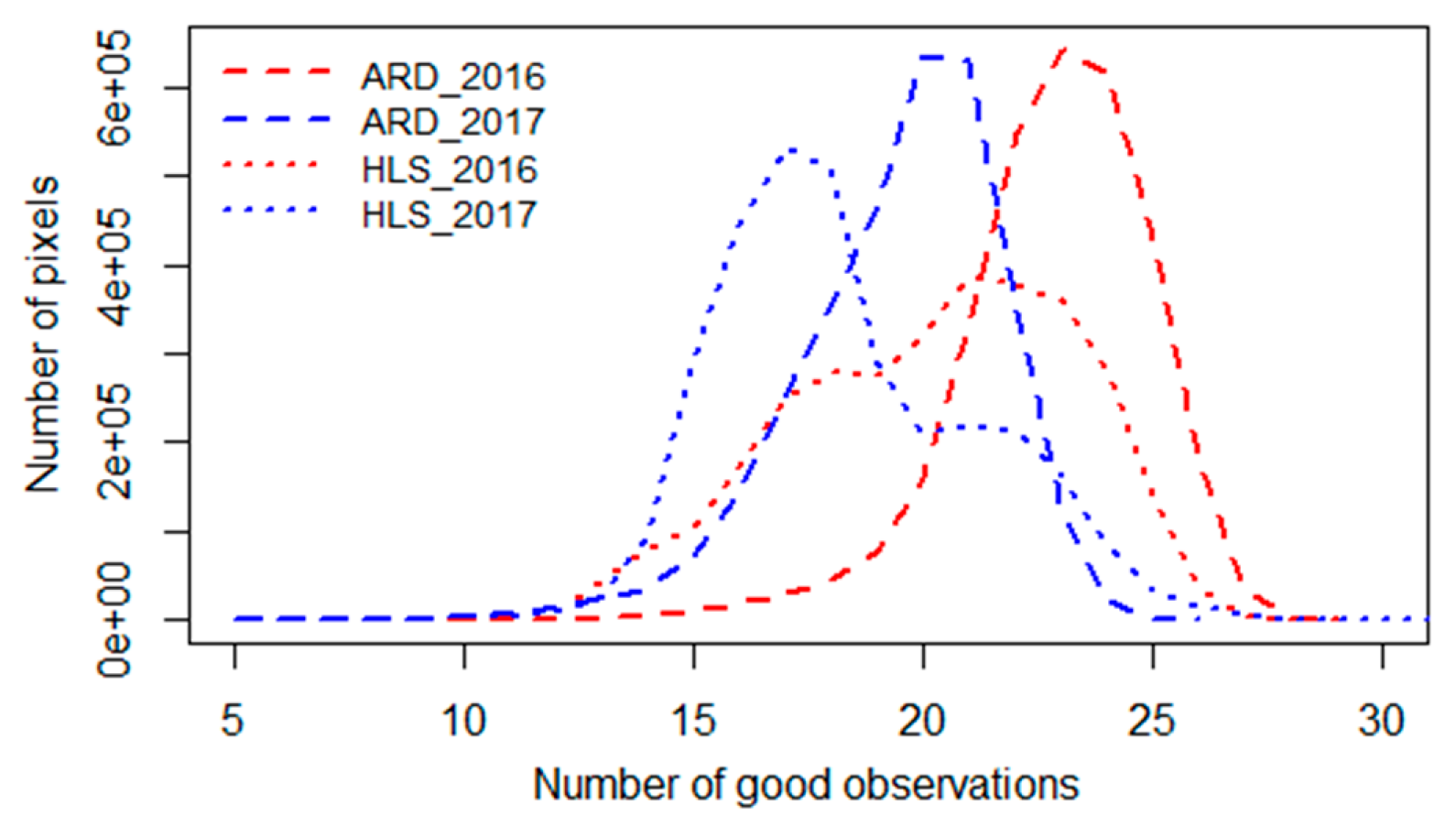
Land surface phenology modeling requires a substantial number of good quality observations over a year [
49]; thus, it may be less suitable for areas with persistent cloud cover if only optical data are available to characterize the LSP. However, the prospect of using phenometrics to enhance land use/land cover classification is very promising. First, our results proved that the use of phenometrics and spectral variables together yielded the most accurate classification and overcame limitations of both phenometrically-based and spectrally-based classifications. Second, seasonality information from all spectral band and ratio time series could be extracted to enhance classification accuracy (e.g., [
50]). Finally, the temporal resolution of satellite data can be improved by using comparable sensor datastreams, e.g., Landsat and Sentinel-2, but substantial pre-processing is required to achieve compatibility.