Investigating the Feasibility of Multi-Scan Terrestrial Laser Scanning to Characterize Tree Communities in Southern Boreal Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Materials
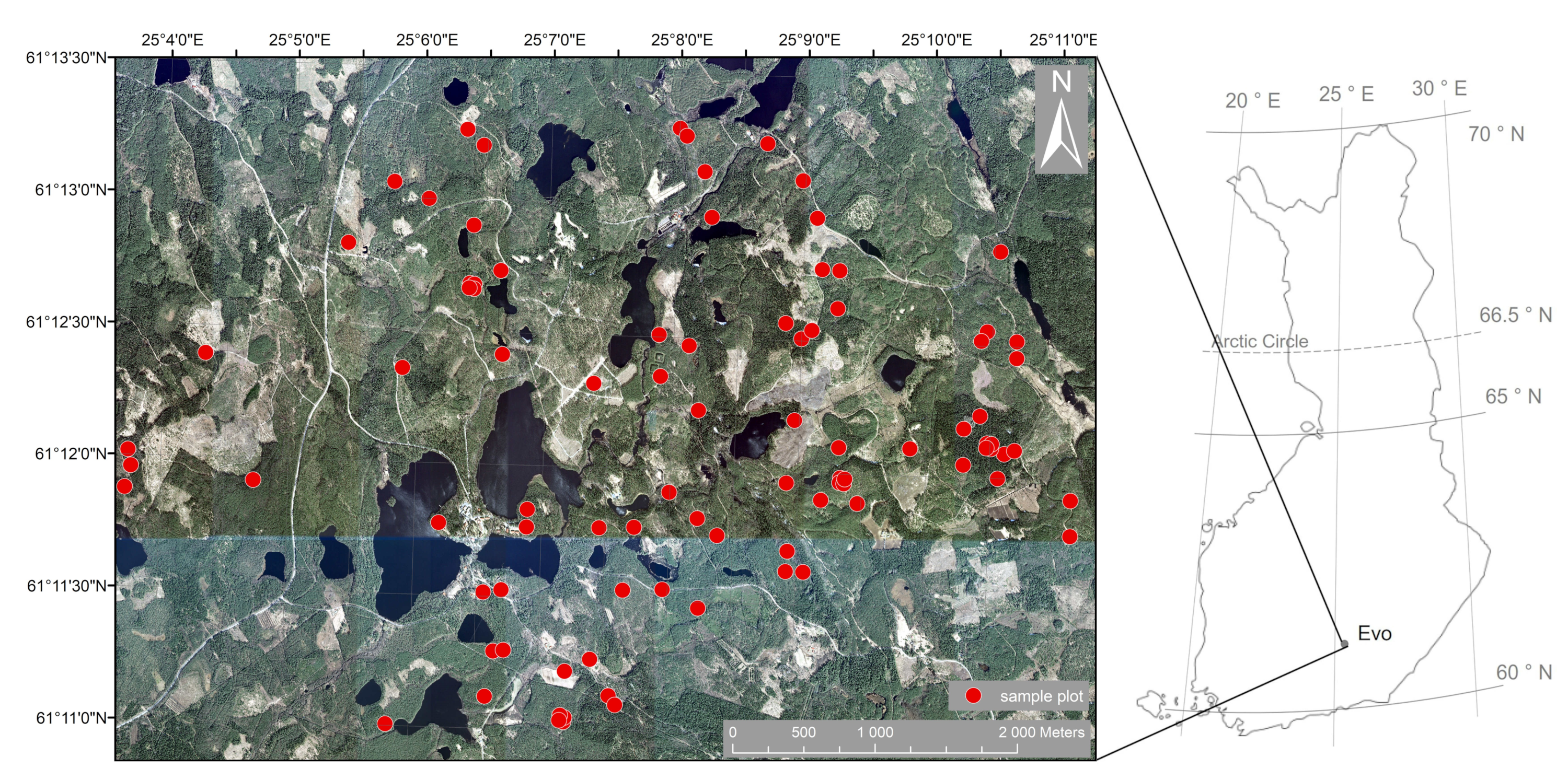
2.1.1. Study Area
2.1.2. Terrestrial Laser Scanning Data Acquisition
2.1.3. Field Inventory
2.2. Automatic Point Cloud Processing Method to Obtain Plot-Level Forest Characteristics
2.2.1. Point Cloud Normalization
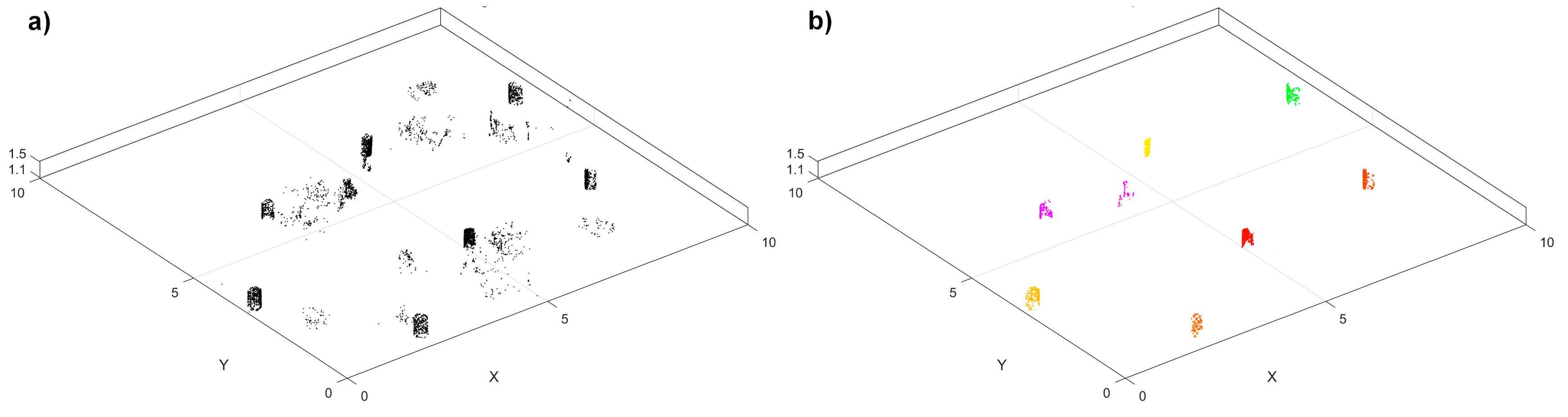
2.2.2. Stage-One Tree Detection
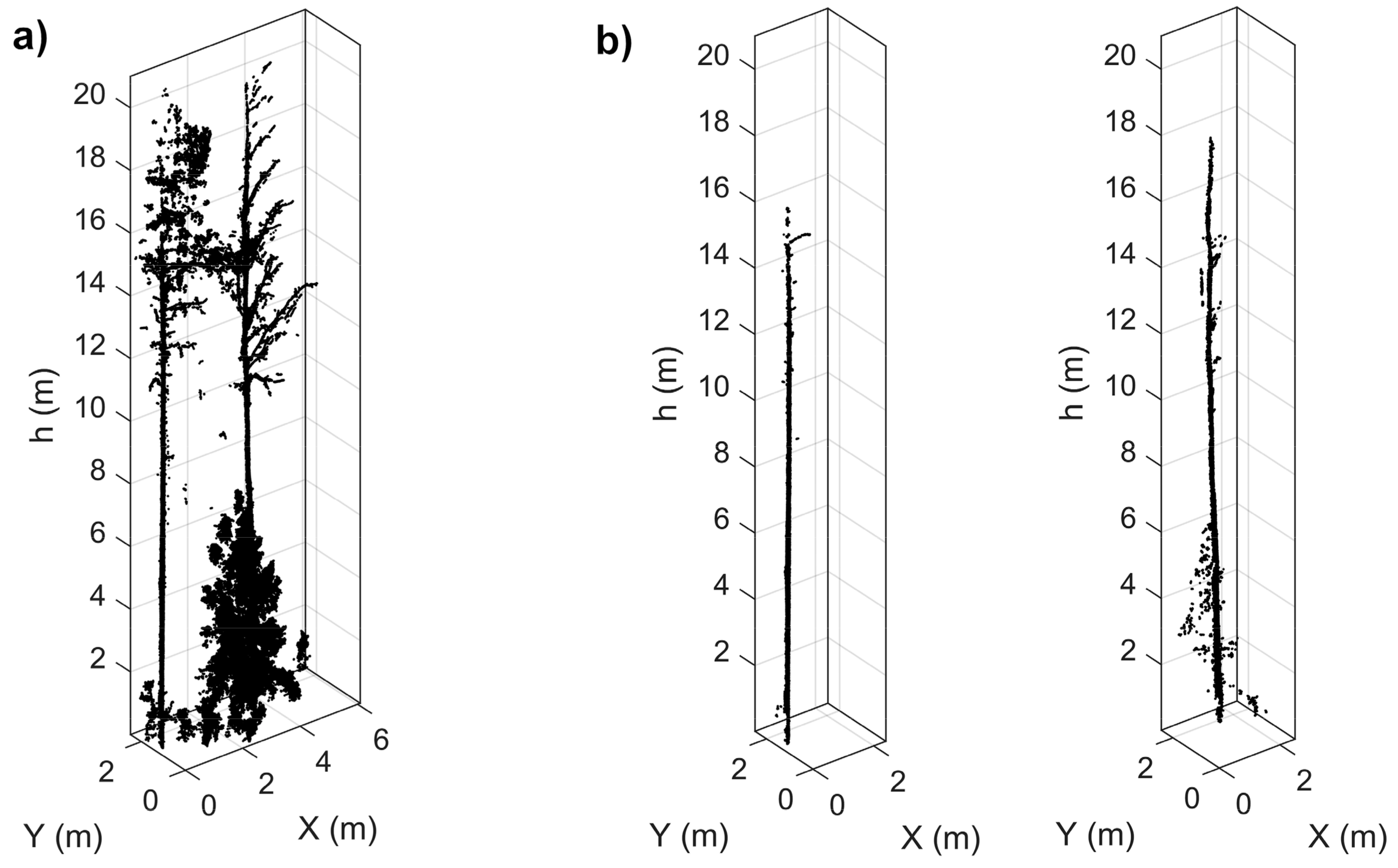
2.2.3. Tree-Wise Point Cloud Extraction
2.2.4. Stage-Two Tree Detection
2.2.5. Tree Metrics Extraction
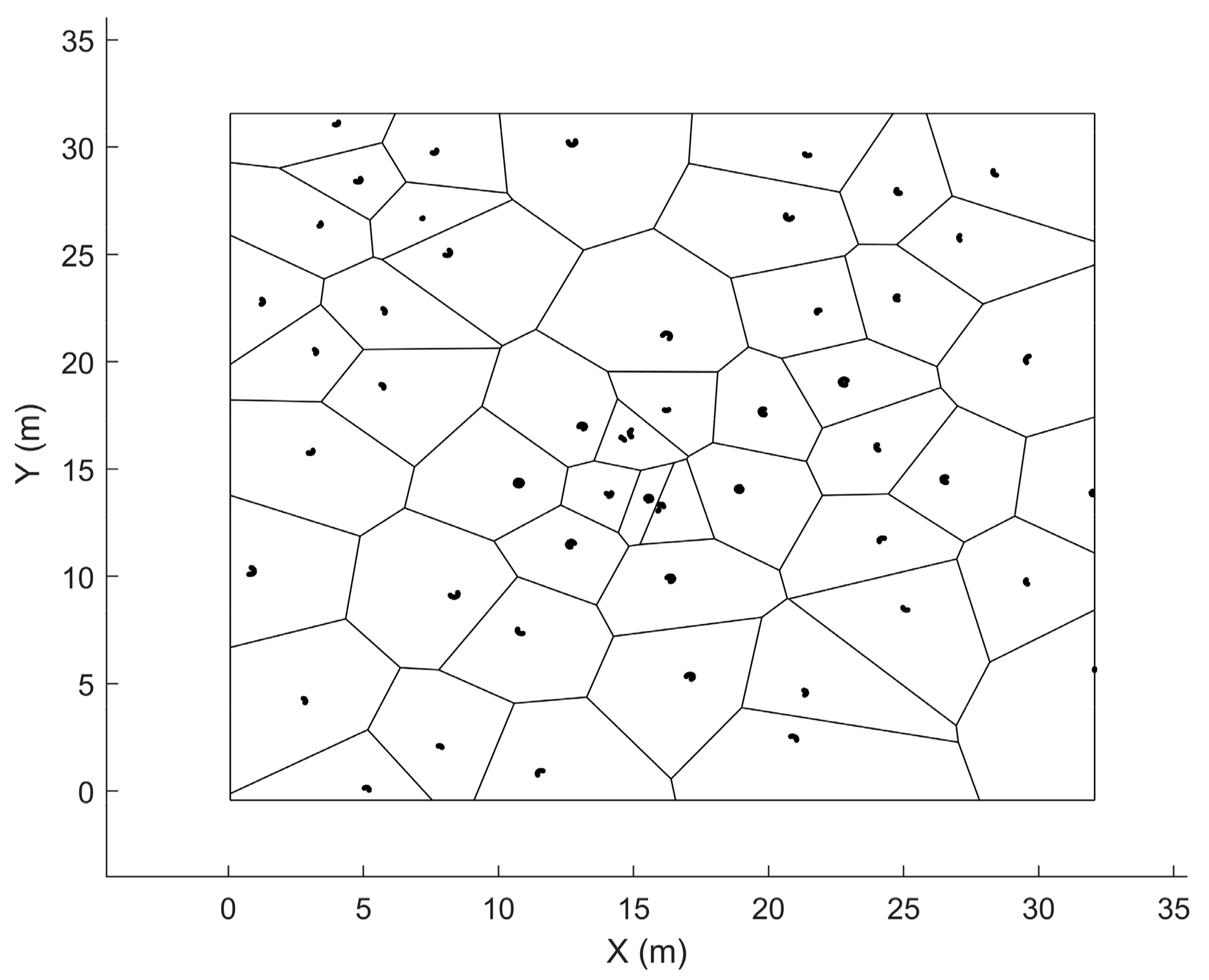
2.2.6. Plot Metrics Extraction
2.3. Evaluating Accuracy and Performance of the TLS-Based Method
2.3.1. Analyzing the Effect of Sample Plot Size on the Estimation Accuracy of Plot-Level Forest Inventory Attributes
2.3.2. Analyzing the Effect of Stand Heterogeneity on the Estimation Accuracy of Plot-Level Forest Inventory Attributes
2.3.3. Analyzing Tree Detection Accuracy
3. Results
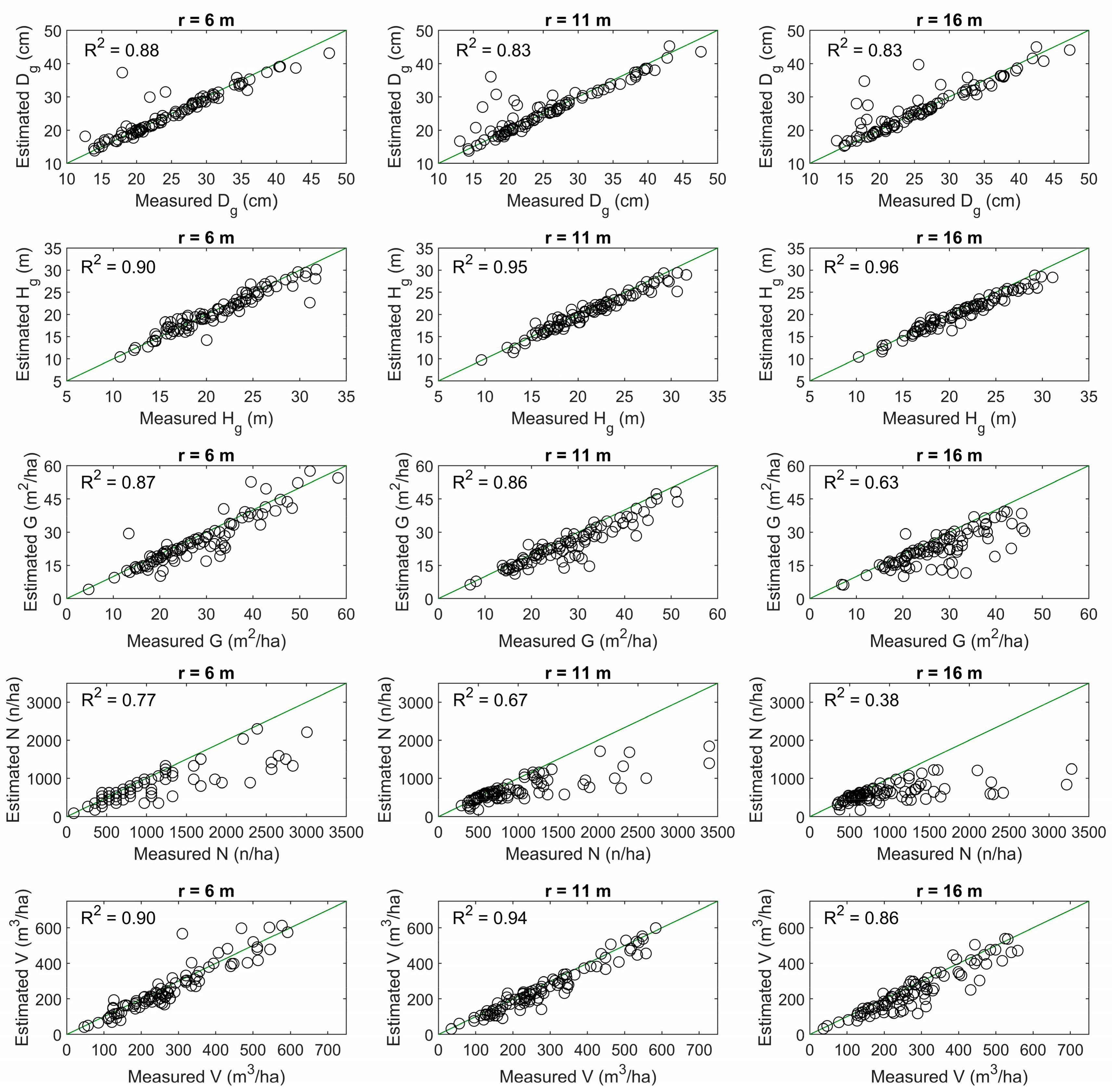
3.1. Effect of the Sample Plot Size on the Estimation Accuracy of Plot-Level Forest Inventory Attributes
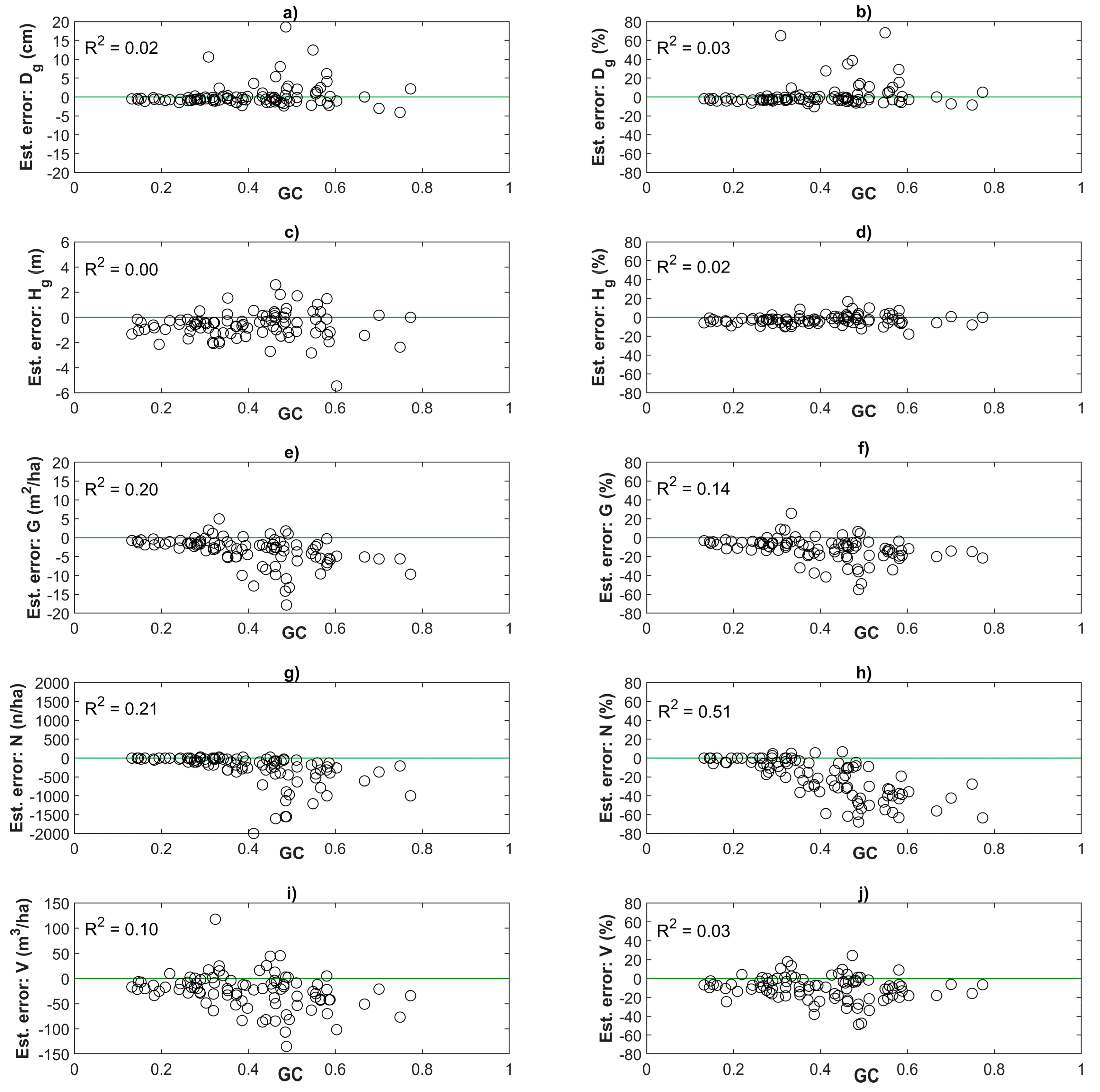
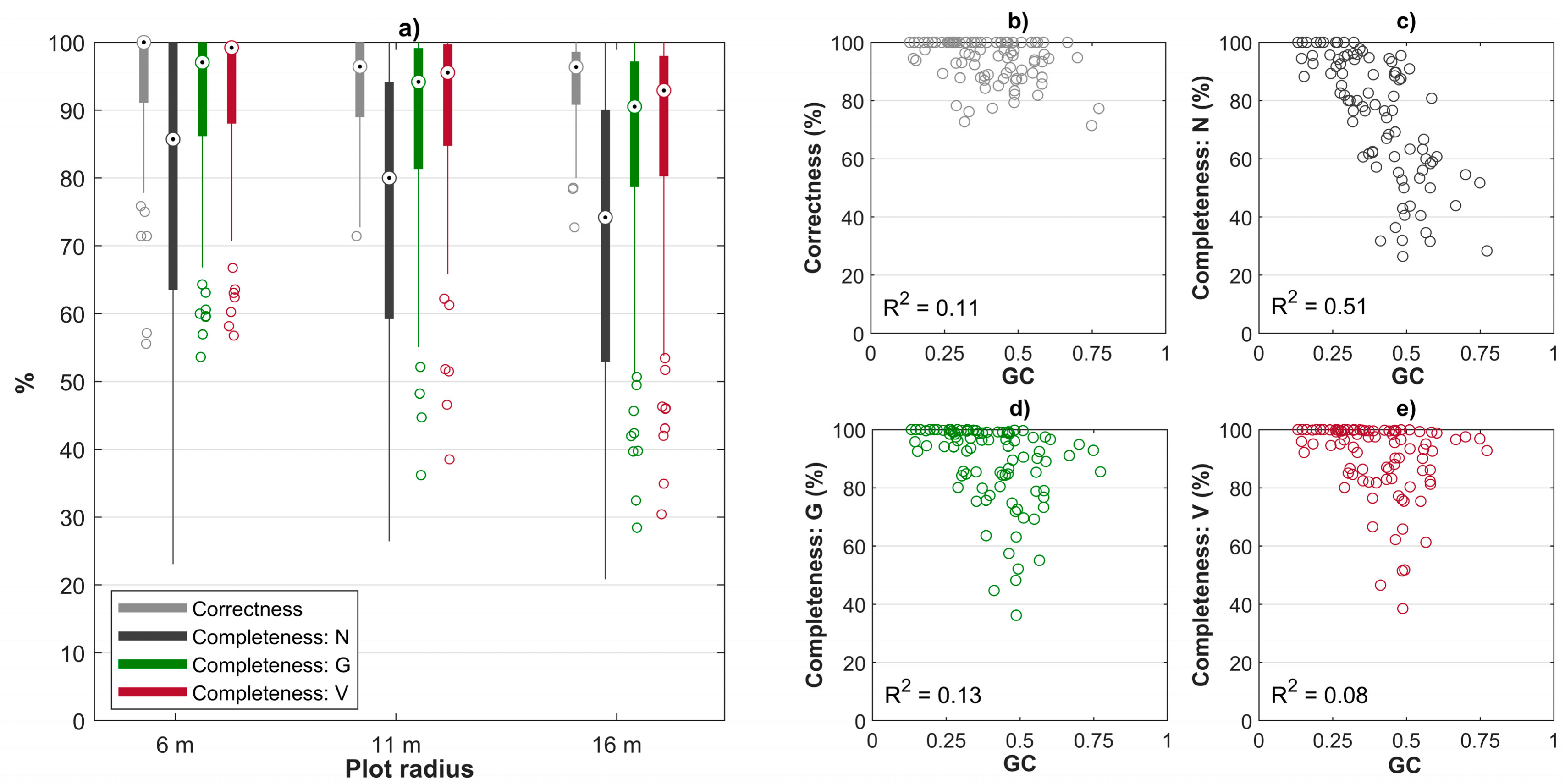
3.2. Effect of Stand Heterogeneity on Estimation Accuracy of Plot-Level Forest Inventory Attributes
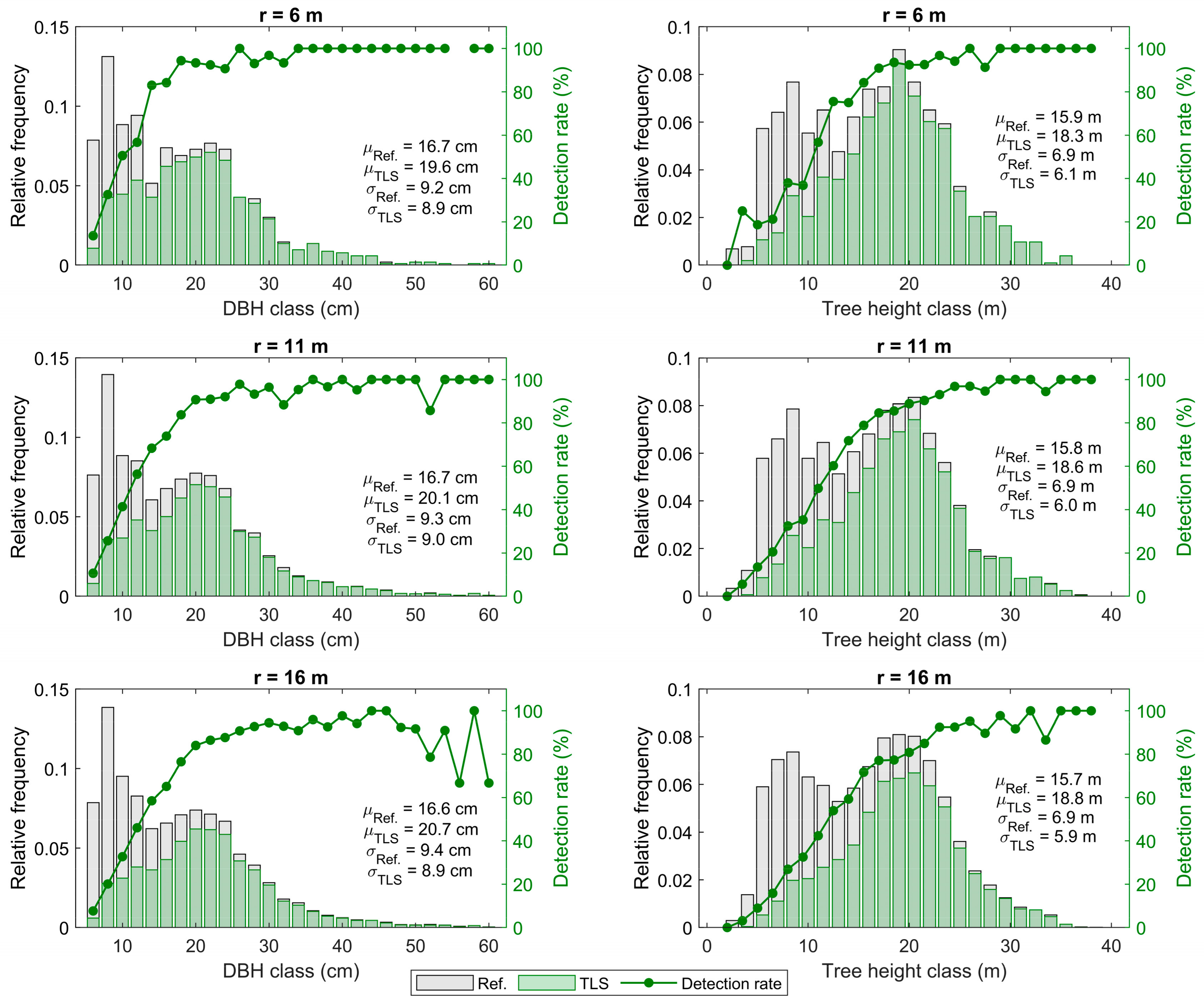
3.3. Tree Detection Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dixon, R.K.; Brown, S.; Houghton, R.A.; Solomon, A.M.; Trexler, M.C.; Wisniewski, J. Carbon Pools and Flux of Global Forest Ecosystems. Science 1994, 263, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.D.; Birdsey, R.A.; Fang, J.Y.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A Large and Persistent Carbon Sink in the World’s Forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Tilman, D.; Isbell, F.; Cowles, J.M. Biodiversity and Ecosystem Functioning. Annu. Rev. Ecol. Evol. S 2014, 45, 471–493. [Google Scholar] [CrossRef]
- Crowther, T.W.; Glick, H.B.; Covey, K.R.; Bettigole, C.; Maynard, D.S.; Thomas, S.M.; Smith, J.R.; Hintler, G.; Duguid, M.C.; Amatulli, G.; et al. Mapping tree density at a global scale. Nature 2015, 525, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughn, N.R.; Llactayo, W. FOREST CONSERVATION Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Dassot, M.; Constant, T.; Fournier, M. The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Ann. For. Sci. 2011, 68, 959–974. [Google Scholar] [CrossRef]
- Liang, X.L.; Kankare, V.; Hyyppa, J.; Wang, Y.S.; Kukko, A.; Haggren, H.; Yu, X.W.; Kaartinen, H.; Jaakkola, A.; Guan, F.Y.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Kangas, A.; Maltamo, M. Forest Inventory: Methodology and Applications; Springer Science & Business Media: Berlin, Germany, 2006; Volume 10. [Google Scholar]
- West, P.W. Stand Measurement. In Tree and Forest Measurement; Springer: Berlin, Germany, 2015; pp. 71–95. [Google Scholar]
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Danson, F.M. Terrestrial Laser Scanning for Plot-Scale Forest Measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef]
- Liang, X.L.; Hyyppa, J.; Kaartinen, H.; Lehtomaki, M.; Pyorala, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Pirotti, F.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Maas, H.G.; Bienert, A.; Scheller, S.; Keane, E. Automatic forest inventory parameter determination from terrestrial laser scanner data. Int. J. Remote Sens. 2008, 29, 1579–1593. [Google Scholar] [CrossRef]
- Liang, X.L.; Hyyppa, J. Automatic Stem Mapping by Merging Several Terrestrial Laser Scans at the Feature and Decision Levels. Sensors Basel 2013, 13, 1614–1634. [Google Scholar] [CrossRef] [PubMed]
- Heinzel, J.; Huber, M.O. Detecting Tree Stems from Volumetric TLS Data in Forest Environments with Rich Understory. Remote Sens. 2017, 9, 9. [Google Scholar] [CrossRef]
- Cabo, C.; Ordonez, C.; Lopez-Sanchez, C.A.; Armesto, J. Automatic dendrometry: Tree detection, tree height and diameter estimation using terrestrial laser scanning. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 164–174. [Google Scholar] [CrossRef]
- Zhang, W.M.; Wan, P.; Wang, T.J.; Cai, S.S.; Chen, Y.M.; Jin, X.L.; Yan, G.J. A Novel Approach for the Detection of Standing Tree Stems from Plot-Level Terrestrial Laser Scanning Data. Remote Sens. 2019, 11, 211. [Google Scholar] [CrossRef]
- Liang, X.L.; Kankare, V.; Yu, X.W.; Hyyppa, J.; Holopainen, M. Automated Stem Curve Measurement Using Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2014, 52, 1739–1748. [Google Scholar] [CrossRef]
- Olofsson, K.; Holmgren, J. Single Tree Stem Profile Detection Using Terrestrial Laser Scanner Data, Flatness Saliency Features and Curvature Properties. Forests 2016, 7, 207. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, X.L.; Liang, Z.Y.; Welham, C.; Li, W.Z. Deriving Merchantable Volume in Poplar through a Localized Tapering Function from Non-Destructive Terrestrial Laser Scanning. Forests 2016, 7, 87. [Google Scholar] [CrossRef]
- Pitkanen, T.P.; Raumonen, P.; Kangas, A. Measuring stem diameters with TLS in boreal forests by complementary fitting procedure. ISPRS J. Photogramm. Remote Sens. 2019, 147, 294–306. [Google Scholar] [CrossRef]
- Pyörälä, J.; Liang, X.; Saarinen, N.; Kankare, V.; Wang, Y.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Assessing branching structure for biomass and wood quality estimation using terrestrial laser scanning point clouds. Can. J. Remote Sens. 2018, 44, 1–14. [Google Scholar] [CrossRef]
- Kankare, V.; Holopainen, M.; Vastaranta, M.; Puttonen, E.; Yu, X.W.; Hyyppa, J.; Vaaja, M.; Hyyppa, H.; Alho, P. Individual tree biomass estimation using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2013, 75, 64–75. [Google Scholar] [CrossRef]
- Yu, X.W.; Liang, X.L.; Hyyppa, J.; Kankare, V.; Vastaranta, M.; Holopainen, M. Stem biomass estimation based on stem reconstruction from terrestrial laser scanning point clouds. Remote Sens. Lett. 2013, 4, 344–353. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Stovall, A.E.L.; Vorster, A.G.; Anderson, R.S.; Evangelista, P.H.; Shugart, H.H. Non-destructive aboveground biomass estimation of coniferous trees using terrestrial LiDAR. Remote Sens. Environ. 2017, 200, 31–42. [Google Scholar] [CrossRef]
- Polewski, P.; Yao, W.; Heurich, M.; Krzystek, P.; Stilla, U. A voting-based statistical cylinder detection framework applied to fallen tree mapping in terrestrial laser scanning point clouds. ISPRS J. Photogramm. Remote Sens. 2017, 129, 118–130. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Saarinen, N.; Luoma, V.; Tanhuanpää, T.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Detecting and characterizing downed dead wood using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2019, 151, 76–90. [Google Scholar] [CrossRef]
- Harmon, M.E.; Franklin, J.F.; Swanson, F.J.; Sollins, P.; Gregory, S.V.; Lattin, J.D.; Anderson, N.H.; Cline, S.P.; Aumen, N.G.; Sedell, J.R.; et al. Ecology of Coarse Woody Debris in Temperate Ecosystems. Adv. Ecol. Res. 1986, 15, 133–302. [Google Scholar] [CrossRef]
- Esseen, P.-A.; Ehnström, B.; Ericson, L.; Sjöberg, K. Boreal forests. Ecol. Bull. 1997, 46, 16–47. [Google Scholar]
- Wang, Y.S.; Lehtomaki, M.; Liang, X.L.; Pyorala, J.; Kukko, A.; Jaakkola, A.; Liu, J.B.; Feng, Z.Y.; Chen, R.Z.; Hyyppa, J. Is field-measured tree height as reliable as believed A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Abegg, M.; Kukenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial Laser Scanning for Forest Inventories Tree Diameter Distribution and Scanner Location Impact on Occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef]
- Trochta, J.; Kral, K.; Janik, D.; Adam, D. Arrangement of terrestrial laser scanner positions for area-wide stem mapping of natural forests. Can. J. Remote Sens. 2013, 43, 355–363. [Google Scholar] [CrossRef]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; de Tanago, J.G.; Bartholomeus, H.; Brede, B.; Herold, M. Data acquisition considerations for Terrestrial Laser Scanning of forest plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- Yu, X.W.; Hyyppa, J.; Karjalainen, M.; Nurminen, K.; Karila, K.; Vastaranta, M.; Kankare, V.; Kaartinen, H.; Holopainen, M.; Honkavaara, E.; et al. Comparison of Laser and Stereo Optical, SAR and InSAR Point Clouds from Air- and Space-Borne Sources in the Retrieval of Forest Inventory Attributes. Remote Sens. Basel 2015, 7, 15933–15954. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest Inventory with Terrestrial LiDAR: A Comparison of Static and Hand-Held Mobile Laser Scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef]
- Holopainen, M.; Kankare, V.; Vastaranta, M.; Liang, X.L.; Lin, Y.; Vaaja, M.; Yu, X.W.; Hyyppa, J.; Hyyppa, H.; Kaartinen, H.; et al. Tree mapping using airborne, terrestrial and mobile laser scanning—A case study in a heterogeneous urban forest. Urban For. Urban Green 2013, 12, 546–553. [Google Scholar] [CrossRef]
- Kankare, V.; Liang, X.L.; Vastaranta, M.; Yu, X.W.; Holopainen, M.; Hyyppa, J. Diameter distribution estimation with laser scanning based multisource single tree inventory. ISPRS J. Photogramm. Remote Sens. 2015, 108, 161–171. [Google Scholar] [CrossRef]
- Liu, G.J.; Wang, J.L.; Dong, P.L.; Chen, Y.; Liu, Z.Y. Estimating Individual Tree Height and Diameter at Breast Height (DBH) from Terrestrial Laser Scanning (TLS) Data at Plot Level. Forests 2018, 9, 398. [Google Scholar] [CrossRef]
- Liu, J.B.; Liang, X.L.; Hyyppa, J.; Yu, X.W.; Lehtomaki, M.; Pyorala, J.; Zhu, L.L.; Wang, Y.S.; Chen, R.Z. Automated matching of multiple terrestrial laser scans for stem mapping without the use of artificial references. Int. J. Appl. Earth Obs. Geoinf. 2017, 56, 13–23. [Google Scholar] [CrossRef]
- Murphy, G.E.; Acuna, M.A.; Dumbrell, I. Tree value and log product yield determination in radiata pine (Pinus radiata) plantations in Australia: comparisons of terrestrial laser scanning with a forest inventory system and manual measurements. Can. J. For. Res. 2010, 40, 2223–2233. [Google Scholar] [CrossRef]
- Reddy, R.S.; Rakesh; Jha, C.S.; Rajan, K.S. Automatic estimation of tree stem attributes using terrestrial laser scanning in central Indian dry deciduous forests. Curr. Sci. India 2018, 114, 201–206. [Google Scholar] [CrossRef]
- Ritter, T.; Schwarz, M.; Tockner, A.; Leisch, F.; Nothdurft, A. Automatic Mapping of Forest Stands Based on Three-Dimensional Point Clouds Derived from Terrestrial Laser-Scanning. Forests 2017, 8, 265. [Google Scholar] [CrossRef]
- Watt, P.J.; Donoghue, D.N.M. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Yang, B.S.; Dai, W.X.; Dong, Z.; Liu, Y. Automatic Forest Mapping at Individual Tree Levels from Terrestrial Laser Scanning Point Clouds with a Hierarchical Minimum Cut Method. Remote Sens. 2016, 8, 372. [Google Scholar] [CrossRef]
- Luoma, V.; Saarinen, N.; Wulder, M.A.; White, J.C.; Vastaranta, M.; Holopainen, M.; Hyyppa, J. Assessing Precision in Conventional Field Measurements of Individual Tree Attributes. Forests 2017, 8, 38. [Google Scholar] [CrossRef]
- Laasasenaho, J. Taper Curve and Volume Functions for Pine, Spruce and Birch; Metsäntutkimuslaitos: Rovaniemi, Finland, 1982. [Google Scholar]
- Aschoff, T.; Spiecker, H. Algorithms for the automatic detection of trees in laser scanner data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 36, W2. [Google Scholar]
- Liang, X.; Kukko, A.; Hyyppä, J.; Lehtomäki, M.; Pyörälä, J.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Wang, Y. In-situ measurements from mobile platforms: An emerging approach to address the old challenges associated with forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 143, 97–107. [Google Scholar] [CrossRef]
- Saarinen, N.; Kankare, V.; Vastaranta, M.; Luoma, V.; Pyorala, J.; Tanhuanpaa, T.; Liang, X.L.; Kaartinen, H.; Kukko, A.; Jaakkola, A.; et al. Feasibility of Terrestrial laser scanning for collecting stem volume information from single trees. ISPRS J. Photogramm. Remote Sens. 2017, 123, 140–158. [Google Scholar] [CrossRef]
- Isenburg, M. LAStools—Efficient LiDAR Processing Software, (version 170511 academic); rapidlasso GmbH: Gilching, Germany. Available online: http://rapidlasso.com/LAStools (accessed on 18 August 2017).
- Lexerod, N.L.; Eid, T. An evaluation of different diameter diversity indices based on criteria related to forest management planning. For. Ecol. Manag. 2006, 222, 17–28. [Google Scholar] [CrossRef]

| Study Site Location | Number of Plots | Plot Size (m2) | Stem Density 1 (n/ha) | Reference |
|---|---|---|---|---|
| Finland | 24 | 1024 | 381–2871 | [12] |
| Germany | 5 | 707 | 212–410 | [13] |
| Finland | 5 | 314 | 605–1210 | [14] |
| Switzerland | 9 | 500 | 200–800 | [15] |
| Spain / Mexico | 3 | 500–600 | 300–2100 | [16] |
| Finland / China | 7 | 1024 | 366–2304 | [17] |
| Sweden | 7 | 1257 | ~1241 | [19] |
| China | 8 | 707 | ~350 | [29] |
| Belgium | 10 | 707 | 114–1344 | [36] |
| Finland | 1 | 27,000 | ~162 | [37] |
| Finland | 27 | 300 | 334–1167 | [38] |
| China | 39 | 1257 | - | [39] |
| Finland | 10 | 1024 | 342–1191 | [40] |
| Australia | 33 | 300–1300 | 153–570 | [41] |
| India | 4 | 1257 | 400–500 | [42] |
| Austria | 1 | 40,800 | ~438 | [43] |
| UK | 2 | 200 | 600–2800 | [44] |
| Finland | 5 | 1024 | 507–928 | [45] |
| Forest Inventory Attribute | Minimum | Mean | Maximum | Standard Deviation |
|---|---|---|---|---|
| Dg (cm) | 13.9 | 25.8 | 46.4 | 7.5 |
| Hg (m) | 10.0 | 21.1 | 31.1 | 4.4 |
| G (m2/ha) | 6.6 | 26.9 | 43.2 | 7.9 |
| N (n/ha) | 342 | 943 | 3076 | 556 |
| V (m3/ha) | 34.5 | 271.5 | 518.4 | 110.7 |
| Stage | Techniques | Parameters |
|---|---|---|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
| Plot Size | Accuracy Measure | Dg (cm) | Hg (m) | G (m2/ha) | N (n/ha) | V (m3/ha) |
|---|---|---|---|---|---|---|
| r = 6 m | Bias | −0.1 (−0.2%) | −0.5 (−2.4%) | −2.4 (−8.3%) | −235.1 (−23.5%) | −17.6 (−6.0%) |
| RMSE | 2.7 (10.6%) | 1.6 (7.6%) | 5.0 (17.6%) | 459.0 (45.9%) | 55.7 (19.1%) | |
| r = 11 m | Bias | 0.3 (1.3%) | −0.6 (−2.8%) | −3.5 (−12.5%) | −281.6 (−29.2%) | −24.7 (−8.8%) |
| RMSE | 3.1 (12.3%) | 1.3 (5.9%) | 5.1 (18.4%) | 498.3 (51.7%) | 43.1 (15.3%) | |
| r = 16 m | Bias | 0.5 (1.9%) | −0.9 (−4.2%) | −5.0 (−18.2%) | −349.4 (−36.4%) | −37.6 (−13.5%) |
| RMSE | 3.2 (12.3%) | 1.3 (6.3%) | 7.3 (26.4%) | 596.1 (62.1%) | 59.5 (21.3%) | |
| 32 m × 32 m | Bias | 0.8 (3.1%) | −1.1 (−5.0%) | −5.4 (−20.1%) | −369.0 (−39.1%) | −41.8 (−15.4%) |
| RMSE | 3.6 (13.8%) | 1.5 (7.1%) | 7.7 (28.5%) | 613.7 (65.1%) | 64.8 (23.9%) |
| Sample Plot Size | Correctness (%) | Completeness (%) | ||
|---|---|---|---|---|
| N | G | V | ||
| r = 6 m | 93.0 | 71.1 | 90.8 | 93.4 |
| r = 11 m | 93.6 | 66.2 | 88.3 | 91.3 |
| r = 16 m | 94.1 | 59.8 | 83.2 | 86.6 |
| 32 m × 32 m | 93.9 | 57.0 | 80.6 | 84.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yrttimaa, T.; Saarinen, N.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Investigating the Feasibility of Multi-Scan Terrestrial Laser Scanning to Characterize Tree Communities in Southern Boreal Forests. Remote Sens. 2019, 11, 1423. https://doi.org/10.3390/rs11121423
Yrttimaa T, Saarinen N, Kankare V, Liang X, Hyyppä J, Holopainen M, Vastaranta M. Investigating the Feasibility of Multi-Scan Terrestrial Laser Scanning to Characterize Tree Communities in Southern Boreal Forests. Remote Sensing. 2019; 11(12):1423. https://doi.org/10.3390/rs11121423
Chicago/Turabian StyleYrttimaa, Tuomas, Ninni Saarinen, Ville Kankare, Xinlian Liang, Juha Hyyppä, Markus Holopainen, and Mikko Vastaranta. 2019. "Investigating the Feasibility of Multi-Scan Terrestrial Laser Scanning to Characterize Tree Communities in Southern Boreal Forests" Remote Sensing 11, no. 12: 1423. https://doi.org/10.3390/rs11121423
APA StyleYrttimaa, T., Saarinen, N., Kankare, V., Liang, X., Hyyppä, J., Holopainen, M., & Vastaranta, M. (2019). Investigating the Feasibility of Multi-Scan Terrestrial Laser Scanning to Characterize Tree Communities in Southern Boreal Forests. Remote Sensing, 11(12), 1423. https://doi.org/10.3390/rs11121423







