1. Introduction
Urban trees play an important role in city planning and urban environment preservation. They can absorb airborne pollutants, improve local water quality, mitigate urban heat islands’ effects and reduce energy consumption associated with cooling buildings [
1,
2,
3]. Thus, precise forest management and planning are also important tasks in city management and planning. It is essential to derive and update the single tree information such as tree distribution, diameter at breast height(DBH), stems volume, biomass and so on, whereas tree detection and species recognition are fundamental. Vitality describes tree’s growing status which is useful for precise forest management. The large number of trees makes the manual updating process very laborious. Remote sensing has become the main technique to monitoring forest because of its low cost and highly efficient data acquisition capability at a large scale, especially the LiDAR which can acquire 3D measurements and is particularly suitable for observing the forests. Use of LiDAR in urban tree management can lead to savings in maintenance actions and costs, prevention of urban heat island effect, purified air and water, and increasing livability of neighborhoods etc.
Previous studies focused mainly on forest trees, though some studies have taken urban trees as research objects. However, those trees are located within the forest environment without suffering from the influences of the complex man-made objects, and these studies also mostly relied on using LiDAR data acquired from an airborne platform to recognize the geometric structure of tree crowns [
4,
5,
6,
7].
Compared to the relatively simple extraction of tree points in forested areas, it is very interesting to investigate whether the vehicle-borne mobile LiDAR technique can conduct single tree inventories in a very complex urban street environment. According to the used platforms, the LiDAR system used in surveys can be categorized into terrestrial laser scanning( TLS), MLS and ALS, which collect data from different view perspectives, platforms and data resolutions (i.e., point cloud density).
Regarding the combined use of different sensors for vegetation monitoring, the studies can be divided into two types: one uses the heterogeneous data from different sensors such as LiDAR with optical sensors [
8,
9,
10,
11], synthetic aperture radar(SAR) with optical sensors [
12,
13] and high geometrical resolution images with hyper/multi spectral images [
14]; the other category is through the synthetic use of similar sensors from different platforms, such as TLS with ALS [
15], airborne with space-borne and so on.
To the best of our knowledge, the combined use of these LiDAR data sets acquired from two different platforms/perspectives for tree monitoring is still lacking, especially for tree species and vitality classification in the complicated urban street environment. To a great extent, complementary information is supposed to exist between the MLS and ALS data, since both platforms can collect information from different perspectives, area coverage, and collection efficiency and data resolutions.
Kankare et al. [
5] mapped the tree diameter distribution of a Finland urban forest by synergistic use of TLS and ALS data. In [
5], TLS was used to derive precise DBH and potential undergrowth which are difficult to implement using ALS alone; meanwhile, the ALS data was used to obtain tree species information based on both the stem location derived from TLS data and metrics extracted from ALS data. The main tree populations are coniferous with 4 species. The authors of [
6] conducted the mapping of street roadside trees using the ALS only, which achieved detection accuracy of 88.8% and estimation root mean square error (RMSE) of 1.27 m and 6.9 cm for tree height and DBH respectively. In [
7], TLS, MLS and ALS were compared for tree mapping. The authors concluded that challenges to be solved in further studies include ALS individual tree detection (ITD) auto over-segmentation as well as MLS auto processing methodologies and data collection for tree detection.
Most recently, the authors of [
15] presented a data fusion approach to extract tree crowns from TLS by exploiting the complementary perspective of ALS. The combined segmentation results from co-registered TLS scan and the ALS data using adapted tree boundaries from ALS are fused into single-tree point clouds, from which canopy-level structural parameters can be extracted.
Besides LiDAR data, optical images collected from overhead to ground were also adopted to detect and classify individual trees using photogrammetric techniques [
16]. In particular, Google Street View (GSV, available through
http://www.google.com/streetview) was found to be well suited for assessing street-level greenery [
17]. GSV provides many free street tree photos, and processing these free available pictures through manual interpretation [
18] or deep learning method [
19] can generate some types of street tree information such as DBH and tree specifics.
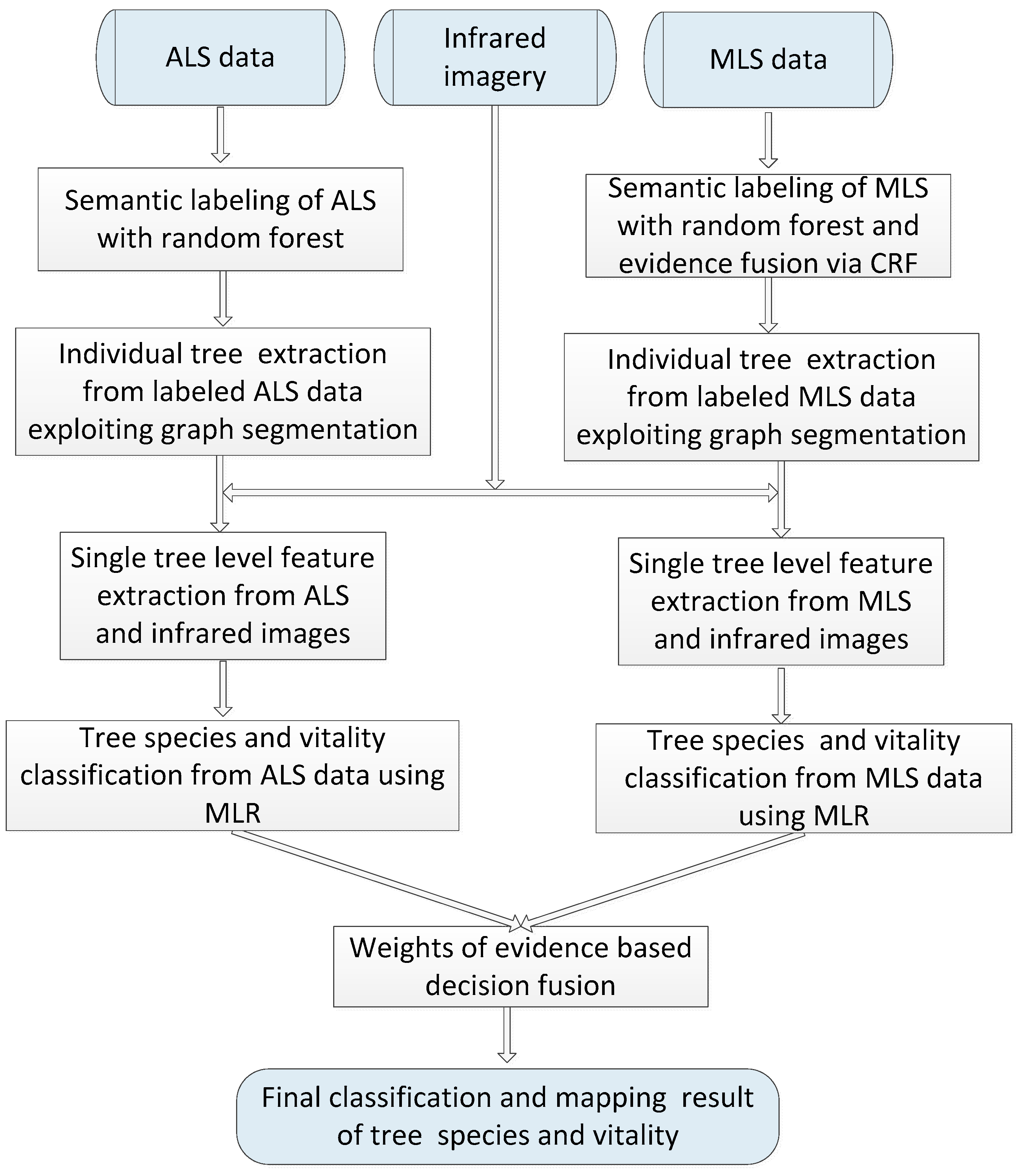
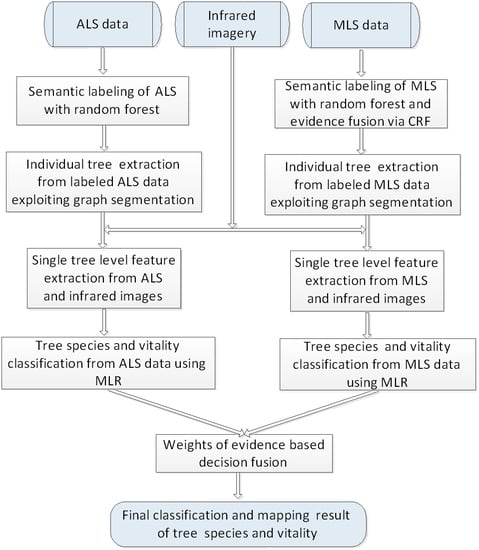
This paper analyzes the potential of MLS-based methods for tree species mapping and vitality monitoring in a complex urban road environment using high-density point clouds and provides a comparison with the ALS-based methods. It is therefore naturally expected in the next step that we propose to combine MLS with ALS to characterize the tree attributes.
As a case study, we aim to map roadside trees in a European city and distinguish fine deciduous tree species and vitality conditions by synergistic use of laser scanning data and infrared images. This case is challenging since individual trees have diverse distribution patterns, and physiological status but less varied shapes and sizes. In addition, separating trees from neighboring man-made objects in LiDAR point clouds of urban road corridors is complex for non-experts and for automatic classification methods since the surrounding plants and the scene background strongly differ and interact with each other. In particular, the contributions of this work are summarized as:
- (1)
Developing an accurate and efficient context-based classification method for vegetation mask mapping using ultra-high density MLS point clouds;
- (2)
Comparing MLS-based methods with ALS-based methods in terms of performance in detecting trees and recognizing their species and vitality;
- (3)
Designing a decision fusion scheme that combines outputs of ALS models with MLS model for tree attribute recognition;
- (4)
Demonstrating that the sole use of ALS datasets for tree detection and characterization in urban road corridors can lead to satisfactory results that can be further enhanced by incorporating MLS data using decision fusion techniques.
4. Discussion
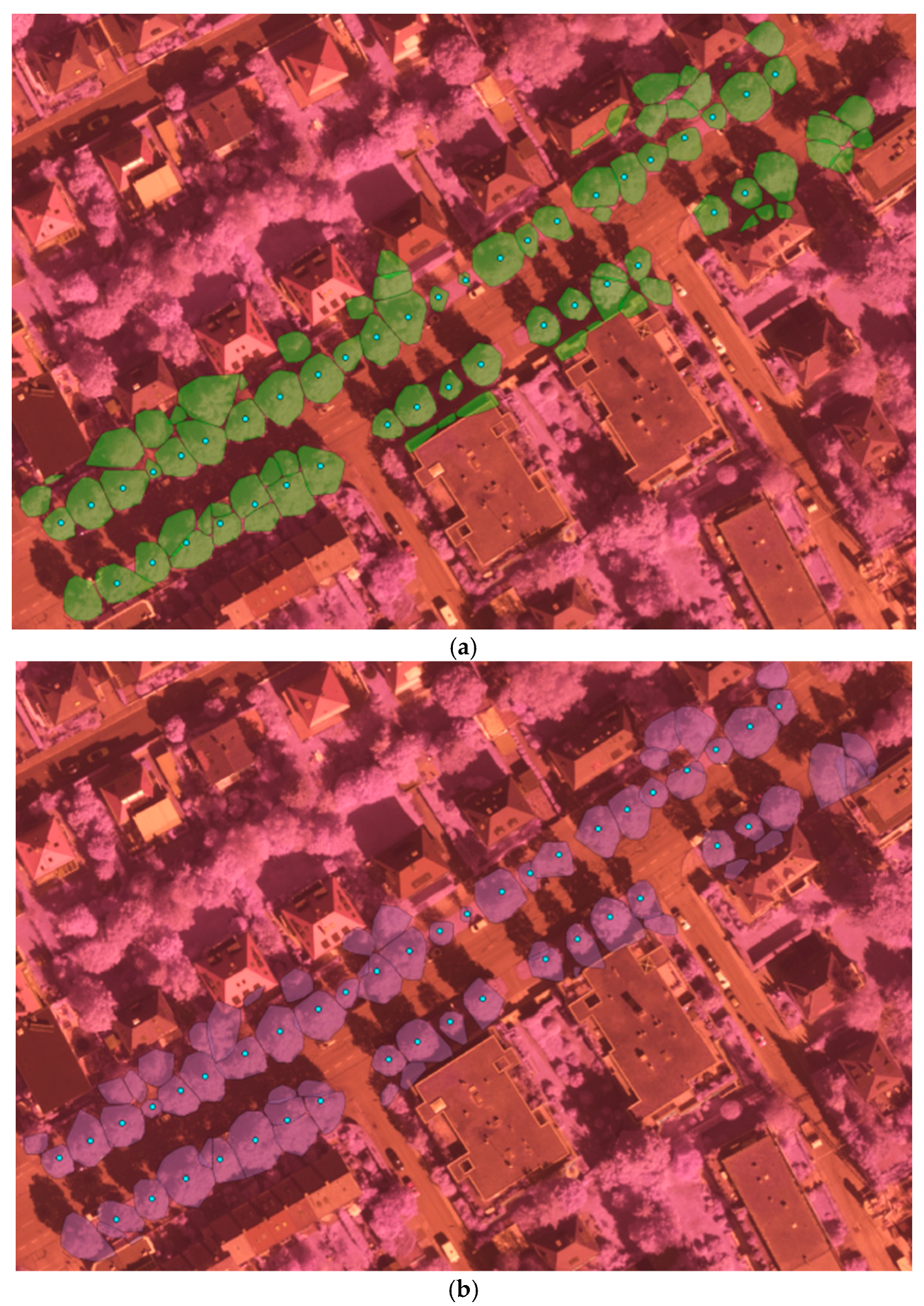
As we can see from the tree detection results in
Table 1, ALS achieved better detection accuracy than MLS: 83.63% vs. 77.27%. About 80 more trees were detected from ALS in the same covering test area. Compared to [
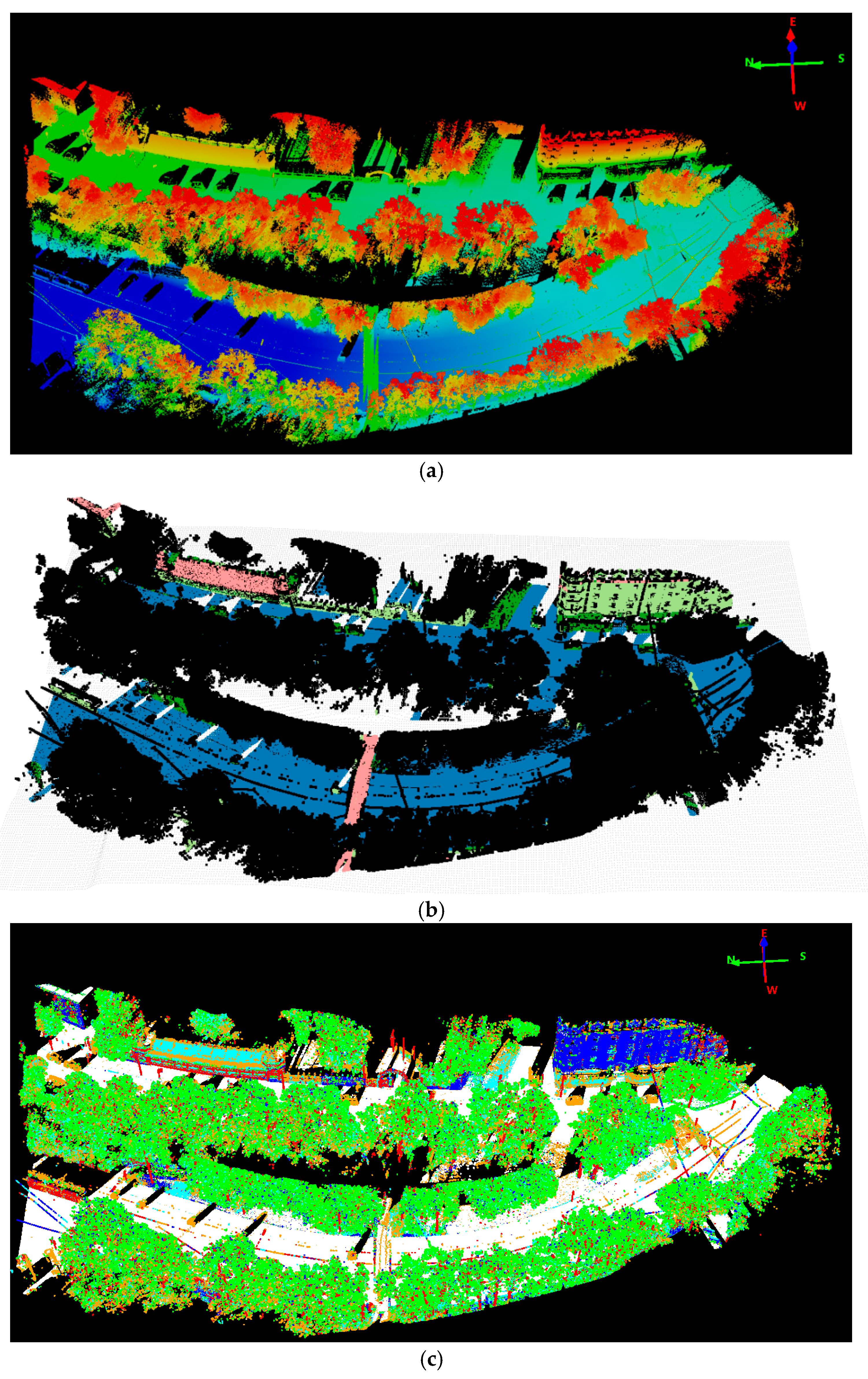
7], we achieved comparable detection accuracy while in a more complicated environment. One example of such a complicated scene is depicted in
Figure 6, in which trees appeared largely mixed with man-made object especially the buildings. The overall classification accuracy of both species and vitality was depicted in
Table 11. From
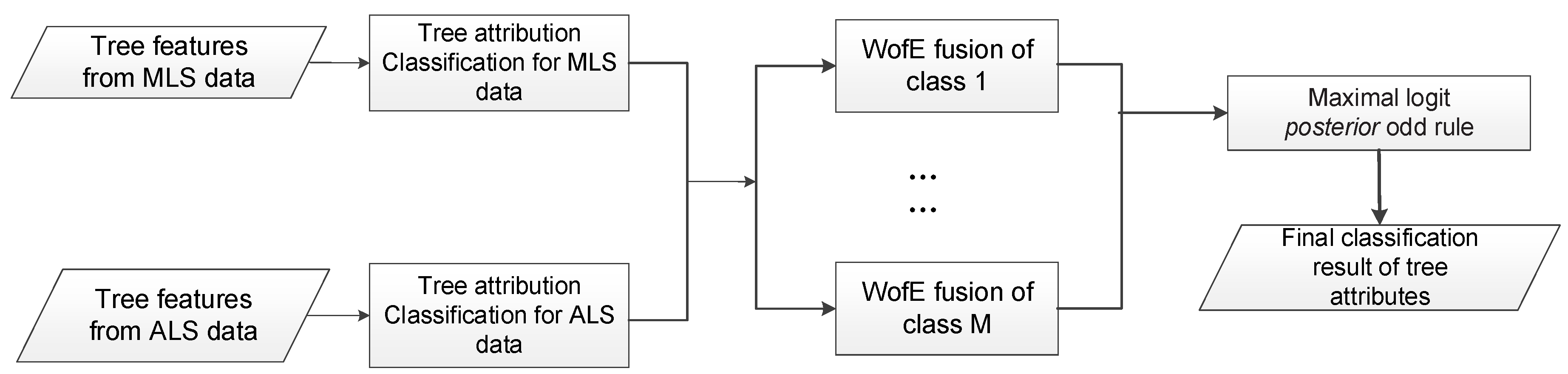
Table 11 we can conclude that ALS achieved better classification accuracy than MLS: 72.19% vs. 65.16% for species classification, and 68.89% vs. 57.50% for vitality classification. Through decision level fusion of the ALS and MLS classification results, we achieved better accuracy both on determining trees’ species and health status: 77.89% and 73.63%, which constituted an improvement of 5.7 and 4.64 percentage points compared to the ALS result respectively. The obtained accuracy improvement through fusion demonstrates information from different scanning perspectives is complementary for tree mapping and information extraction.
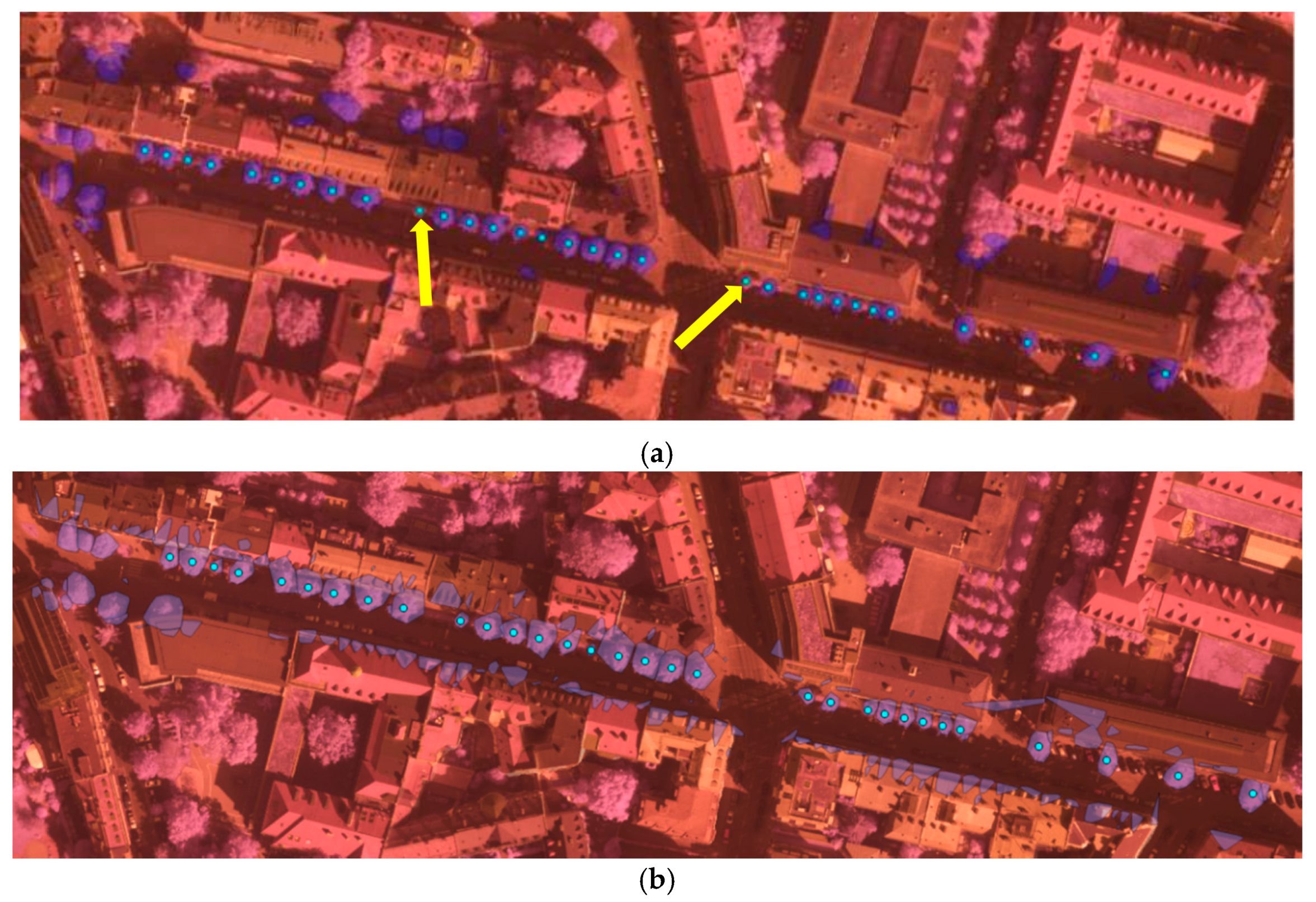
From the detection results in
Figure 6, we can see that our method detected almost all the trees with ground truth. Through comparing the results of ALS and MLS, we can state that some trees’ contours derived from MLS were too large due to adjacent building points’ influence. As MLS collects data from the ground platform in a side-view perspective with a dense scanning of elevated surface, MLS data can’t cover upper part of high objects. It is greatly affected by the visibility along the scanning vehicle’s trajectory, in particular occlusions due to the presence of moving objects, buildings and trees etc. Thus, for the complicated scene in
Figure 6, tree points with adjacent building points are prone to be segmented into larger trees and mixed small trees might be detected as one larger tree in the MLS data. Besides, some small structures of building tops and edges are also falsely detected as trees. On the other hand, the ALS collects data in a nadir (top-down) view perspective with aerial platform and less dense scanning at a high altitude, thus ALS data can achieve wider area coverage and represent the non-occluded top surface well. Thus, the tree crowns were extracted more accurately, which can be observed from
Figure 6a. The point density is also an important factor in tree detection. It’s difficult and almost impossible to detect trees from sparse points. In the experiment two small ground truth trees(in
Figure 6a, the two yellow arrows direct the two undetected small trees) were not correctly detected as depicted in
Figure 6a in the relatively less dense ALS data, while they were both correctly detected in the MLS data. Overall, the detection accuracy from MLS data was worse than ALS data, though the MLS data was with much higher density. Higher density is not equivalent to better detection accuracy, the more complete and better data coverage of trees can be more important.
Comparing the tree attributes classification results from ALS and MLS data summarized in
Table 11, it shows the ALS dataset obtained better accuracy for both the species and vitality using the MLR classifier. Thus, features extracted from the data collected in different view perspectives mainly caused the differences in attribute classification accuracy. In the experiment, features were extracted and selected with same approach from the detected tree points collected in different scanning perspectives: nadir (top-down ALS) view versus side view (MLS). As analysed and discussed above, view perspectives for LiDAR data collection have impact on the extraction accuracy of tree crowns, which further influences the correctness and reliability of the feature extraction for single trees. The selected features were arranged and listed in
Table 2 according to the feature importance for classification. From
Table 2, we can derive: (1) fewer features were selected from the ALS data, which means fewer features from nadir view collected data are essential to describe the tree attributes; (2) fewer features were selected for species classification, which means it needs more essential features to determine health status than classify tree species with LiDAR remote sensing data; (3) intensity feature S1_I and S2_I from ALS data were selected as the most important LiDAR features for determining tree species and vitality respectively in this experiment, while no intensity feature from MLS data was selected. The MLS intensity from side-view mode suffers from strongly varying incident angles and ranging distances, causing the intensity feature’s inconsistency; (4) compared to species classification, more LiDAR geometric features were selected for classifying tree vitality especially from the MLS data. The MLS scans the trees from a side-view perspective and can model their vertical structure in much more detail with dense point clouds, thus more internal geometric features from MLS can contribute to the tree vitality classification in the experiment; (5) as we used the infrared imagery which is very useful for vegetation classification, the image intensity related features for singles trees detected by LiDAR point clouds played an important role in tree attributes classification.
Better classification accuracy was achieved through fusing MLS and ALS classification probability outputs, which demonstrated that there exists complementary information useful for tree attributes classification. This is in compliance with the fact that MLS and ALS data were collected in different view perspectives, recording information describing the tree structure in different ways. Similarly, tree crown structure was accurately reconstructed through fusing the complementary perspective of ALS with the ultra-dense scanning of TLS in [
15].
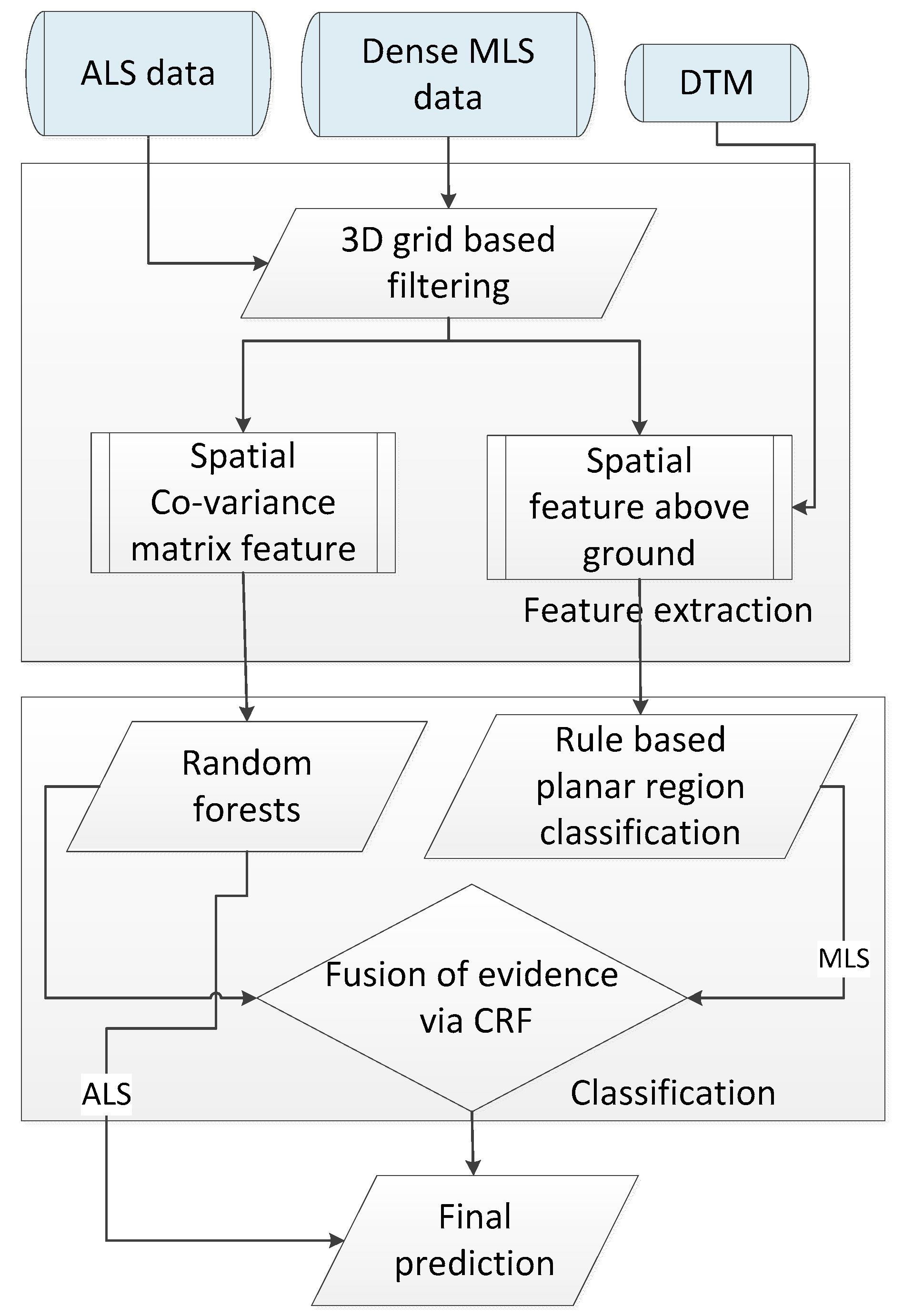
The different scanning perspectives also result in the difference of data collection cost and efficiency: ALS collects data more quickly and efficiently but less flexibly than the MLS as ALS is more likely to be constrained by weather conditions; it also costs more for ALS than MLS to enable the data collection in specific areas. However, more intensive computation arises for the MLS data. In our approach, a new method for the semantic labeling of ultra-high point density MLS data is developed based on combining a CRF for the context-based classification of 3D point clouds with shape priors, which makes the final point labeling results look smoother for the MLS data as depicted in
Figure 4d. To cope with the ultra-high point density and maintain a graph structure of rational size for CRF solution, a plane-based region growing method combined with a rule-based classifier is applied to first generate semantic labels for man-made objects. Once these kind of points, that usually account for up to 60% proportion of the entire data set, are pre-labelled, the CRF classifier can be solved by optimizing the discriminative probability for nodes within a subgraph structure excluded from those nodes assigned with pre-fixed labels. Moreover, through the pre-labeling of planar structures, the scene can often be partitioned into disjoint regions whose labels no longer interact, and hence can be optimized independently.
Compared to traditional field work and laboratory measurement methods [
35,
36,
37], tree vitality classification by remote sensing is a relatively new and challenging task, especially at the single tree level. Within our experiment, the infrared imagery features contributed more than point cloud features, which could be induced from
Table 2. Almost all existing studies from remote sensing community mainly adopted spectral and image features for tree vitality or condition classification with aerial or space-born platform [
38,
39,
40,
41,
42,
43,
44], and studies using LiDAR related features for tree vitality classification are seldom reported. Our experiments demonstrated that the infrared features are more powerful than both the MLS and ALS data for tree vitality determination. On the other hand, laser scanning can acquires tree structure more accurately which can be used to assess the tree health status [
45]. From
Table 2 it can also be deduced that the MLS geometric features contribute more than that of ALS to tree vitality classification, which is attributed to the capability of MLS to scan the tree vertical structure in much more details from side-view.
Although a re-balancing process was conducted to adjust great differences in training sample size per class, it should be noted that the imbalanced sample sizes in our experimental data still influenced the classification accuracy: the number of Robinia trees was much smaller than other species, and training samples for trees of growing health level 2 were too few compared to other tree growing health levels, which can be observed from
Table 3,
Table 4,
Table 5 and
Table 6. Tree species and vitality classes with similar sample sizes could even improve the classification results. Through analyzing
Table 8 and
Table 10 for confusion-matrix of tree vitality classification, it can be observed that there was a relatively higher confusion between 1+ and 1- category than that between 1+ and 1 or between 1 and 1−. The reason is that almost all the tree samples with growth health level 1 belong to Dutch linden, while tree samples with 1+ and 1- category are evenly distributed across all 4 species.
As ALS and MLS data were collected in two different platforms, fine registration is essential for their data level fusion. In our experiment, only accurate georeferencing process was conducted on the ALS and MLS data respectively, which achieved good geolocation accuracies. However, no mutual fine registration was applied to the ALS and MLS data which impeded a data-level fusion at the beginning. To combine the tree attribute classification results of ALS and MLS data, we selected the nearest trees extracted from MLS data as the corresponding trees to ALS data. This relatively simple process may encounter nonalignment problem which will cause fusion errors in the complicated environment. Single tree level attributes should be studied as the constraints to find corresponding trees between ALS and MLS data in the further work. It consists of many processing steps for single tree level mapping. Accurate semantic labeling of the LiDAR data is the basis of good tree detection results. Tree detection error can reduce classification accuracy: the adjacent trees with clumped crowns are easy to be detected as one tree which will cause wrong classification results. Besides, the accuracy of detected trees’ contour also influences the extraction of trees’ features, which has direct impact on the classification accuracy.
5. Conclusions and Future Work
In this work, we studied and compared the urban tree mapping potential of ALS and MLS data in terms of performance in detecting trees and recognizing their species and vitality. A complete approach was presented, including semantic labeling for point cloud, single tree detection and feature extraction, classification of tree species and vitality, and decision fusion of ALS and MLS classification outputs to enhance final mapping results. To make better point-wise semantic labeling for the highly dense MLS data, an improved framework for combining the point-wise class labeling results of RF and local contexts was proposed, which reduces the highly intensive computation efficiently and makes it possible to accurately label ultra-dense point clouds through CRF smoothing.
Through comparing ALS based and MLS based methods, we can conclude: (1) LiDAR scanning perspective is a greatly influential factor for tree detection rate and classification accuracy of tree species and vitality. In our experiment, ALS based methods achieved better accuracy of both the tree detection and classification of the two tree attributes. ALS can cover the tree top surface more completely with nadir scanning perspective, which is very useful for tree detection and tree feature extraction; in contrast, MLS collects data in a side-view perspective and collected tree points are always affected by the adjacent man-made objects like buildings. Contours of tree crowns extracted from ALS are more accurate than that from MLS, which contribute to extraction of more accurate tree features; (2) Higher density is helpful for deriving better geometric features but not equivalent to yielding better mapping accuracy. MLS achieves much higher density data and can retrieve vertical structure information of trees in much more details. In our experiment, more geometric features from MLS were selected for determining tree health status than that from ALS. However, MLS still obtained worse accuracy than ALS for determining tree vitality; (3) Only use of ALS data can lead to satisfactory results for tree detection and characterization in urban road corridors, and even better accuracy can be achieved through fusing the complementary information of the ALS and MLS data. Applying the approach presented in this work to the ALS data only, we achieved tree detection accuracy of 83.6%, species classification accuracy of 72.19% and vitality classification accuracy of 68.89% under the complicated urban road environment. To make use of the complementary information of the ALS and MLS data, a weight of evidence based decision method was adopted to improve the tree attributes classification accuracy. In our experiment, the fusion methods achieved an overall accuracy of 77.89% for tree species classification and 73.53% for tree vitality classification, which was better than the method based on a single data source for both ALS and MLS.
Since there exists good complementarity between ALS and MLS in terms of different scanning perspectives and platforms, point density, ranging distance and data coverage, ALS and MLS data can be further combined to solve more challenging tasks such as fine tree structure modeling, accurate carbon stock estimation and so on in the future work. As these two data sources are collected from different platforms, the fine registration of ALS and MLS data needs further study to handle the redundancy for various applications.