Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Reflectance Measurements and Digital Images
2.3. Spectral and Image Data Analysis for Disease Detection
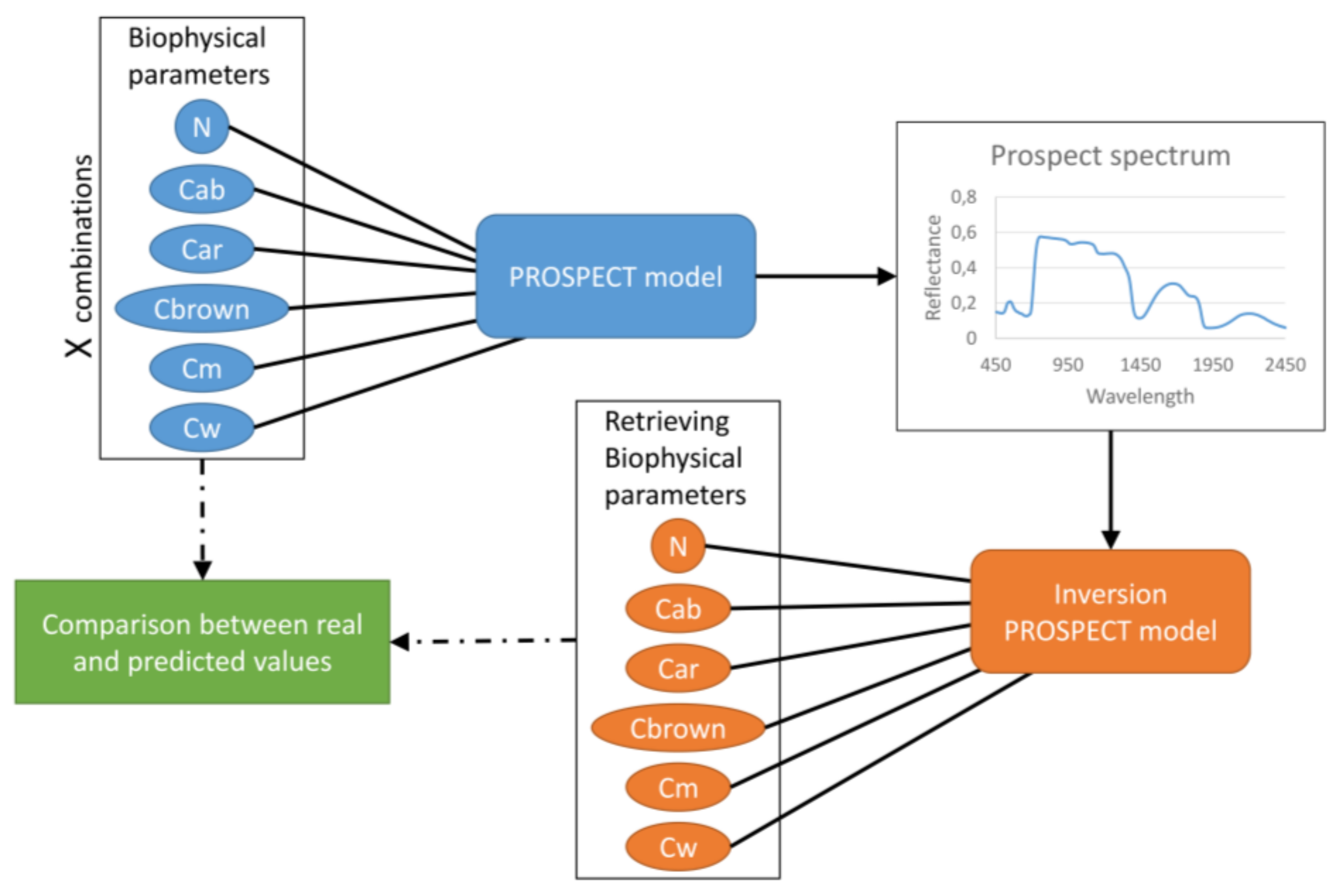
2.3.1. PROSPECT Model
2.3.2. Biophysical Parameters (BPs) Calculation
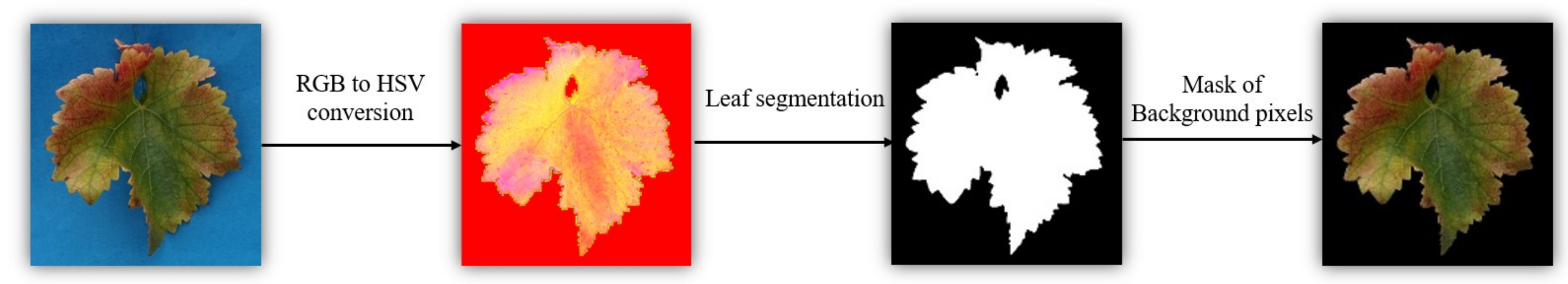
2.3.3. Pre-Processing of Digital Images
2.3.4. Co-Occurrence Matrices
2.3.5. Texture Parameters (TPs) Calculation
2.3.6. Classification: Neural Networks
3. Results
3.1. Best Method and Function for the PROSPECT Model Inversion
3.2. Classification of Data Using BPNN
3.2.1. Classification of Esca Disease
3.2.2. Classification of Yellowing Disease
4. Discussion
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lehrer, A.T.; Moore, P.H.; Komor, E. Impact of sugarcane yellow leaf virus (ScYLV) on the carbohydrate status of sugarcane: Comparison of virus-free plants with symptomatic and asymptomatic virus-infected plants. Physiol. Mol. Plant Pathol. 2007, 70, 180–188. [Google Scholar] [CrossRef]
- Matthews, R.E.F.; Hull, R.; Matthews, R.E.F. Matthews’ Plant Virology; Academic Press: San Diego, CA, USA, 2002; ISBN 978-0-08-053599-9. [Google Scholar]
- Mahlein, A.-K.; Steiner, U.; Dehne, H.-W.; Oerke, E.-C. Spectral signatures of sugar beet leaves for the detection and differentiation of diseases. Precis. Agric. 2010, 11, 413–431. [Google Scholar] [CrossRef]
- Gazala, I.F.S.; Sahoo, R.N.; Pandey, R.; Mandal, B.; Gupta, V.K.; Singh, R.; Sinha, P. Spectral reflectance pattern in soybean for assessing yellow mosaic disease. Indian J. Virol. 2013, 24, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liao, H.; Zhu, Y.; Sun, J.; Sun, Q.; Liu, X. Hyperspectral detection of rice damaged by rice leaf folder (Cnaphalocrocis medinalis). Comput. Electron. Agric. 2012, 82, 100–107. [Google Scholar] [CrossRef]
- Atzberger, C.; Darvishzadeh, R.; Immitzer, M.; Schlerf, M.; Skidmore, A.; le Maire, G. Comparative analysis of different retrieval methods for mapping grassland leaf area index using airborne imaging spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 19–31. [Google Scholar] [CrossRef]
- Al-Hiary, H.; Bani-Ahmad, S.; Reyalat, M.; Braik, M.; ALRahamneh, Z. Fast and accurate detection and classification of plant diseases. Mach. Learn. 2011, 14, 31–38. [Google Scholar] [CrossRef]
- Camargo, A.; Smith, J.S. Image pattern classification for the identification of disease causing agents in plants. Comput. Electron. Agric. 2009, 66, 121–125. [Google Scholar] [CrossRef]
- Majumdar, D.; Kole, D.K.; Chakraborty, A.; Dutta, D. Detection and diagnosis of plant leaf disease using integrated image processing approach. Int. J. Comput. Eng. Appl. 2014, 6, 10–16. [Google Scholar]
- Xie, C.; He, Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Shao, Y.; Li, X.; He, Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Carisse, O.; Canada, Agriculture and Agri-Food Canada, Québec (Province); Ministère de l’agriculture, des pêcheries et de l’alimentation. Identification Guide to the Major Diseases of Grapes; Agriculture and Agri-Food Canada: Ottawa, ON, Canada, 2006; ISBN 978-0-662-43594-5.
- Li, S.; Bonneu, F.; Chadoeuf, J.; Picart, D.; Gégout-Petit, A.; Guérin-Dubrana, L. Spatial and Temporal Pattern Analyses of Esca Grapevine Disease in Vineyards in France. Phytopathology 2017, 107, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, P.; Darrieutort, G.; Liminana, J.-M.; Comont, G.; Muruamendiaraz, A.; Legorburu, F.-J.; Choueiri, E.; Jreijiri, F.; El Amil, R.; Fermaud, M. New insights into esca of grapevine: The development of foliar symptoms and their association with xylem discoloration. Plant Dis. 2012, 96, 924–934. [Google Scholar] [CrossRef]
- Grapevine Measles. Available online: http://articles.extension.org/pages/64365/grapevine-measles (accessed on 6 March 2018).
- Esca (Black Measles). Available online: http://ipm.ucanr.edu/PMG/r302100511.html (accessed on 15 March 2018).
- Mugnai, L.; Graniti, A.; Surico, G. Esca (black measles) and brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis. 1999, 83, 404–418. [Google Scholar] [CrossRef]
- Grapevine Yellows Information—Is There A Treatment For Grapevine Yellows. Available online: https://www.gardeningknowhow.com/edible/fruits/grapes/grapevine-yellows-information.htm (accessed on 27 March 2018).
- Chuche, J.; Thiéry, D. Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: A review. Agron. Sustain. Dev. 2014, 34, 381–403. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Feret, J.-B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.P.R.; Ustin, S.L.; le Maire, G.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Duan, S.-B.; Li, Z.-L.; Wu, H.; Tang, B.-H.; Ma, L.; Zhao, E.; Li, C. Inversion of the PROSAIL model to estimate leaf area index of maize, potato, and sunflower fields from unmanned aerial vehicle hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 12–20. [Google Scholar] [CrossRef]
- Romero, A.; Aguado, I.; Yebra, M. Estimation of dry matter content in leaves using normalized indexes and PROSPECT model inversion. Int. J. Remote Sens. 2012, 33, 396–414. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F.; Myneni, R.; Pragnère, A.; Knyazikhin, Y. Investigation of a model inversion technique to estimate canopy biophysical variables from spectral and directional reflectance data. Agronomie 2000, 20, 3–22. [Google Scholar] [CrossRef]
- Sehgal, V.K.; Chakraborty, D.; Sahoo, R.N. Inversion of radiative transfer model for retrieval of wheat biophysical parameters from broadband reflectance measurements. Inf. Process. Agric. 2016, 3, 107–118. [Google Scholar] [CrossRef]
- Atzberger, C. Object-based retrieval of biophysical canopy variables using artificial neural nets and radiative transfer models. Remote Sens. Environ. 2004, 93, 53–67. [Google Scholar] [CrossRef]
- Berjón, A.J.; Cachorro, V.E.; Zarco-Tejada, P.J.; de Frutos, A. Retrieval of biophysical vegetation parameters using simultaneous inversion of high resolution remote sensing imagery constrained by a vegetation index. Precis. Agric. 2013, 14, 541–557. [Google Scholar] [CrossRef]
- Li, P.; Wang, Q. Retrieval of Leaf Biochemical Parameters Using PROSPECT Inversion: A New Approach for Alleviating Ill-Posed Problems. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2499–2506. [Google Scholar] [CrossRef]
- Nelder, J.A.; Mead, R. A Simplex Method for Function Minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
- Deborah, H.; Richard, N.; Hardeberg, J.Y. A Comprehensive Evaluation of Spectral Distance Functions and Metrics for Hyperspectral Image Processing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3224–3234. [Google Scholar] [CrossRef]
- Zhu, C.; Byrd, R.; Lu, P.; Nocedal, J. LBFGS-B Fortran Subroutines for Large-Scale Bound Constrained Optimization; Report NAM-11 EECS Department, Northwestern University: Evanston, IL, USA, 31 December 1994; pp. 1–17. [Google Scholar]
- Sequential Quadratic Programming. Numerical Optimization; Springer: New York, NY, USA, 2006; pp. 529–562. ISBN 978-0-387-30303-1. [Google Scholar]
- Nash, S.G. Preconditioning of Truncated-Newton Methods. SIAM J. Sci. Stat. Comput. 1985, 6, 599–616. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Pydipati, R.; Burks, T.F.; Lee, W.S. Identification of citrus disease using color texture features and discriminant analysis. Comput. Electron. Agric. 2006, 52, 49–59. [Google Scholar] [CrossRef]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Sannakki, S.S.; Rajpurohit, V.S.; Nargund, V.B.; Kulkarni, P. Diagnosis and classification of grape leaf diseases using neural networks. In Proceedings of the 2013 Fourth International Conference on Computing, Communications and Networking Technologies (ICCCNT), Tiruchengode, India, 4–6 July 2013; pp. 1–5. [Google Scholar]
- Torheim, T.; Malinen, E.; Kvaal, K.; Lyng, H.; Indahl, U.G.; Andersen, E.K.F.; Futsaether, C.M. Classification of Dynamic Contrast Enhanced MR Images of Cervical Cancers Using Texture Analysis and Support Vector Machines. IEEE Trans. Med. Imaging 2014, 33, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Tanougast, C.; Dandache, A.; Bouridane, A. Extracted haralick’s texture features and morphological parameters from segmented multispectrale texture bio-images for classification of colon cancer cells. WSEAS Trans. Biol. Biomed. 2011, 8, 39–50. [Google Scholar]
- Rojas, R. Neural Networks: A Systematic Introduction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1996; ISBN 978-3-642-61068-4. [Google Scholar]
- Miller, W.M.; Drouillard, G.P. Multiple Feature Analysis for Machine Vision Grading of Florida Citrus. Appl. Eng. Agric. 2001, 17. [Google Scholar] [CrossRef]
- Durbha, S.S.; King, R.L.; Younan, N.H. Support vector machines regression for retrieval of leaf area index from multiangle imaging spectroradiometer. Remote Sens. Environ. 2007, 107, 348–361. [Google Scholar] [CrossRef]
- Price, J.C. How unique are spectral signatures? Remote Sens. Environ. 1994, 49, 181–186. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.-B.; Dedieu, G. Detection of Flavescence dorée Grapevine Disease Using Unmanned Aerial Vehicle (UAV) Multispectral Imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflectance modeling: the SAIL model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef]
- Ramakrishnan, M. Groundnut leaf disease detection and classification by using back probagation algorithm. In Proceedings of the Communications and Signal Processing (ICCSP), Melmaruvathur, India, 2–4 April 2015; pp. 0964–0968. [Google Scholar]
- Al Bashish, D.; Braik, M.; Bani-Ahmad, S. Detection and Classification of Leaf Diseases using K-means-based Segmentation and Neural-networks-based Classification. Inf. Technol. J. 2011, 10, 267–275. [Google Scholar] [CrossRef]
- Waghmare, H.; Kokare, R.; Dandawate, Y. Detection and classification of diseases of Grape plant using opposite colour Local Binary Pattern feature and machine learning for automated Decision Support System. In Proceedings of the Signal Processing and Integrated Networks (SPIN), Noida, India, 11–12 February 2016; pp. 513–518. [Google Scholar]
- Mirik, M.; Michels, G.J.; Kassymzhanova-Mirik, S.; Elliott, N.C.; Catana, V.; Jones, D.B.; Bowling, R. Using digital image analysis and spectral reflectance data to quantify damage by greenbug (Hemitera: Aphididae) in winter wheat. Comput. Electron. Agric. 2006, 51, 86–98. [Google Scholar] [CrossRef]


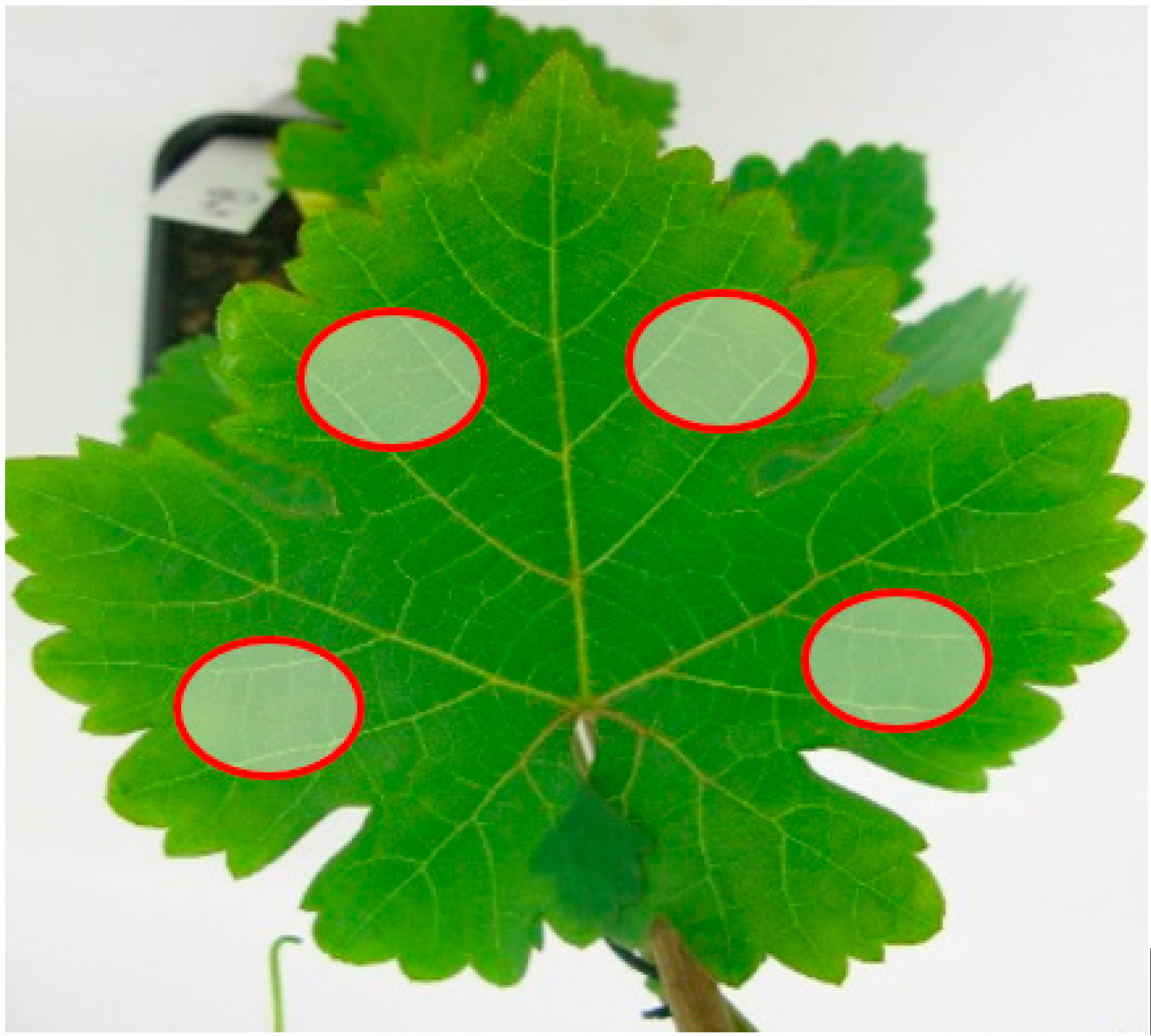


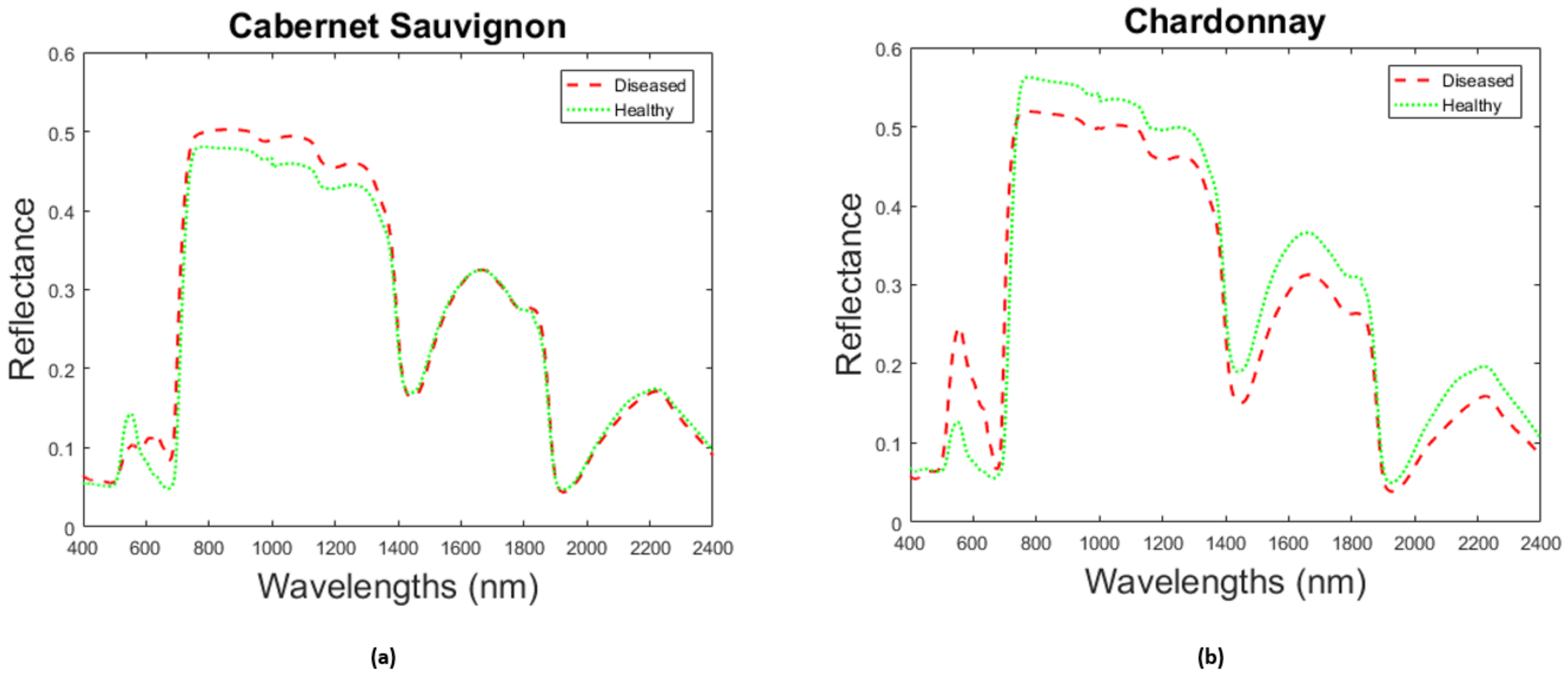
| Parameters | Abbreviation | Unit | Min | Max | Step |
|---|---|---|---|---|---|
| Leaf structure coefficient | N | No dimension | 0.5 | 3.5 | 0.5 |
| Leaf chlorophyll content | Cab | μg/cm2 | 0.00001 | 120 | 0.5 |
| Leaf carotenoid content | Car | μg/cm2 | 0.00001 | 30 | 0.5 |
| Brown pigment content | Cbrown | Arbitrary units | 0.00001 | 0.8 | 0.1 |
| Equivalent water thickness | Cw | cm | 0.00001 | 0.08 | 0.01 |
| Dry matter content | Cm | g/cm2 | 0.00001 | 0.04 | 0.01 |
| Methods | Function | N | Cab | Car | Cbrown | Cw | Cm | Global Accuracy |
|---|---|---|---|---|---|---|---|---|
| L-BFGS-B | RMSE | 0.9706 | 0.8636 | 0.4673 | 0.7293 | 0.9090 | 0.8447 | 0.2193 |
| SLSQP | 0.9694 | 0.7285 | 0.3082 | 0.7293 | 0.9483 | 0.7044 | 0.1060 | |
| TNC | 0.8765 | 0.6731 | 0.4360 | 0.5201 | 0.9380 | 0.5068 | 0.0636 | |
| Nelder-Mead | 0.0238 | 0.0268 | 0.0219 | 0.0290 | 0.0117 | 0.0363 | 0.0000 | |
| L-BFGS-B | SCM | 0.2671 | 0.4826 | 0.2502 | 0.7719 | 0.8405 | 0.5660 | 0.0118 |
| SLSQP | 0.0366 | 0.1166 | 0.0331 | 0.5081 | 0.5190 | 0.1623 | 0.0000 | |
| TNC | 0.2342 | 0.4844 | 0.2418 | 0.5817 | 0.7677 | 0.4984 | 0.0061 | |
| Nelder-Mead | 0.0094 | 0.0366 | 0.0465 | 0.0446 | 0.0483 | 0.0213 | 0.0000 | |
| L-BFGS-B | SAM | 0.7006 | 0.7857 | 0.5597 | 0.8372 | 0.9853 | 0.8953 | 0.2275 |
| SLSQP | 0.9742 | 0.9916 | 0.6186 | 0.9944 | 0.9964 | 0.9872 | 0.5845 | |
| TNC | 0.1746 | 0.3294 | 0.1132 | 0.2992 | 0.7550 | 0.3540 | 0.0005 | |
| Nelder-Mead | 0.0032 | 0.0316 | 0.0266 | 0.0066 | 0.0516 | 0.0198 | 0.0000 | |
| L-BFGS-B | SAM + RMSE | 0.9742 | 0.6538 | 0.5485 | 0.7510 | 0.7604 | 0.7832 | 0.1563 |
| SLSQP | 0.9996 | 0.9952 | 0.6874 | 0.9938 | 0.9986 | 0.9962 | 0.6761 | |
| TNC | 0.7303 | 0.3797 | 0.0687 | 0.1913 | 0.8616 | 0.3387 | 0.0011 | |
| Nelder-Mead | 0.0169 | 0.0284 | 0.0206 | 0.0265 | 0.0391 | 0.0135 | 0.0000 |
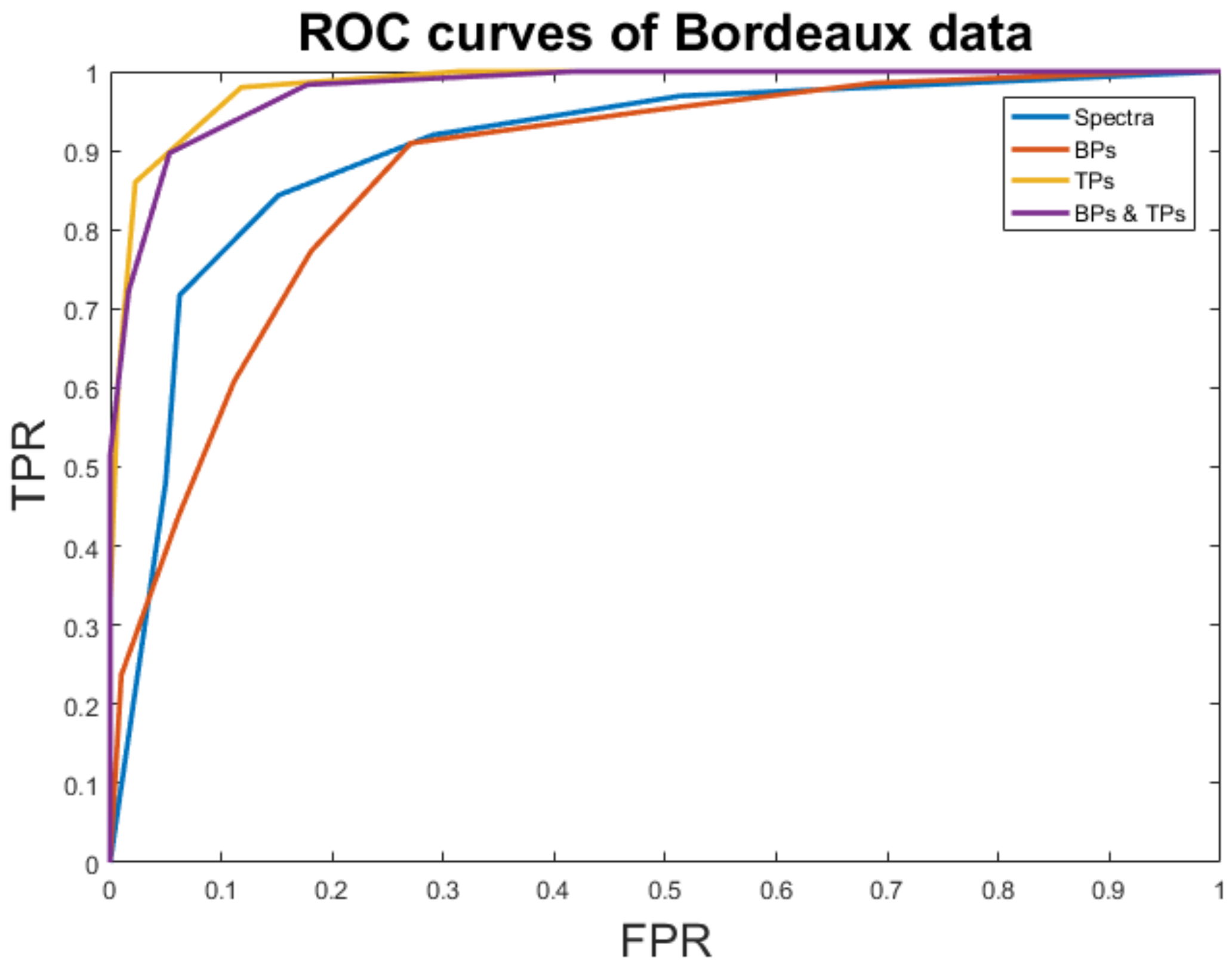
| Accuracy | FM | AUC | |
|---|---|---|---|
| Spectra | 80.62% | 0.78 | 0.90 |
| BPs | 77.50% | 0.67 | 0.82 |
| TPs | 100% | 1 | 0.98 |
| BPs + TPs | 99.37% | 0.99 | 0.97 |
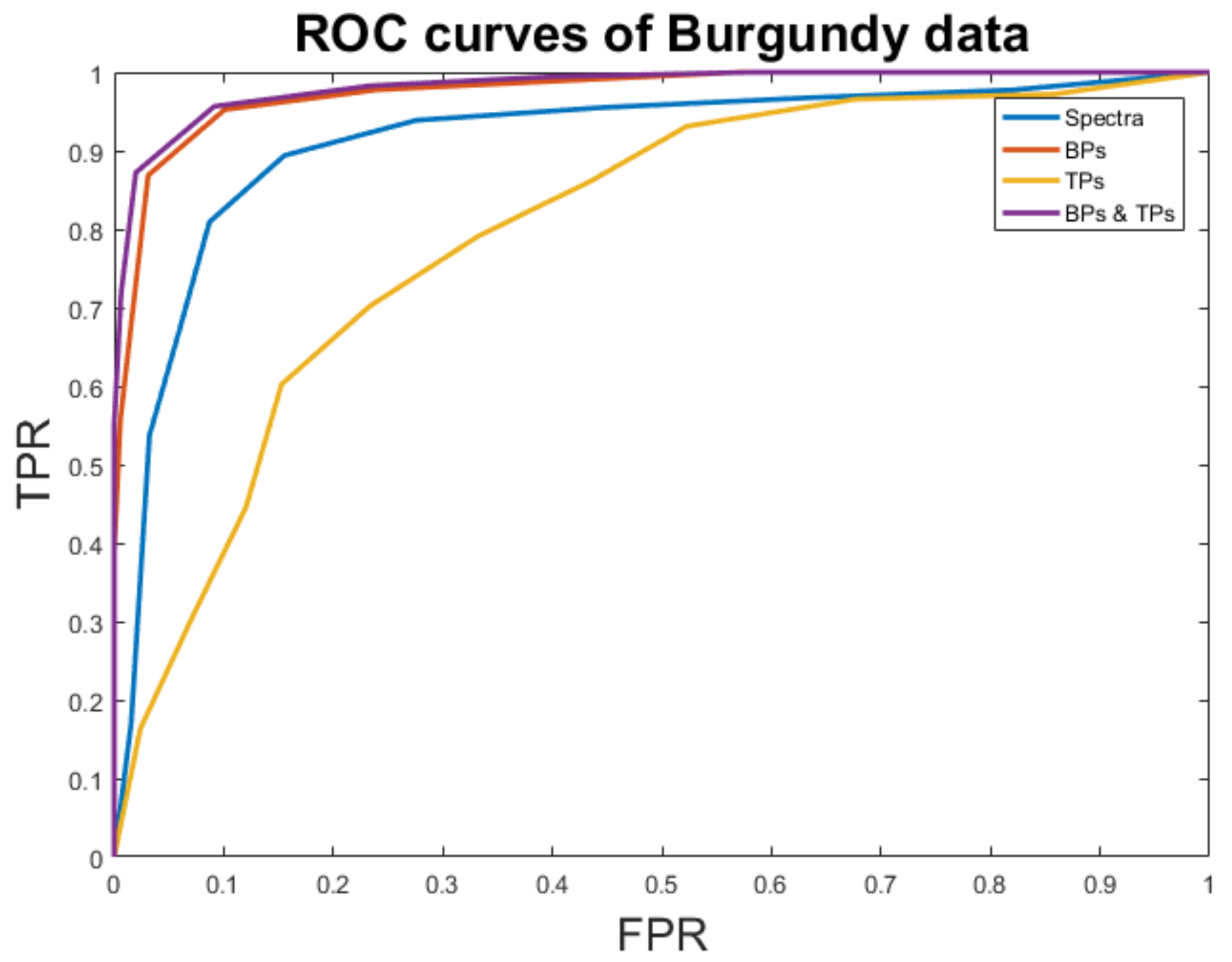
| Accuracy | FM | AUC | |
|---|---|---|---|
| Spectra | 93.63% | 0.92 | 0.91 |
| BPs | 95.45% | 0.95 | 0.97 |
| TPs | 70.45% | 0.69 | 0.80 |
| BPs + TPs | 99.54% | 0.99 | 0.98 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Saddik, H.; Laybros, A.; Billiot, B.; Cointault, F. Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level. Remote Sens. 2018, 10, 618. https://doi.org/10.3390/rs10040618
Al-Saddik H, Laybros A, Billiot B, Cointault F. Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level. Remote Sensing. 2018; 10(4):618. https://doi.org/10.3390/rs10040618
Chicago/Turabian StyleAl-Saddik, Hania, Anthony Laybros, Bastien Billiot, and Frederic Cointault. 2018. "Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level" Remote Sensing 10, no. 4: 618. https://doi.org/10.3390/rs10040618
APA StyleAl-Saddik, H., Laybros, A., Billiot, B., & Cointault, F. (2018). Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level. Remote Sensing, 10(4), 618. https://doi.org/10.3390/rs10040618






