Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques
Abstract
1. Introduction
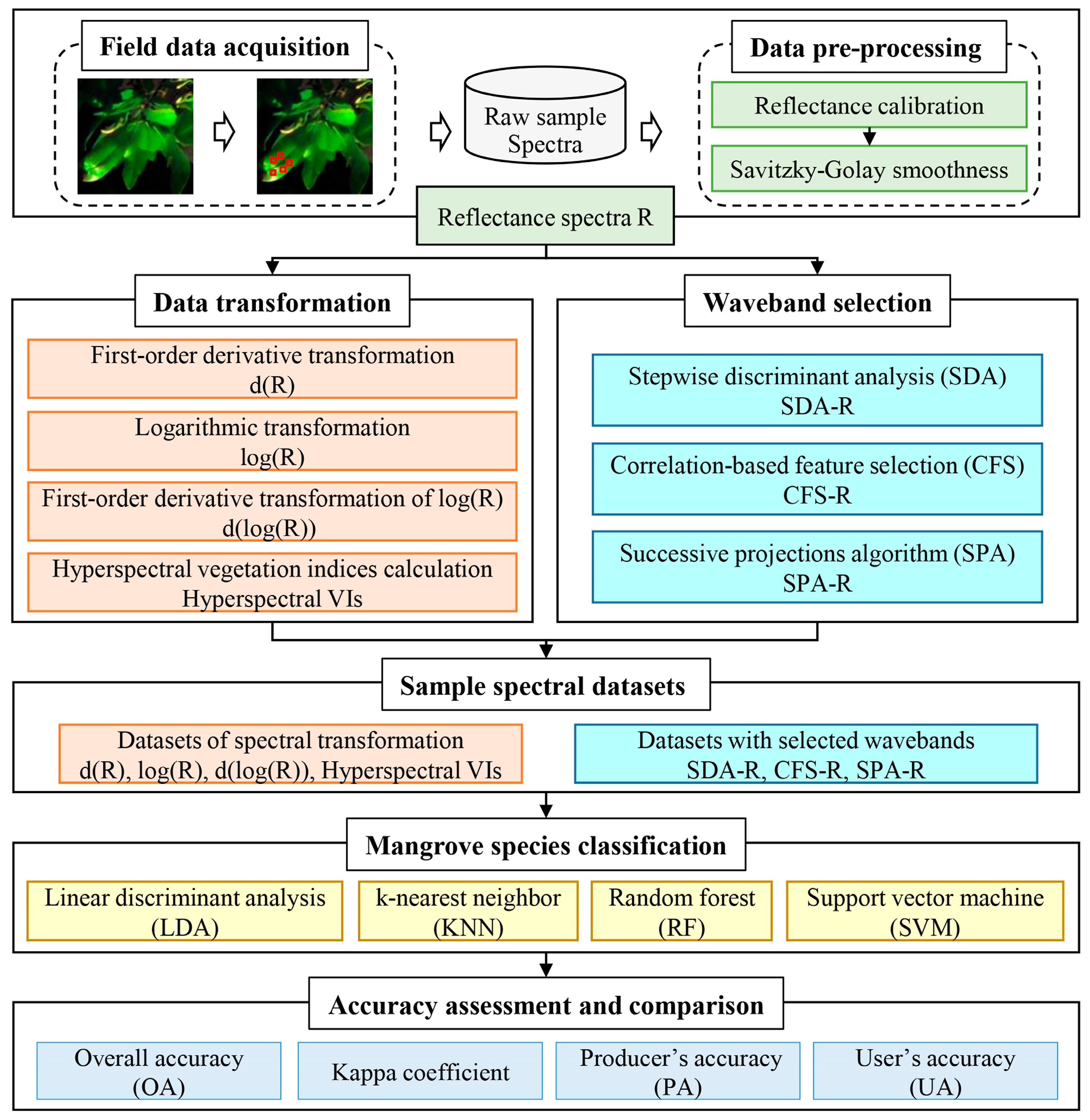
2. Materials and Methods
2.1. Study Area Description
2.2. Data Acquisition and Sample Collection
2.2.1. Field Hyperspectral Measurement
2.2.2. Data Pre-Processing
2.2.3. Sample Spectra Preparation
2.3. Hyperspectral Metrics Extraction
2.3.1. Data Transformation
2.3.2. Vegetation Index Calculation
2.4. Waveband Selection
2.4.1. Stepwise Discriminant Analysis
2.4.2. Correlation-Based Feature Selection
2.4.3. Successive Projections Algorithm
2.5. Mangrove Species Classification
2.5.1. Linear Discriminant Analysis
2.5.2. K-Nearest Neighbor
2.5.3. Random Forest
2.5.4. Support Vector Machine
2.6. Accuracy Assessment
3. Results
3.1. Spectral Properties of Mangrove Species
3.2. Classification Results of the Transformed Datasets
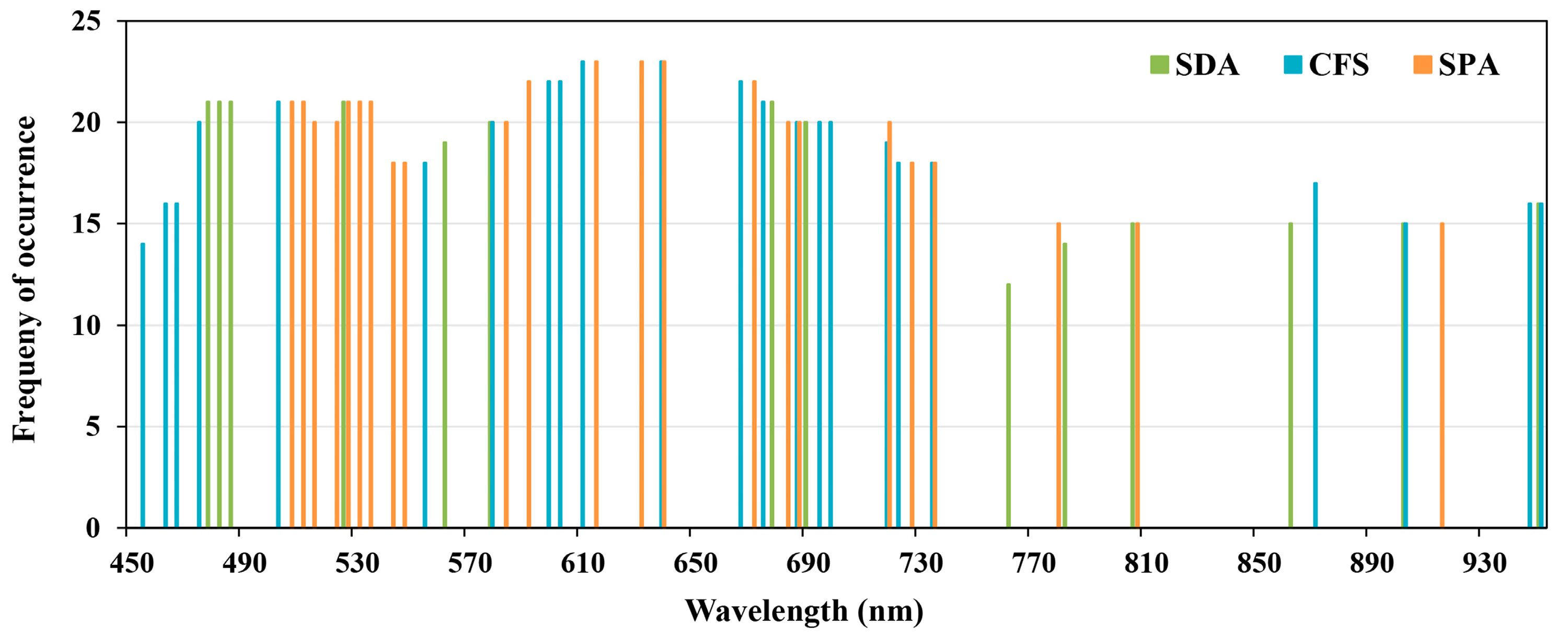
3.3. Optimal Waveband Selection
3.4. Classification Results with the Selected Wavebands
4. Discussion
4.1. Effect of the Optimal Waveband Selection Methods
4.2. Impact of Spectral Datasets With Different Transformations
4.3. Performance of the Machine-Learning Classifiers
4.4. Applicability of Field Close-range Snapshot Hyperspectral Imaging
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuenzer, C.; Bluemel, A.; Gebhardt, S.; Quoc, T.V.; Dech, S. Remote sensing of mangrove ecosystems: A review. Remote Sens. 2011, 3, 878–928. [Google Scholar] [CrossRef]
- Giri, C. Observation and monitoring of mangrove forests using remote sensing: Opportunities and challenges. Remote Sens. 2016, 8, 783. [Google Scholar] [CrossRef]
- Bahuguna, A.; Nayak, S.; Roy, D. Impact of the tsunami and earthquake of 26th December 2004 on the vital coastal ecosystems of the Andaman and Nicobar islands assessed using RESOURCESAT AWiFS data. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 229–237. [Google Scholar] [CrossRef]
- Duke, N.C.; Meynecke, J.-O.; Dittmann, S.; Ellison, A.M.; Anger, K.; Berger, U.; Cannicci, S.; Diele, K.; Ewel, K.C.; Field, C.D. A world without mangroves? Science 2007, 317, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization (FAO). The World’s Mangroves 1980–2005; FAO: Rome, Italy, 2007. [Google Scholar]
- Zhang, H.; Wang, T.; Liu, M.; Jia, M.; Lin, H.; Chu, L.; Devlin, A.T. Potential of combining optical and dual polarimetric SAR data for improving mangrove species discrimination using Rotation Forest. Remote Sens. 2018, 10, 467. [Google Scholar] [CrossRef]
- Pham, T.D.; Bui, D.T.; Yoshino, K.; Le, N.N. Optimized rule-based logistic model tree algorithm for mapping mangrove species using ALOS PALSAR imagery and GIS in the tropical region. Environ. Earth Sci. 2018, 77, 159. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Myint, S.; Wang, S.; Liu, H.; He, Z. Exploring the potential of WorldView-2 red-edge band-based vegetation indices for estimation of mangrove leaf area index with machine learning algorithms. Remote Sens. 2017, 9, 1060. [Google Scholar] [CrossRef]
- Jia, M.; Zhang, Y.; Wang, Z.; Song, K.; Ren, C. Mapping the distribution of mangrove species in the Core Zone of Mai Po Marshes Nature Reserve, Hong Kong, using hyperspectral data and high-resolution data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 226–231. [Google Scholar] [CrossRef]
- Cao, J.; Leng, W.; Liu, K.; Liu, L.; He, Z.; Zhu, Y. Object-based mangrove species classification using unmanned aerial vehicle hyperspectral images and digital surface models. Remote Sens. 2018, 10, 89. [Google Scholar] [CrossRef]
- Ruwaimana, M.; Satyanarayana, B.; Otero, V.; Muslim, A.M.; Syafiq, M.; Ibrahim, S.; Raymaekers, D.; Koedam, N.; Dahdouh-Guebas, F. The advantages of using drones over space-borne imagery in the mapping of mangrove forests. PLoS ONE 2018, 13, e0200288. [Google Scholar] [CrossRef] [PubMed]
- Wannasiri, W.; Nagai, M.; Honda, K.; Santitamnont, P.; Miphokasap, P. Extraction of mangrove biophysical parameters using airborne LiDAR. Remote Sens. 2013, 5, 1787–1808. [Google Scholar] [CrossRef]
- Tong, Q.; Zhang, B.; Zheng, L. Hyperspectral Remote Sensing: Principles, Techniques and Applications; Higher Education Press: Beijing, China, 2006. [Google Scholar]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Kumar, T.; Panigrahy, S.; Kumar, P.; Parihar, J.S. Classification of floristic composition of mangrove forests using hyperspectral data: Case study of Bhitarkanika National Park, India. J. Coast. Conserv. 2013, 17, 121–132. [Google Scholar] [CrossRef]
- Chakravortty, S.; Sinha, D. Analysis of multiple scattering of radiation amongst end members in a mixed pixel of hyperspectral data for identification of mangrove species in a mixed stand. J. Indian Soc. Remote Sens. 2015, 43, 559–569. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Z. Data fusion and classifier ensemble techniques for vegetation mapping in the coastal everglades. Geocarto Int. 2014, 29, 228–243. [Google Scholar] [CrossRef]
- Green, E.P.; Clark, C.D.; Mumby, P.J.; Edwards, A.J.; Ellis, A. Remote sensing techniques for mangrove mapping. Int. J. Remote Sens. 1998, 19, 935–956. [Google Scholar] [CrossRef]
- Jensen, R.; Mausel, P.; Dias, N.; Gonser, R.; Yang, C.; Everitt, J.; Fletcher, R. Spectral analysis of coastal vegetation and land cover using AISA+ hyperspectral data. Geocarto Int. 2007, 22, 17–28. [Google Scholar] [CrossRef]
- Prasad, K.A.; Gnanappazham, L. Multiple statistical approaches for the discrimination of mangrove species of using transformed field and laboratory hyperspectral data. Geocarto Int. 2016, 31, 891–912. [Google Scholar] [CrossRef]
- Li, S.; Tian, Q. Mangrove canopy species discrimination based on spectral features of Geoeye-1 imagery. Spectrosc. Spectr. Anal. 2013, 33, 136–141. (In Chinese) [Google Scholar] [CrossRef]
- Vaiphasa, C.; Ongsomwang, S.; Vaiphasa, T.; Skidmore, A.K. Tropical mangrove species discrimination using hyperspectral data: A laboratory study. Estuar. Coast. Shelf Sci. 2005, 65, 371–379. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M.; Liu, Y.; Flores-Verdugo, F.; Flores-de-Santiago, F. Separating mangrove species and conditions using laboratory hyperspectral data: A case study of a degraded mangrove forest of the Mexican Pacific. Remote Sens. 2014, 6, 11673–11688. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, F.; Liu, Q.; Li, D.; Zhao, D. Analysis of typical mangrove spectral reflectance characteristics. Spectrosc. Spectr. Anal. 2013, 33, 454–458. (In Chinese) [Google Scholar] [CrossRef]
- Yu, X.; Zhao, D.; Zhang, F.; Xiao, Z. Research on mangrove hyperspectrum analysis technology. J. Binzhou Univ. 2006, 22, 53–56. (In Chinese) [Google Scholar]
- Weng, Q.; Lu, C. Research on mangrove canopy apparent spectral reflectance characteristics. J. Fujian For. Sci. Technol. 2006, 33, 14–19. (In Chinese) [Google Scholar]
- Tian, M. Monitoring Winter Wheat Growth Conditions in the Northwest Region of China by Using Hyperspectral Remote Sensing. Ph.D. Dissertation, Northwest A&F University, Shanxi, China, 2017. (In Chinese). [Google Scholar]
- Shang, K.; Zhang, X.; Sun, Y.; Zhang, L.; Wang, S.; Zhuang, Z. Sophisticated vegetation classification based on feature band set using hyperspectral image. Spectrosc. Spectr. Anal. 2015, 35, 1669–1676. [Google Scholar] [CrossRef]
- Xiao, B.; Mao, W.; Liang, X.; Zhang, L.; Han, L. Study on varieties identification of Kentucky bluegrass using hyperspectral imaging and discriminant analysis. Spectrosc. Spectr. Anal. 2012, 32, 1620–1623. (In Chinese) [Google Scholar] [CrossRef]
- Gao, J.; Nuyttens, D.; Lootens, P.; He, Y.; Pieters, J.G. Recognising weeds in a maize crop using a random forest machine-learning algorithm and near-infrared snapshot mosaic hyperspectral imagery. Biosyst. Eng. 2018, 170, 39–50. [Google Scholar] [CrossRef]
- Pu, R.; Gong, P. Hyperspectral Remote Sensing and Its Applications; Higher Education Press: Beijing, China, 2000. [Google Scholar]
- Pu, R.; Gong, P. Hyperspectral remote sensing of vegetation bioparameters. In Advances in Environmental Remote Sensing: Sensors, Algorithms, and Applications; CRC Press: Boca Raton, FL, USA, 2011; pp. 101–142. [Google Scholar]
- Giri, S.; Mukhopadhyay, A.; Hazra, S.; Mukherjee, S.; Roy, D.; Ghosh, S.; Ghosh, T.; Mitra, D. A study on abundance and distribution of mangrove species in Indian Sundarban using remote sensing technique. J. Coast. Conserv. 2014, 18, 359–367. [Google Scholar] [CrossRef]
- Barrett, B.; Nitze, I.; Green, S.; Cawkwell, F. Assessment of multi-temporal, multi-sensor radar and ancillary spatial data for grasslands monitoring in Ireland using machine learning approaches. Remote Sens. Environ. 2014, 152, 109–124. [Google Scholar] [CrossRef]
- Liao, B.; Wei, G.; Zhang, J.; Tang, G.; Lei, Z.; Yang, X. Studies on dynamic development of mangrove communities on Qi’ao Island, Zhuhai. J. South China Agric. Univ. 2008, 29, 59–64. (In Chinese) [Google Scholar]
- Tang, H.; Liu, K.; Zhu, Y.; Wang, S.; Liu, L.; Song, S. Mangrove community classification based on WorldView-2 image and SVM method. Acta Sci. Nat. Univ. Sunyatseni 2015, 54, 102–111. (In Chinese) [Google Scholar]
- Zhou, F.; Kuang, D.; Jian, Y.; Huang, Q.; Ding, L. Primary study on the composition of mangrove community in Qi’ao Island, Zhuhai. Ecol. Sci. 2003, 22, 237–241. (In Chinese) [Google Scholar]
- Liu, K.; Liu, L.; Liu, H.; Li, X.; Wang, S. Exploring the effects of biophysical parameters on the spatial pattern of rare cold damage to mangrove forests. Remote Sens. Environ. 2014, 150, 20–33. [Google Scholar] [CrossRef]
- Ye, X.; Li, J.; Wang, A. Sedimentary environment and its response to anthropogenic impacts in the coastal wetland of the Qi’ao Island, Zhujiang River Estuary. Haiyang Xuebao 2018, 40, 79–89. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Bareth, G. Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS-J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Vaiphasa, C. Consideration of smoothing techniques for hyperspectral remote sensing. ISPRS-J. Photogramm. Remote Sens. 2006, 60, 91–99. [Google Scholar] [CrossRef]
- Zhao, A.; Tang, X.; Zhang, Z.; Liu, J. Optimizing Savitzky-Golay parameters and its smoothing pretreatment for FTIR gas spectra. Spectrosc. Spectr. Anal. 2016, 36, 1340–1344. (In Chinese) [Google Scholar] [CrossRef]
- Lopatin, J.; Fassnacht, F.E.; Kattenborn, T.; Schmidtlein, S. Mapping plant species in mixed grassland communities using close range imaging spectroscopy. Remote Sens. Environ. 2017, 201, 12–23. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Sánchez-Azofeifa, A.; Castro-Esau, K. Intra- and inter-class spectral variability of tropical tree species at La Selva, Costa Rica: Implications for species identification using HYDICE imagery. Remote Sens. Environ. 2006, 105, 129–141. [Google Scholar] [CrossRef]
- Qian, Y.; Yu, J.; Jia, Z.; Yang, F.; Palidan, T. Extraction and analysis of hyperspectral data from typical desert grassland in Xinjiang. Acta Pratacult. Sin. 2013, 22, 157–166. (In Chinese) [Google Scholar]
- Prasad, K.A.; Gnanappazham, L. Species discrimination of mangroves using derivative spectral analysis. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, 2, 45. [Google Scholar] [CrossRef]
- Becker, B.L.; Lusch, D.P.; Qi, J. Identifying optimal spectral bands from in situ measurements of Great Lakes coastal wetlands using second-derivative analysis. Remote Sens. Environ. 2005, 97, 238–248. [Google Scholar] [CrossRef]
- Pearson, R.L.; Miller, L.D. Remote mapping of standing crop biomass for estimation of the productivity of the shortgrass prairie. In Proceedings of the Eighth International Symposium on Remote Sensing of Environment, Ann Arbor, MI, USA, 2–6 October 1972; p. 1355. [Google Scholar]
- Sun, Y.; Gong, H. Quantitative Research on Wetland Plants Based on Hyperspectral Remote Sensing: A Case Study in Honghe Nature Reserve; China Environmental Science Press: Beijing, China, 2015. (In Chinese) [Google Scholar]
- Jain, N.; Ray, S.S.; Singh, J.P.; Panigrahy, S. Use of hyperspectral data to assess the effects of different nitrogen applications on a potato crop. Precis. Agric. 2007, 8, 225–239. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Berjón, A.; López-Lozano, R.; Miller, J.R.; Martín, P.; Cachorro, V.; González, M.R.; De Frutos, A. Assessing vineyard condition with hyperspectral indices: Leaf and canopy reflectance simulation in a row-structured discontinuous canopy. Remote Sens. Environ. 2005, 99, 271–287. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; Paper-A20; National Aeronautics and Space Administration (NASA): Washington, DC, USA, 1974; pp. 309–317.
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Ar, H. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Baret, F.; Guyot, G. Potentials and limits of vegetation indices for LAI and APAR assessment. Remote Sens. Environ. 1991, 35, 161–173. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; Colstoun, E.B.D.; McMurtrey, J.E.I. Estimating corn leaf chlorophyll concentration from leaf and canopy reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Suárez, L.; Morales, F.; Zarco-Tejada, P.J. Assessing structural effects on PRI for stress detection in conifer forests. Remote Sens. Environ. 2011, 115, 2360–2375. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2000, 76, 156–172. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Zarcotejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1491–1507. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, K.; Liu, Y.; Kovacs, J.M.; Flores-Verdugo, F.; Santiago, F.J.F.D. Spectral response to varying levels of leaf pigments collected from a degraded mangrove forest. J. Appl. Remote Sens. 2012, 6, 339–355. [Google Scholar] [CrossRef]
- Maire, G.L.; François, C.; Dufrêne, E. Towards universal broad leaf chlorophyll indices using prospect simulated database and hyperspectral reflectance measurements. Remote Sens. Environ. 2004, 89, 1–28. [Google Scholar] [CrossRef]
- Koedsin, W.; Vaiphasa, C. Discrimination of tropical mangroves at the species level with EO-1 Hyperion data. Remote Sens. 2013, 5, 3562–3582. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Skidmore, A.K.; de Boer, W.F.; Vaiphasa, T. A hyperspectral band selector for plant species discrimination. ISPRS-J. Photogramm. Remote Sens. 2007, 62, 225–235. [Google Scholar] [CrossRef]
- Fung, T.; Yan Ma, H.F.; Siu, W.L. Band selection using hyperspectral data of subtropical tree species. Geocarto Int. 2003, 18, 3–11. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Meer, B.V.D. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Hall, M.A. Feature selection for discrete and numeric class machine learning. In Proceedings of the Seventeenth International Conference on Machine Learning, Stanford, CA, USA, 29 June–2 July 2000; pp. 359–366. [Google Scholar]
- Wollmer, M.; Schuller, B.; Eyben, F.; Rigoll, G. Combining long short-term memory and dynamic Bayesian networks for incremental emotion-sensitive artificial listening. IEEE J. Sel. Top. Signal Process. 2010, 4, 867–881. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Ma, H.; Ji, H.; Lee, W.S. Identification of the citrus greening disease using spectral and textural features based on hyperspectral imaging. Spectrosc. Spectr. Anal. 2016, 36, 2344–2350. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Feret, J.B.; Asner, G.P. Tree species discrimination in tropical forests using airborne imaging spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Prospere, K.; McLaren, K.; Wilson, B. Plant species discrimination in a tropical wetland using in situ hyperspectral data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Kotsiantis, S.B. Supervised machine learning: A review of classification techniques. In Proceedings of the 2007 Conference on Emerging Artificial Intelligence Applications in Computer Engineering: Real Word Ai Systems with Applications in Ehealth, Hci, Information Retrieval and Pervasive Technologies; IOS Press: Amsterdam, The Netherlands, 2007; pp. 3–24. ISBN 978-1-58603-780-2. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS-J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Féret, J.-B.; Asner, G.P. Semi-supervised methods to identify individual crowns of lowland tropical canopy species using imaging spectroscopy and LiDAR. Remote Sens. 2012, 4, 2457–2476. [Google Scholar] [CrossRef]
- Tan, Y.; Xia, W.; Xu, B.; Bai, L. Multi-feature classification approach for high spatial resolution hyperspectral images. J. Indian Soc. Remote Sens. 2017, 46, 9–17. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. Libsvm: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Stehman, S.V. Selecting and interpreting measures of thematic classification accuracy. Remote Sens. Environ. 1997, 62, 77–89. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Yang, L.; Li, X.; Tang, W. The Principle and Method of Analysis of Remote Sensing Application; Science Press: Beijing, China, 2003. [Google Scholar]
- Abdel-Rahman, E.M.; Mutanga, O.; Adam, E.; Ismail, R. Detecting sirex noctilio grey-attacked and lightning-struck pine trees using airborne hyperspectral data, random forest and support vector machines classifiers. ISPRS-J. Photogramm. Remote Sens. 2014, 88, 48–59. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Cohen, J. Weighted kappa: Nominal scale agreement provision for scaled disagreement or partial credit. Psychol. Bull. 1968, 70, 213. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Gong, Z.; Zhao, W.; Fan, L. Identifying typical plant ecological types based on spectral characteristic variables: A case study in Wild Duck Lake wetland, Beijing. Acta Ecol. Sin. 2013, 33, 1172–1185. [Google Scholar] [CrossRef]
- Huang, J.; Sun, Y. Sifting of hyperspectral vegetation indices applying to classification of 5 kinds of plants in Honghe wetlands. Wetl. Sci. 2016, 14, 888–894. (In Chinese) [Google Scholar]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and machine-learning classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.E.; Sousa, W.P. Distinguishing mangrove species with laboratory measurements of hyperspectral leaf reflectance. Int. J. Remote Sens. 2009, 30, 1267–1281. [Google Scholar] [CrossRef]
- Pham, L.T.H.; Brabyn, L. Monitoring mangrove biomass change in Vietnam using spot images and an object-based approach combined with machine learning algorithms. ISPRS-J. Photogramm. Remote Sens. 2017, 128, 86–97. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Li, C.; Li, Z.; Wang, Y.; Feng, H.; Xu, B. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Ishida, T.; Kurihara, J.; Viray, F.A.; Namuco, S.B.; Paringit, E.C.; Perez, G.J.; Takahashi, Y.; Marciano, J.J., Jr. A novel approach for vegetation classification using UAV-based hyperspectral imaging. Comput. Electron. Agric. 2018, 144, 80–85. [Google Scholar] [CrossRef]
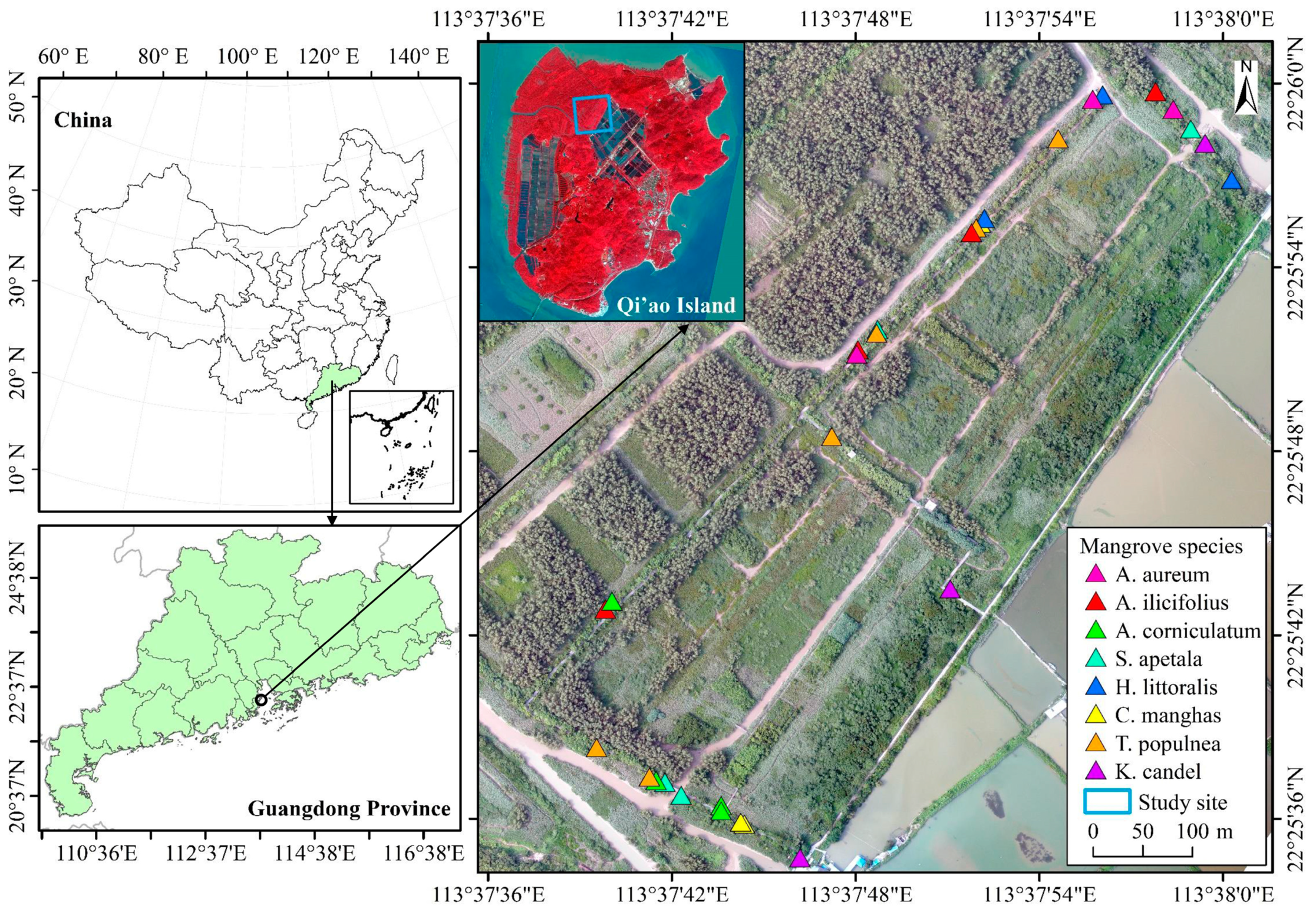

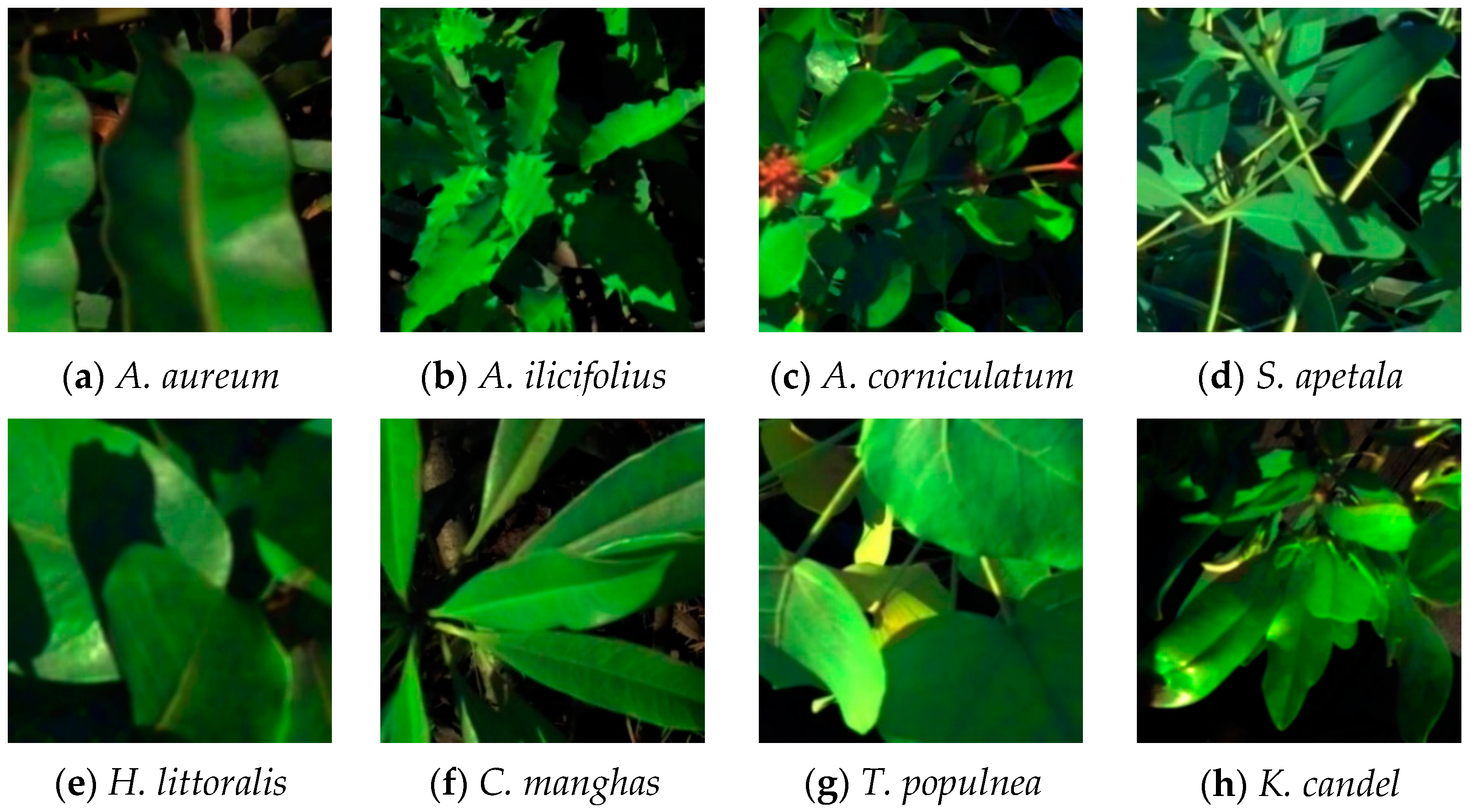

| Mangrove Species Name | Species Code | Functional Group | Ground Survey Points | Samples |
|---|---|---|---|---|
| Acrostichum aureum (A. aureum) | AA | Herbage | 4 | 60 |
| Acanthus ilicifolius (A. ilicifolius) | AI | Frutex | 4 | 60 |
| Aegiceras corniculatum (A. corniculatum) | AC | Arbor | 4 | 60 |
| Sonneratia apetala (S. apetala) | SA | Arbor | 4 | 60 |
| Heritiera littoralis (H. littoralis) | HL | Arbor | 4 | 60 |
| Cerbera manghas (C. manghas) | CM | Arbor | 4 | 60 |
| Therspesia populnea (T. populnea) | TP | Arbor | 6 | 60 |
| Kandelia candel (K. candel) | KC | Arbor/Frutex | 3 | 60 |
| Vegetation Indices | Definition | Commonly Related to | References |
|---|---|---|---|
| Blue Green Pigment Index 2 (BGI 2) | Chlorophylls, Carotenoids | [52] | |
| Normalized Difference Vegetation Index (NDVI) | LAI, biomass, vegetation cover | [53] | |
| Reformed Difference Vegetation Index (RDVI) | LAI | [54,55] | |
| Soil-Adjusted Vegetation Index(SAVI) | Biomass | [56] | |
| Adjusted Transformed Soil-Adjusted VI (ATSAVI) | where X = 0.08, a = 1.22, b = 0.03 | LAI, biomass, soil variation | [57] |
| Modified SAVI (MSAVI) | Biomass, soil variation | [58] | |
| Transformed Chlorophyll Absorption in Reflectance Index (TCARI) | Chlorophylls | [59] | |
| Optimized Soil-Adjusted Vegetation Index (OSAVI) | Biomass, soil variation | [60] | |
| TCARI/OSAVI | Chlorophylls | [59] | |
| Modified Chlorophyll Absorption in Reflectance Index (MCARI) | Chlorophylls | [61] | |
| Modified Chlorophyll Absorption Ratio Index 1 (MCARI1) | LAI | [55] | |
| Modified Chlorophyll Absorption Ratio Index 2 (MCARI 2) | Chlorophylls | [55] | |
| Photochemical Reflectance Index (PRI) | Water content | [40,62] | |
| Triangular Vegetation Index (TVI) | LAI | [63] | |
| Modified Triangular VI 1 (MTVI 1) | LAI | [55] | |
| Modified Triangular VI 2 (MTVI 2) | LAI | [55] | |
| Simple Ratio (SR) | Chlorophylls | [51,64] | |
| Modified Simple Ratio (MSR) | LAI | [55,65] | |
| Zarco Tejada and Miller (ZTM) | Red edge | [66] | |
| Vogelmann Red Edge Index 1 (VOG1) | Red edge | [67] | |
| Vogelmann Red Edge Index 2 (VOG2) | Red edge | [66] | |
| Vogelmann Red Edge Index 3 (VOG3) | Red edge | [66] | |
| Red Edge NDVI (RENDVI) | Red edge | [68] | |
| Vogelmann’s Index (VOI) | Red edge | [67,69] | |
| Modified Simple Ratio of Derivatives (DMSR) | Chlorophylls | [70] |
| Spectral Datasets | Performance Metrics | Classifiers | |||
|---|---|---|---|---|---|
| LDA | KNN | RF | SVM | ||
| Reflectance spectra R | OA(%) | 84.17 (5.22) | 87.50 (5.29) | 87.92 (4.03) | 93.54 (2.68) |
| Kappa | 0.8179 (0.0595) | 0.8562 (0.0607) | 0.8609 (0.0460) | 0.9256 (0.0308) | |
| Hyperspectral VIs | OA(%) | 85.00 (4.79) | 84.58 (5.12) | 85.83 (5.62) | 93.54 (2.07) |
| Kappa | 0.8270 (0.0544) | 0.8223 (0.0585) | 0.8366 (0.0652) | 0.9253 (0.0244) | |
| First-order derivative spectra d(R) | OA(%) | 84.58 (6.45) | 90.63 (3.84) | 92.71 (3.57) | 95.83 (2.20) |
| Kappa | 0.8218 (0.0736) | 0.8917 (0.0437) | 0.9156 (0.0410) | 0.9518 (0.0252) | |
| Log-transformed spectra log(R) | OA(%) | 89.79 (3.73) | 88.54 (4.20) | 86.04 (4.40) | 93.75 (4.39) |
| Kappa | 0.8823 (0.0431) | 0.8681 (0.0478) | 0.8396 (0.0505) | 0.9281 (0.0505) | |
| First-order derivative of log(R) d[log(R)] | OA(%) | 85.83 (7.07) | 95.00 (3.43) | 92.29 (3.55) | 96.46 (2.42) |
| Kappa | 0.8367 (0.0812) | 0.9421 (0.0398) | 0.9112 (0.0408) | 0.9591 (0.0280) | |
| Mangrove Species | Classifiers | |||||||
|---|---|---|---|---|---|---|---|---|
| LDA | KNN | RF | SVM | |||||
| PA(%) | UA(%) | PA(%) | UA(%) | PA(%) | UA(%) | PA(/%) | UA(%) | |
| A. aureum (AA) | 81.86 (20.30) | 92.46 (10.59) | 93.65 (8.40) | 94.31 (10.89) | 93.15 (9.40) | 90.06 (11.67) | 98.75 (3.95) | 100.00 (0.00) |
| A. ilicifolius (AI) | 90.25 (14.26) | 88.65 (15.81) | 94.67 (11.67) | 93.89 (11.25) | 93.83 (10.12) | 89.08 (8.20) | 96.33 (7.77) | 95.76 (10.61) |
| A. corniculatum (AC) | 85.33 (17.26) | 86.11 (16.47) | 93.00 (16.36) | 88.99 (8.19) | 84.71 (14.72) | 85.50 (13.83) | 95.89 (9.45) | 93.00 (12.01) |
| S. apetala (SA) | 98.33 (5.27) | 92.00 (9.76) | 98.57 (4.52) | 94.72 (9.11) | 97.50 (5.27) | 97.50 (5.27) | 98.57 (4.52) | 98.89 (3.51) |
| H. littoralis (HL) | 82.88 (15.53) | 93.99 (7.85) | 89.40 (16.61) | 97.32 (5.66) | 87.64 (9.06) | 92.05 (10.88) | 90.24 (17.83) | 97.14 (6.02) |
| C. manghas (CM) | 69.30 (21.82) | 67.90 (21.61) | 91.65 (9.08) | 98.00 (6.32) | 89.23 (12.82) | 95.00 (8.05) | 93.24 (8.83) | 96.07 (8.66) |
| T. populnea (TP) | 97.32 (5.66) | 90.74 (11.40) | 100.00 (0.00) | 98.75 (3.95) | 98.75 (3.95) | 92.08 (13.95) | 100.00 (0.00) | 97.50 (5.27) |
| K. candel (KC) | 83.17 (19.44) | 87.64 (14.97) | 96.00 (8.43) | 96.67 (10.54) | 94.75 (8.70) | 98.33 (5.27) | 98.00 (6.32) | 96.33 (7.77) |
| OA(%) | 85.83 (7.07) | 95.00 (3.43) | 92.29 (3.55) | 96.46 (2.42) | ||||
| Kappa | 0.8367 (0.0812) | 0.9421 (0.0398) | 0.9112 (0.0408) | 0.9591 (0.0280) | ||||
| Methods | Selected Wavebands (nm) |
|---|---|
| SDA | 14 bands: 478–482, 486, 526, 562, 578, 678, 690, 762, 782, 806, 862, 902, 950 |
| CFS | 23 bands: 454, 462–466, 474, 502, 554, 578, 598–602, 610, 638, 666, 674, 686, 694–698, 718–722, 734, 870, 902, 946–950 |
| SPA | 23 bands: 506–514, 522–534, 542–546, 582, 590, 614, 630, 638, 670, 682–686, 718, 726, 734, 778, 806, 914 |
| Selected Wavebands | Performance Metrics | Classifiers | |||
|---|---|---|---|---|---|
| LDA | KNN | RF | SVM | ||
| 125 wavebands | OA(%) | 84.17 (5.22) | 87.50 (5.29) | 87.92 (4.03) | 93.54 (2.68) |
| Kappa | 0.8179 (0.0595) | 0.8562 (0.0607) | 0.8609 (0.0460) | 0.9256 (0.0308) | |
| 14 wavebands (SDA) | OA(%) | 74.79 (7.04) | 86.46 (5.31) | 86.04 (5.38) | 91.46 (4.10) |
| Kappa | 0.7104 (0.0812) | 0.8442 (0.0609) | 0.8394 (0.0617) | 0.9017 (0.0471) | |
| 23 wavebands (CFS) | OA(%) | 81.67 (7.47) | 87.92 (4.59) | 87.71 (4.33) | 92.29 (2.79) |
| Kappa | 0.7896 (0.0852) | 0.8608 (0.0527) | 0.8586 (0.0498) | 0.9112 (0.0321) | |
| 23 wavebands (SPA) | OA(%) | 83.75 (3.51) | 86.88 (4.29) | 84.58 (4.93) | 93.13 (1.98) |
| Kappa | 0.8133 (0.0400) | 0.8490 (0.0489) | 0.8226 (0.0565) | 0.9208 (0.0227) | |
| Mangrove Species | Classifiers | |||||||
|---|---|---|---|---|---|---|---|---|
| LDA | KNN | RF | SVM | |||||
| PA(%) | UA(%) | PA(%) | UA(%) | PA(%) | UA(%) | PA(%) | UA(%) | |
| A. aureum (AA) | 84.88 (12.70) | 77.84 (15.44) | 93.99 (7.85) | 76.31 (16.02) | 86.37 (11.50) | 77.55 (14.20) | 93.89 (8.11) | 84.43 (13.84) |
| A. ilicifolius (AI) | 87.75 (14.55) | 92.14 (15.95) | 84.57 (15.14) | 82.17 (15.95) | 75.81 (16.34) | 77.89 (11.32) | 90.40 (14.09) | 89.87 (9.47) |
| A. corniculatum (AC) | 96.67 (10.54) | 69.55 (15.29) | 72.08 (16.43) | 90.50 (12.96) | 71.33 (20.25) | 80.75 (18.93) | 80.71 (15.02) | 96.57 (7.35) |
| S. apetala (SA) | 76.62 (21.44) | 98.33 (5.27) | 98.33 (5.27) | 93.57 (8.33) | 97.08 (6.23) | 96.90 (6.55) | 100.00 (0.00) | 98.57 (4.52) |
| H. littoralis (HL) | 83.68 (17.68) | 80.25 (14.68) | 90.07 (14.31) | 87.57 (13.23) | 84.95 (17.91) | 79.92 (13.45) | 94.57 (12.95) | 88.50 (10.64) |
| C. manghas (CM) | 59.79 (31.60) | 73.40 (28.72) | 80.90 (22.09) | 90.07 (14.31) | 83.57 (22.23) | 87.64 (12.49) | 91.67 (21.15) | 96.14 (9.23) |
| T. populnea (TP) | 98.57 (4.52) | 98.57 (4.52) | 96.67 (7.03) | 90.05 (11.09) | 95.42 (7.47) | 93.17 (9.90) | 100.00 (0.00) | 98.33 (5.27) |
| K. candel (KC) | 83.99 (15.28) | 90.07 (14.31) | 75.92 (13.46) | 94.25 (9.72) | 82.32 (16.24) | 92.98 (9.53) | 89.17 (14.72) | 97.50 (7.91) |
| OA(%) | 83.75 (3.51) | 86.88 (4.29) | 84.58 (4.93) | 93.13 (1.98) | ||||
| Kappa | 0.8133 (0.0400) | 0.8490 (0.0489) | 0.8226 (0.0565) | 0.9208 (0.0227) | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Liu, K.; Liu, L.; Zhu, Y.; Li, J.; He, Z. Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques. Remote Sens. 2018, 10, 2047. https://doi.org/10.3390/rs10122047
Cao J, Liu K, Liu L, Zhu Y, Li J, He Z. Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques. Remote Sensing. 2018; 10(12):2047. https://doi.org/10.3390/rs10122047
Chicago/Turabian StyleCao, Jingjing, Kai Liu, Lin Liu, Yuanhui Zhu, Jun Li, and Zhi He. 2018. "Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques" Remote Sensing 10, no. 12: 2047. https://doi.org/10.3390/rs10122047
APA StyleCao, J., Liu, K., Liu, L., Zhu, Y., Li, J., & He, Z. (2018). Identifying Mangrove Species Using Field Close-Range Snapshot Hyperspectral Imaging and Machine-Learning Techniques. Remote Sensing, 10(12), 2047. https://doi.org/10.3390/rs10122047










