Abstract
Heavy metals are natural constituents of the environment, but indiscriminate use for human purposes has altered their geochemical cycles and biochemical balance. This results in excess release of heavy metals such as cadmium, copper, lead, nickel, zinc etc. into natural resources like the soil and aquatic environments. Prolonged exposure and higher accumulation of such heavy metals can have deleterious health effects on human life and aquatic biota. The role of microorganisms and plants in biotransformation of heavy metals into nontoxic forms is well-documented, and understanding the molecular mechanism of metal accumulation has numerous biotechnological implications for bioremediation of metal-contaminated sites. In view of this, the present review investigates the abilities of microorganisms and plants in terms of tolerance and degradation of heavy metals. Also, advances in bioremediation technologies and strategies to explore these immense and valuable biological resources for bioremediation are discussed. An assessment of the current status of technology deployment and suggestions for future bioremediation research has also been included. Finally, there is a discussion of the genetic and molecular basis of metal tolerance in microbes, with special reference to the genomics of heavy metal accumulator plants and the identification of functional genes involved in tolerance and detoxification.
1. Introduction
With the growth of industry, there has been a considerable increase in the discharge of industrial waste to the environment, chiefly soil and water, which has led to the accumulation of heavy metals, especially in urban areas. Slow depletion of heavy metals also takes place through leaching, plant uptake, erosion and deflation. The indiscriminate release of heavy metals into the soil and waters is a major health concern worldwide, as they cannot be broken down to non-toxic forms and therefore have long-lasting effects on the ecosystem. Many of them are toxic even at very low concentrations; arsenic, cadmium, chromium, copper, lead, mercury, nickel, selenium, silver, zinc etc. are not only cytotoxic but also carcinogenic and mutagenic in nature [1]. Some metals are required by plants in very small amounts for their growth and optimum performance. However, the increasing concentration of several metals in soil and waters due to industrial revolution has created an alarming situation for human life and aquatic biota. This is evident from various reports citing harmful effects of heavy metals on human health (Table 1).
In order to make the environment healthier for human beings, contaminated water bodies and land need to be rectified to make them free from heavy metals and trace elements. There are several techniques to remove these heavy metals, including chemical precipitation, oxidation or reduction, filtration, ion-exchange, reverse osmosis, membrane technology, evaporation and electrochemical treatment. But most of these techniques become ineffective when the concentrations of heavy metals are less than 100 mg/L [2]. Most heavy metal salts are water-soluble and get dissolved in wastewater, which means they cannot be separated by physical separation methods [3]. Additionally, physico-chemical methods are ineffective or expensive when the concentration of heavy metals is very low. Alternately, biological methods like biosorption and/or bioaccumulation for removal of heavy metals may be an attractive alternative to physico-chemical methods [4]. Use of microorganisms and plants for remediation purposes is thus a possible solution for heavy metal pollution since it includes sustainable remediation technologies to rectify and re-establish the natural condition of soil. However, introduction of heavy metals into the soil causes considerable modification of the microbial community, despite their vital importance for the growth of microorganisms at relatively low concentrations [5]. The modification of the microbial make up is mainly brought about by exerting an inhibitory action through blockage of essential functional groups, displacement of essential metal ions or modification of active conformations of biological molecules [6,7]. The response of microbial communities to heavy metals depends on the concentration and availability of heavy metals and is a complex process which is controlled by multiple factors, such as type of metal, the nature of the medium, and microbial species [8].

Table 1.
Toxic effect of some heavy metals on human health.
| Heavy Metal | EPA Regulatory Limit (ppm) [9] | Toxic Effects | Ref. |
|---|---|---|---|
| Ag | 0.10 | Exposure may cause skin and other body tissues to turn gray or blue-gray, breathing problems, lung and throat irritation and stomach pain. | [10] |
| As | 0.01 | Affects essential cellular processes such asoxidative phosphorylation and ATP synthesis | [11] |
| Ba | 2.0 | Cause cardiac arrhythmias, respiratory failure, gastrointestinal dysfunction, muscle twitching and elevated blood pressure | [12] |
| Cd | 5.0 | Carcinogenic, mutagenic, endocrine disruptor, lung damage and fragile bones, affects calcium regulation in biological systems | [1,13] |
| Cr | 0.1 | Hair loss | [1] |
| Cu | 1.3 | Brain and kidney damage, elevated levels result in liver cirrhosis and chronic anemia, stomach and intestine irritation | [1,14] |
| Hg | 2.0 | Autoimmune diseases, depression, drowsiness, fatigue, hair loss, insomnia, loss of memory, restlessness, disturbance of vision, tremors, temper outbursts, brain damage, lung and kidney failure | [15,16,17] |
| Ni | 0.2 (WHO permissible limit) | Allergic skin diseases such as itching, cancer of the lungs, nose, sinuses, throat through continuous inhalation, immunotoxic, neurotoxic, genotoxic, affects fertility, hair loss | [1,18,19,20] |
| Pb | 15 | Excess exposure in children causes impaired development, reduced intelligence, short-term memory loss, disabilities in learning and coordination problems, risk of cardiovascular disease | [1,14,21] |
| Se | 50 | Dietary exposure of around 300 µg/day affects endocrine function, impairment of natural killer cells activity, hepatotoxicity and gastrointestinal disturbaces | [22] |
| Zn | 0.5 | Dizziness, fatigue etc. | [23] |
Bioremediation is an innovative and promising technology available for removal of heavy metals and recovery of the heavy metals in polluted water and lands. Since microorganisms have developed various strategies for their survival in heavy metal-polluted habitats, these organisms are known to develop and adopt different detoxifying mechanisms such as biosorption, bioaccumulation, biotransformation and biomineralization, which can be exploited for bioremediation either ex situ or in situ [24,25,26,27]. A global survey to examine the use of bioremediation technologies for addressing the environmental problems was carried out by Elekwachi et al. [28]. They found that despite aspirations from respondents to apply bioremediation techniques, it should not become the current practice. Developed economies made higher use of low-cost in situ bioremediation technologies such as monitored natural attenuation, while their developing counterparts appeared to focus on occasionally more expensive ex situ technologies. Despite significant investment and widespread availability of online resources, their limited use underlines the need to explore improved training and development of more user-friendly resources. There are many reports about biodegradation and bioremediation strategies being utilized by bacteria or plant species [29,30,31,32], but so far none of these investigations suggest possible drivers in the global use of the said techniques [28]. Among the preferred methods for treatment of contaminated areas, 51% of the respondents preferred environment friendly approaches, including microbial remediation (35%) and phytoremediation (16%) [33,34].
Microorganisms uptake heavy metals actively (bioaccumulation) and/or passively (adsorption) [35]. The microbial cell walls, which mainly consist of polysaccharides, lipids and proteins, offer many functional groups that can bind heavy metal ions, and these include carboxylate, hydroxyl, amino and phosphate groups [36]. Among various microbe-mediated methods, the biosorption process seems to be more feasible for large scale application compared to the bioaccumulation process, because microbes will require addition of nutrients for their active uptake of heavy metals, which increases the biological oxygen demand or chemical oxygen demand in the waste. Further, it is very difficult to maintain a healthy population of microorganisms due to heavy metal toxicity and other environmental factors [37,38]. Fungi of the genera Penicillium, Aspergillus and Rhizopus have been studied extensively as potential microbial agents for the removal of heavy metals from aqueous solutions [39,40]. Xiao et al. reported a novel technology for obtaining highly efficient biosorbents from endophytes, a hyperaccumulator, which is more convenient than the traditional method of obtaining biosorbents [41]. Sun et al. evaluated the genetic diversity of endophytic bacteria from the copper-tolerant species of Elshotzia apliendens and Commelina communis, reporting increased dry weights of roots and aboveground tissues compared to uninoculated plants [42]. Further, they also reported significant amounts of (ranging from 63% to 125%) Cu content in inoculated plants compared to uninoculated ones.
In view of such reports on the use of microorganisms and plants for removal of heavy metals from contaminated sites, the present review focuses on recent developments in bioremediation techniques. Additionally, new approaches such as the designer plant approach and rhizosphere modification to achieve the goal of bioremediation in a cheaper and safer way are also discussed.
2. Sources of Heavy Metal in the Environment
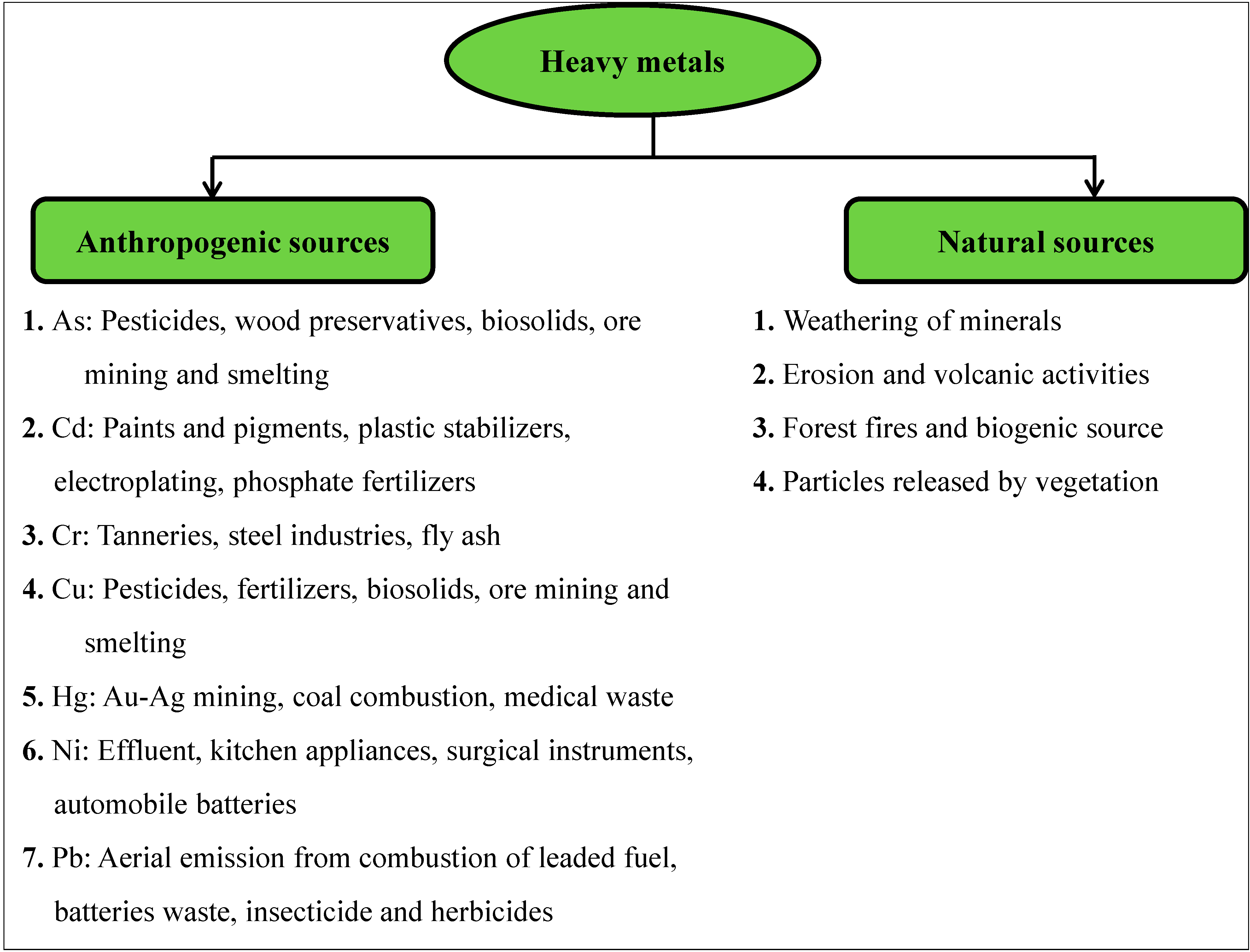
Heavy metals occur naturally in the environment from pedogenetic processes of weathering of parent materials and also through anthropogenic sources (Figure 1). The most significant natural sources are weathering of minerals, erosion and volcanic activity, while the anthropogenic sources depend upon human activities such as mining, smelting, electroplating, use of pesticides and phosphate fertilizer discharge, as well biosolids (e.g., livestock manures, composts, and municipal sewage sludge), atmospheric deposition, etc. [14,43,44,45,46,47]. The disturbance of nature’s slowly occurring geochemical cycle of metals by man results in accumulation of one or more of heavy metals in the soil and waters, and above defined levels, this is enough to cause risk to human health, plants, animals and aquatic biota [48]. The heavy metals essentially become contaminants in the soil and water environment because of their excess generation by natural and man-made activities, transfer from mines to other locations where higher exposure to humans occurs, discharge of high concentration of metal waste through industries, and greater bioavailability.
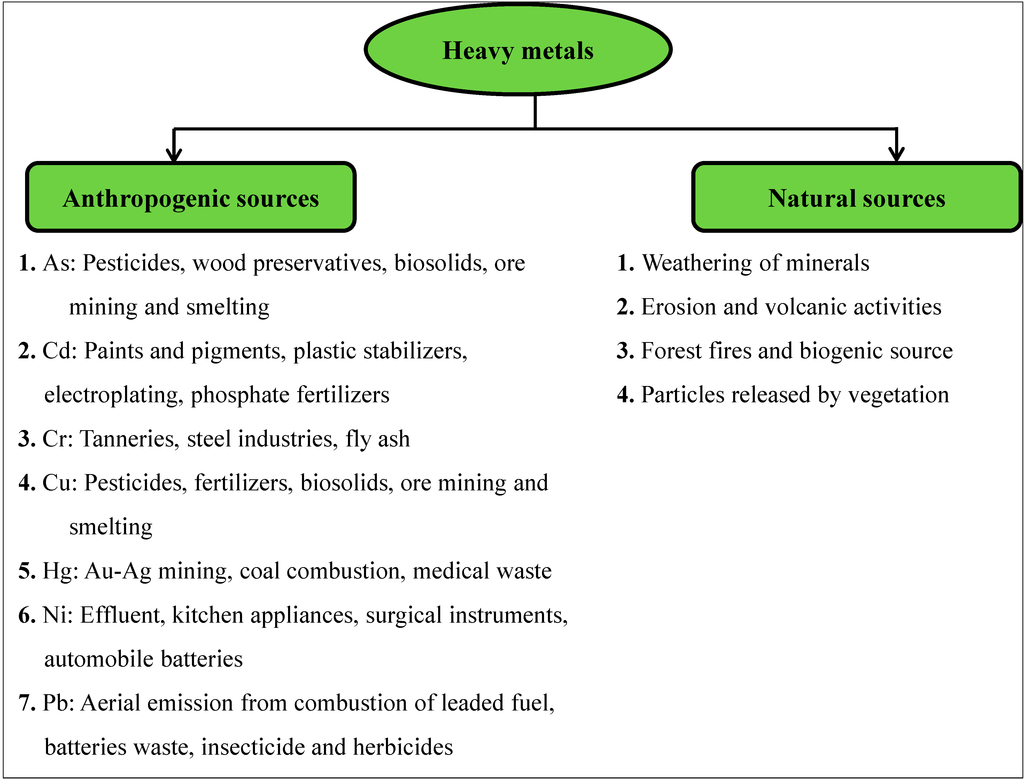
Figure 1.
Sources of heavy metals in the environment.
Figure 1.
Sources of heavy metals in the environment.
A mass balance of heavy metals in the soil environment can be expressed using the formula [49]:
where M is the heavy metal, p is the parent material, a is atmospheric deposition, f is fertilizer source, ag is agrochemical source, ow is organic waste source, ip is inorganic pollutant, cr is crop removal and l is losses by leaching, volatilization and other processes. It is estimated that emission of several heavy metals in atmosphere from anthropogenic sources is one to three orders higher than natural sources [50].
3. Bioremediation: Introducing Microbe Based Clean Up System
Remediation of environment niches such as soil, sediments and water amended with heavy metals can be achieved through biologically encoded changes in the oxidation state. Bioremediation is the microbe-mediated process for clearance or immobilization of the contaminants, including all possible toxins like hydrocarbons, agrochemicals and other organic toxicants. But for inorganic toxic compounds such as heavy metals, microbes are unable to simplify them into harmless compounds, and they should be used according to their specialization for the type of contaminants. Thus the bioremediation strategy for heavy metals depends on the active metabolizing capabilities of microorganisms. Several microorganisms are known to require varying amounts of heavy metals as essential micronutrients for growth and development. For example, Fe3+ is essentially required by all bacteria while Fe2+ is important for anaerobic bacteria [51]. However, the adsorption capacity depends on microbial total biomass and geochemistry of the system. Some oxyanions of metals do not interact with microbes, and their bioremediation is based on their catalyzed redox conversion to insoluble forms. These reduction or oxidation reactions take place due to enzymatic activity and biomass concentration of microbes. Microorganisms have a great deal of undiscovered and unexplored potential for remediation of soil pollutants and increasing the production of agricultural crops with low input. Selection of rhizospheric microbes should be done based on an understanding of mechanisms involved in the adsorption and mobilization of heavy metals and trace elements in the soil to restore soil health. Microorganisms as metal accumulators possess an inherent novel remediation property for toxic metals in the soil. The study of genetics of such metal accumulator microorganisms can help us to transfer the traits in the microbes that are missing through the development of microarrays, which result in differentially expressed microbe genes.
Detoxification and rehabilitation of contaminated soil with the use of microbes has emerged as the most safe, easy and effective technology. Native soil microorganisms have been explored and harnessed for their ability to remove or detoxify toxic products released due to human activities in the environment viz. mining of ores, oil and gas extraction, pesticides, pigments, plastic, organic solvents, fuel and industrial processes [52]. But the lack of information on the cellular responses of microbes towards utilization and interaction with trace element and heavy metal pollutants restricts their successful execution. Studies have been carried out to promote the use of modified microbes designed especially to increase sensitivity towards toxic metals. The biochemical route for the redistribution of the organic pollutant in the soil starts from various physical, chemical and biological processes resulting in adsorption by soil particles and root tissues, volatilization, transport through water and air, microbial degradation and leaching, etc.
3.1. Mechanisms of Bioremediation
Microorganisms are omnipresent that dominate in heavy metal-contaminated soil and can easily convert heavy metals into non-toxic forms. In bioremediation processes, microorganisms mineralize the organic contaminants to end-products such as carbon dioxide and water, or to metabolic intermediates which are used as primary substrates for cell growth. Microorganisms are capable of two-way defense viz. production of degradative enzymes for the target pollutants as well as resistance to relevant heavy metals. Different mechanisms of bioremediation are known, including biosorption, metal-microbe interactions, bioaccumulation, biomineralisation, biotransformation and bioleaching. Microorganisms remove the heavy metals from soil by using chemicals for their growth and development. They are capable of dissolving metals and reducing or oxidizing transition metals. Different methods by which microbes restore the environment are oxidizing, binding, immobilizing, volatizing and transformation of heavy metals. Bioremediation can be made successful in a particular location by the designer microbe approach, and by understanding the mechanism controlling growth and activity of microorganisms in the contaminated sites, their metabolic capabilities and their response to environmental changes. Many contaminants are organic solvents which disrupt membranes, but cells may develop defense mechanisms including formation of outer cell-membrane-protective material, often hydrophobic or solvent efflux pumps [53]. For instance, plasmid-encoded and energy-dependent metal efflux systems involving ATPases and chemiosmotic ion/proton pumps are reported for As, Cr and Cd resistance in many bacteria [54].
3.2. Bioremediation by Adsorption
Heavy metals can be biosorbed by microbes at binding sites present in cellular structure without the involvement of energy. Among the various reactive compounds associated with bacterial cell walls, the extracellular polymeric substances are of particular importance and are well known to have significant effects on acid-base properties and metal adsorption [55]. Studies on the metal binding behavior of extracellular polymeric substances (EPS) revealed a great ability to complex heavy metals through various mechanisms, which include proton exchange and micro-precipitation of metals [56,57]. Recent studies have characterized and quantified the proton and adsorbed metals on bacterial cells and EPS free cells in order to determine the relative importance of EPS molecules in metal removal [58]. Bioremediation research and practice are still hampered in the current scenario due to an incomplete understanding of genetics and genome level characteristics of the organisms used in metal adsorption, the metabolic pathway and their kinetics. This results in an inability to model and predict the process behavior and develop a natural bioremediation process in the field [29,30,32,59,60].
3.3. Bioremediation by Physio-Bio-Chemical Mechanism
Biosorption is the process which involves higher affinity of a biosorbent towards sorbate (metal ions), continued until equilibrium is established between the two components [19]. Saccharomyces cerevisiae acts as a biosorbent for the removal of Zn (II) and Cd (II) through the ion exchange mechanism [61,62]. Cunninghamella elegans emerged as a promising sorbent against heavy metals released by textile wastewater [63]. Heavy metal degradation involves energy for the cell metabolic cycle. The combined active and passive modes of toxic metal bioremediation can be called bioaccumulation [64]. Fungi have emerged as potential biocatalysts to access heavy metals and transform them into less toxic compounds [65]. Some fungi such as Klebsiella oxytoca, Allescheriella sp., Stachybotrys sp., Phlebia sp. Pleurotus pulmonarius, Botryosphaeria rhodina have metal binding potential [66]. Pb (II) contaminated soils can be biodegraded by fungal species like Aspergillus parasitica and Cephalosporium aphidicola with biosorption process [67,68]. Hg resistant fungi (Hymenoscyphus ericae, Neocosmospora vasinfecta and Verticillum terrestre) were able to biotransform a Hg (II) state to a nontoxic state [69]. Many of the contaminants are hydrophobic, and these substances appear to be taken up by microbes through the secretion of some biosurfactant and direct cell-contaminant association. Biosurfactants form stronger ionic bonds with metals and form complexes before being desorbed from soil matrix to water phase due to low interfacial tension [70].
Bioremediation may also involve aerobic or anaerobic microbial activities. Aerobic degradation often involves introduction of oxygen atoms into the reactions mediated by monooxygenases, dioxygenases, hydroxylases, oxidative dehalogenases, or chemically reactive oxygen atoms generated by enzymes such as ligninases or peroxidases. Anaerobic degradations of contaminants involve initial activation reactions followed by oxidative catabolism mediated by anoxic electron acceptors. The technique used to reduce the mobilization of heavy metals from contaminated sites by changing the physical or chemical state of the toxic metals is called immobilization. Solidification treatment involves mixing of chemical agents at the contaminated sites or precipitation of hydroxides [71]. Microbes mobilize the heavy metals from the contaminated sites by leaching, chelation, methylation and redox tansformation of toxic metals. Heavy metals can never be destroyed completely, but the process transforms their oxidation state or organic complex, so that they become water-soluble, less toxic and precipitated [52]. Microorganisms use heavy metals and trace elements as terminal electron acceptors or reduce them through the detoxification mechanism, used for the removal of metals from the contaminated environment. Microorganisms remove heavy metals through the mechanisms which they employ to derive energy from metals redox reactions, to deal with toxic metal through enzymatic and non-enzymatic processes.
Two main mechanisms for development of resistance in bacteria are detoxification (transformation of the toxic metal state and making it unavailable) and active efflux pumping of the toxic metal from cells [72]. The basic redox (oxidation and reduction) reaction takes place in the soil between toxic metals and microorganisms; microorganisms act as an oxidizing agent for heavy metals and cause them to lose electrons, which are accepted by alternative electron acceptors (nitrate, sulphate and ferric oxides). In aerobic conditions, oxygen acts as an electron acceptor, while in anaerobic conditions microbes oxidize organic contaminants by reducing electron acceptors. The microorganism takes energy for growth by oxidizing the organic compound with Fe (III) or Mn (IV) as an electron acceptor [73]. Anaerobic degradation of organic contamination is stimulated with the higher availability of Fe (III) for microbial reduction [74,75]. Metals being used as terminal electron acceptors is called dissimilatory metal reduction [76]. Biodegradation of chlorines from contaminants takes place through reductive dechlorination, where contaminants as chlorinated solvents acts as an electron acceptors in respiration. Microorganisms reduce the state of metals and change their solubility, like the Geobaccter species, and reduce the Uranium soluble state (U6+) to insoluble state (U4+) [77].
Different defense systems (exclusion, compartmentalization, complex formation and synthesis of binding protein and peptides) reduce the stress developed by toxic metals [78]. Heavy metal accumulation by microorganisms can be studied by the expression of metal binding protein and peptides (phytochelatins and metallothionein) [79]. These metal binding protein transcription factors are known to mediate in hormone and redox signaling process in the context of toxic metal (Cd, Zn, Hg, Cu, Au, Ag, Co, Ni and Bi) exposure [80]. Synechococcus sp. (cynobacterial strains) has been reported with the expression of the smtA gene and production of metal-binding protein [81]. Ralstonia eutropha has been genetically modified to express mouse metallothionein on the cell surface and decrease the toxic effect of the Cd (II) in the contaminated sites [82]. Expression of different proteins and peptides by the Escherichia coli regulates the range of accumulation of cadmium [83]. Co-expression of precursor glutathione (GSH) along with phytochelatins (PC) resulted in the 10 fold increase in PC that finally increased cadmium accumulation twofold [84]. Natural resistant pathways for heavy metals (Hg and Ar) in microorganisms have been regulated by metalloregulatory protein [85].
3.4. Molecular Mechanisms Involved in Bioremediation Process
Various mechanisms involved in the removal of heavy metals by microorganisms are known. In a genetically engineered bacterium Deinococcus geothemalis, Hg reduction has been reported at high temperatures due to the expression of mer operon from E. coli coded for Hg2+ reduction [86]. Mercury resistant bacteria Cupriavidus metallidurans strain MSR33 was modified genetically by introducing a pTP6 plasmid that provided genes (merB and merG) regulating Hg biodegradation along with the synthesis of organomercurial lyase protein (MerB) and mercuric reductase (MerA) [87]. Modification of Pseudomonas strain with the pMR68 plasmid with novel genes (mer) made that strain resistant to mercury [88]. Two different mechanisms for Hg degradation by bacteria (Klebsiella pneumonia M426) are mercury volatilization by reduction of Hg (II) to Hg (0) and mercury precipitation as insoluble Hg due to volatile thiol (H2S) [89]. Genetic engineering of Deinococcus radiodurans (radiation resistant bacterium) which naturally reduces Cr (IV) to Cr (III) has been done for complete toluene (fuel hydrocarbon) degradation by cloned genes of tod and xyl operons of Pseudomonas putida [90]. Microbial metabolites like metal bound coenzymes and siderophores mainly involved the degradation pathway [91].
4. Phytoremediation
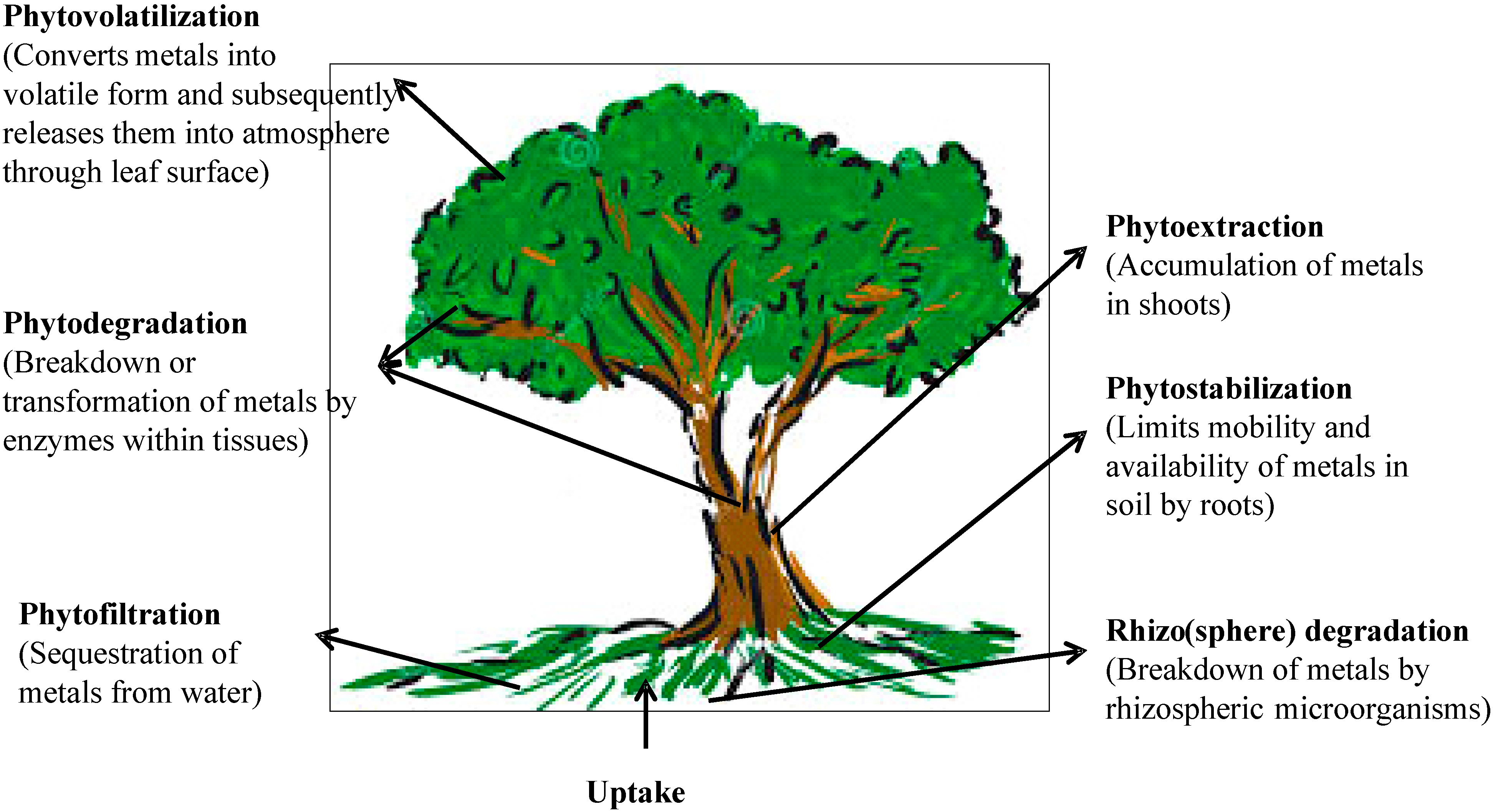
Phytoremediation basically refers to the use of plants and associated microorganisms to partially or completely remediate selected contaminants from soil, sludge, sediments, wastewater and ground water. It can be used for removal of radionuclides, organic pollutants as well as heavy metals [92]. Phytoremediation utilizes a variety of plant processes and the physical characteristics of plants to aid in remediation of contaminated sites. Over the recent years, a special emphasis has been placed on phytoremediation since this property can be exploited for remediation of heavy metal polluted soils [93,94]. It is a cost-effective, efficient and eco-friendly in situ remediation technology driven by solar energy. The technique of phytoremediation includes a number of different processes such as phytoextraction, phytofiltration, phytostabilization, phytovolatilization and phytodegradation [95]. A summary of various processes involved in the phytoremediation of heavy metals is shown in Figure 2. The initial step of phytoremediation is phytoextraction, the uptake of contaminants from soil or water by plant roots and their translocation to and accumulation in biomass, i.e., shoots [96]. Translocation of metals to shoots is an important biochemical process and is desirable in an effective phytoextraction. The next important process of phytoremediation is phytofiltration, which includes rhizofiltration (use of plant roots), blastofiltration (use of seedlings) or caulofiltration (use of excised plant shoots) [97]. In this, the metals are absorbed or adsorbed and thus their movement in underground water is minimized. In addition to the above process, phytostabilization or phytoimmobilization occurs, which reduces the mobility and bioavailability of metals in the environment and thus prevents their migration into groundwater or the food chain [98]. Plants perform the immobilization of heavy metals in soils by sorption through roots, precipitation, complex formation or metal valence reduction in the rhizosphere [99]. Organic pollutants taken by plants are metabolized by enzymes such as dehalogenase and oxygenase, which are not dependent on rhizospheric microorganisms [100]. However, several heavy metals absorbed by plants get converted into volatile forms and subsequently released into the atmosphere by the process called phytovolatilization. This process has been used for removal of some volatile heavy metals like Hg and Se from polluted soils [101]. However, this is limited by the fact that it does not remove the metals completely but rather transfers them from one medium (soil or water) to another (atmosphere) from which they can reenter soil and water.
Recently, removal of heavy metals through phytoremediation, especially hyperaccumulators to degrade and detoxify contaminants, has received wide attention due to its efficacy and cost efficiency [51]. The criteria used for hyperaccumulation varies according the metal, ranging from 100 mg·kg−1 dry mass for Cd to 1000 mg·kg−1 for Cu, Co, Cr and Pb. These values exhibit a shoot-to-soil ratio of metal concentration and the factor for bioaccumulation is higher than 1 [102].
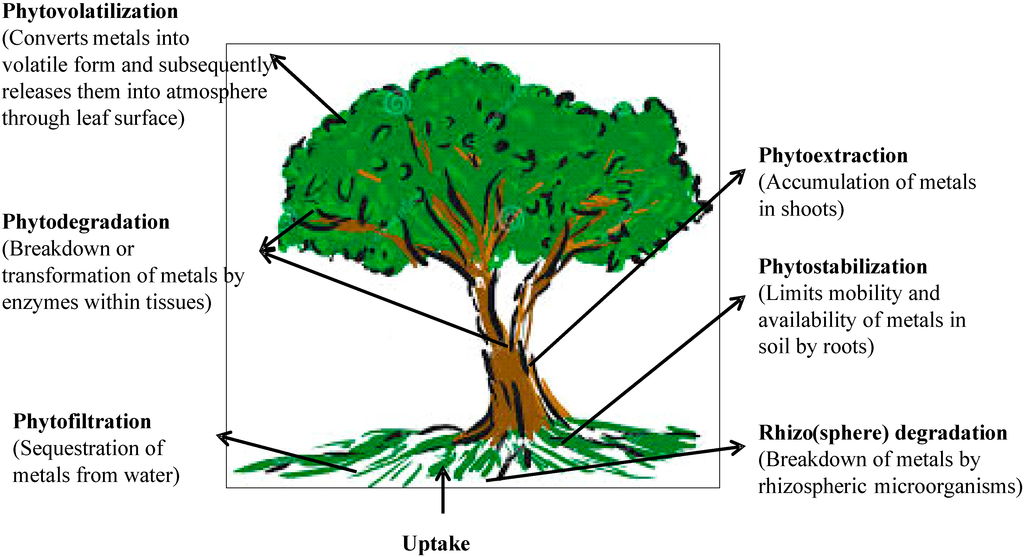
Figure 2.
Various processes involved in the phytoremediation of heavy metals.
Figure 2.
Various processes involved in the phytoremediation of heavy metals.
Hyperaccumulators have been found to exhibit higher heavy metal tolerance and accumulating abilities compared to other plants [103]. Many such plants like Arabidopsis halleri [104] and Solanum nigrum L. [105] have been utilized for phytoremediation of cadmium. Table 2 summarizes the list of different plants reported for remediation of heavy metals. However, the disadvantages that limit the use of hyperaccumulators include difficulty in finding heavy metal hyperaccumulators, slow growth and lower biomass yield. This makes the process quite time-consuming and therefore not feasible for rapidly contaminated sites or sewage treatments [41]. However, different rhizospheric microorganisms that may play important roles in plant growth and/or metal tolerance via different mechanisms are known, and these can be beneficial for the design of a phytoremediation plan to select appropriate multifunctional microbial combinations from the rhizosphere, which may include arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria. It is suggested that the remediation role of rhizosphere is the main part of phytoremediation and one of the main basic theories for removing contaminants by the combined activity of plants and microorganisms [106]. The main reason for the enhanced removal of metals in the rhizosphere is likely the increase in the number and metabolic activities of microorganisms. In the rhizospheric degradation process, the metal toxicity to plants can be reduced by the use of plant growth-promoting bacteria, free-living soil microorganisms that exert beneficial effects on plant growth. In this process, plants can stimulate microbial activity about 10–100 times by the secretion of exudates which contain carbohydrates, amino acids, flavonoids etc. [107]. In return, the rhizosphere bacteria that contain ACC deaminase may act to insure that the ethylene level does not impair root development and to facilitate the generation of larger roots which enhance seedling survival [108]. It is reported that nickel-resistant soil bacterium Kluyvera ascorbata SUD 165 promoted the growth of Brassica campestris in the presence of high concentration of nickel due to its ability to lower the level of ethylene stress in the seedlings [109].

Table 2.
List of selected plants reported for phytoremediation of heavy metals.
| Heavy Metal | Plant Species | Ref. |
|---|---|---|
| Cd, Cu, Pb, Zn | Salix spp. (Salix viminalis, Salix fragilis) | [110,111,112] |
| Cd | Castor (Ricinus communis) | [113] |
| Cd, Pb, Zn | Corn (Zea mays) | [114] |
| Cd, Cu, Pb, Zn | Populus spp. (Populus deltoides, Populus nigra, Populus trichocarpa) | [112] |
| Cd, Cu, Ni, Pb | Jatropha (Jatropha curcas L.) | [115,116] |
| Hg | Populus deltoides | [117] |
| Se | Brassica juncea, Astragalus bisulcatus | [118] |
| Zn | Populus canescens | [119] |
5. Biotechnological Intervention/Genetic Engineering in Bioremediation Processes
5.1. “Designer” Microbes Approach
Genetically engineered microorganisms (GEM) are organisms whose genetic material has been altered using recombinant DNA technology to generate a character-specific efficient strain for bioremediation of soil, water and activated sludge by exhibiting enhanced degrading capabilities against a wide range of chemical contaminants [120]. It offers the advantage of constructing microbial strains which can withstand adverse stressful situations and can be used as a bioremediators under various and complex environmental conditions. A list of selected genetically engineered bacteria for removal of heavy metals is presented in Table 3. Genetic engineering has led to the development of “microbial biosensors” to measure the degree of contamination in contaminated sites quickly and accurately. Various biosensors have been designed to evaluate heavy metal concentrations like mercury (Hg), cadmium (Cd), nickel (Ni), copper (Cu) and arsenic (As) [121,122]. Genetic engineering of endophytes and rhizospheric bacteria for plant-associated degradation of pollutants in soil is considered to be one of the most promising new technologies for remediation of metal contaminated sites [123]. Bacteria like Escherichia coli and Moreaxella sp. expressing phytochelatin 20 on the cell surface have been shown to accumulate 25 times more Cd or Hg than the wild-type strains [124,125]. However, one major obstacle for utilizing these GEMs in hostile field conditions is sustaining the recombinant bacteria population in soil, with various environmental conditions and competition from native bacterial populations [126]. Further, the molecular approaches have been applied to only limited bacterial strains like Escherichia coli, Pseudomonas putida, Bacillus subtilis etc. This means other microorganisms need to be explored for their application in heavy metal bioremediation through molecular intervention.

Table 3.
Genetically engineered bacteria for remediation of heavy metals.
| Heavy Metal | Initial Conc. (ppm) | Removal Efficiency (%) | Genetically Engineered Bacteria | Expressed Gene | Ref. |
|---|---|---|---|---|---|
| As | 0.05 | 100 | E. coli strain | Metalloregulatory protein ArsR | [127] |
| Cd2+ | - | - | E .coli strain | SpPCS | [84] |
| Cr6+ | 1.4–1000 | 100 | Methylococcus capsulatus | CrR | [128] |
| Cr | - | - | P. putida strain | Chromate reductase (ChrR) | [129] |
| Cd2+, Hg | - | - | Ralstonia eutropha CH34, Deinococcus radiodurans | merA | [82,130] |
| Hg | - | - | E. coli strain | Organomurcurial lyase | [131] |
| Hg | 7.4 | 96 | E. coli JM109 | Hg2+ transporter | [132] |
| Hg | - | - | Pseudomonas K-62 | Organomercurial lyase | [133] |
| Hg | - | - | Achromobacter sp AO22 | mer | [134] |
| Ni | 145 | 80 | P. fluorescens 4F39 | Phytochelatin synthase (PCS) | [135,136] |
5.2. Designer Plant Approach
Recent advances in omics technologies such as genomics, proteomics, transcriptomics and metabolomics play important roles in identifying traits that maximize the benefits of field remediation technologies. The main constraint of phytoremediation technology is the accumulation of pollutants or their metabolites in plant tissues, which shortens plant life and releases contaminants into the atmosphere via volatilization. This problem can be minimized by manipulation of metal tolerance, accumulation and degradation potential of plants against various inorganic pollutants. Using the designer plant approach, the bacterial genes responsible for metal degradation can be introduced in plant tissues to allow degradation of metals within the plant tissues [137]. Application of genetically engineered (GE) plant-based bioremediation for various heavy metals pollutants is in the forefront due to its eco-friendliness and reduced health hazards compared to physico-chemical based strategies, which are less eco-friendly and more hazardous to human health [138]. Various microbial genes can be harnessed in the transgenic plant for detoxification and accumulation of inorganic contaminants [139]. The metal-detoxifying chelators such as metallothineins and phytochetains can confer resistance to the plant by enhancing uptake, transport and accumulation of various heavy metals [140]. Similarly, transgenic plants carrying bacterial reductase can enhance the volatilization of Hg and Se while accumulating the arsenic in plant shoots [118,139]. Fast-growing as well as high-biomass-yielding plants like poplar, willow and Jatropa could be used for both phytoremediation and energy production. However, transfer of the metals from soil or water does not solve the problem, and burning metal-contaminated plant material for energy production will release the metals into the atmosphere, moving the problem from soil or water to the air. Thus such metal-accumulating biomasses should be stored or disposed of appropriately so that they do not pose any hazard to the environment.
Among the fast-growing and high-biomass-yielding plants, poplar is the most commonly studied because of its rapid growth rate and potential to produce high biomass within a short period of time (5–8 years). Many of the poplar hybrid varieties have been genetically modified with microbial catabolic genes and specific transporters for increased remediation [141,142]. For example, mercuric reductase and γ-glutamylsysteine synthetase genes showed increased resistance to Hg and Cd and Cu, respectively, through accumulation of higher concentrations of these metals [119,142]. As mentioned earlier, manipulation of desired plant species with multiple genes will facilitate complete degradation of pollutants to ensure that the harvested biomass can be utilized completely for additional benefits [143].
5.3. Rhizosphere Engineering
Engineered bioremediation strategies involve either the addition of growth stimulators (electron acceptors/donors) to the rhizosphere for reduction of heavy metals or addition of nutrients to the contaminated soil for enhancement of microbial growth and bioremediation properties of microorganisms or genetically modified plants [144]. Many engineered bacteria with heavy metal reduction capacity through the expression of improved enzymes like chromate and uranyl reductase were applied in a specific rhizosphere to perform a specific function. Similarly, genetically modified plants are also known to produce specific compounds which may support the rhizospheric transformation of heavy metals. Recently, several researchers have applied these approaches to remediate heavy metal-contaminated soils using the rhizosphere ecosystem.
5.4. Manipulation of Plant-Microbe Symbiosis
The main drawbacks of phytoremediation technology are storage and accumulation of pollutants in the plant materials and the remediation process slowing down and often becoming inadequate when the contaminated site has multiple pollutants [145]. The appropriate solution to these problems is to combine the microbe-plant symbiosis within the plant rhizosphere [126] or to introduce microbes as endophytes to allow degradation of pollutants within the plant tissues [137]. The microbial population in the rhizosphere is much higher than present in vegetation-less soil, and this is due to the facilitation provided by the plants through release of substances that are nutrients for microorganisms. This approach has been evaluated under laboratory conditions, and if it succeeds in field conditions, this technology could facilitate accelerated removal of pollutants, which in turn will support high plant biomass production for bioenergy [146]. The major strategies for implementing bioremediation processes include biostimulation and bioaugmentation approaches guided by specific microbes in combination with plants. Biostimulation involves adding supplements to a contaminated site with the objective of stimulating growth of the microbial population already present there, which may be capable of degrading the contaminants. Bioaugmentation refers to addition of selected and acclimated microbial inocula to the environment that do not contain microbes capable of degrading the contaminants.
5.5. Application of Nano-Biotechnology
Apart from the above discussed strategies, the remediation of heavy metals and trace elements can be achieved by an emerging technology i.e., nanotechnology. Various nanoparticles or nanomaterials have been found to be very effective for the removal of a wide range of toxic metals from the environment as compared to conventional methods. Nanoparticles enhancing microbial activity to remove toxic pollutants is called “nanobioremediation.” Nano-based technologies not only reduce the costs of cleaning up contaminated sites at a large scale, but also reduce the process time as well. “Bionanotechnology” or “nanotechnology through biotechnology” is the bio-fabrication of nano-objects or bifunctional macromolecules used as tools to construct or manipulate nano-objects. Wide physiological diversity, small size, genetic manipulability and controlled culturability make microbial cells ideal producers of nanostructures ranging from natural products, such as polymers and magnetosomes, to engineered proteins or protein constructs, such as virus-like proteins (VLP) and tailored metal particles [147]. Deinococcus radiodurans, a radioactive-resistant organism, has the ability to withstand radiation well beyond the naturally occurring levels, thus its application in radioactive waste clean-up initiatives funded by US Department of Energy (DOE) [130,148]. Metal chelating polymers require toxic solvents for their synthesis and ultrafiltration for their separation, and this can be solved by developing metal binding materials that can be recovered by changing the environment surround them like pH, temperature etc. One such material is nanoscale modified biopolymers, which are manufactured by genetic and protein engineering of microorganisms, and their size can be controlled at the molecular level [149]. This innovative technique would be a promising tool to address the escalating problem of heavy metal as well as organic contaminants in the environment.
5.6. Application of Genomics
Genomics has been explored and used mainly in microbial genetics and in agriculture, e.g., GMO crops, but it is now a new tool for bioremediation. Knowledge of genomics in of the context of understanding microbe-mediated remediation provides a view of genes related to the sensitivity of microbes towards toxic metals in the soil. Application of genomics in bioremediation makes it possible to analyze the microorganism on the basis of not only biochemical parameters but also molecular levels related to mechanism.
6. Future Prospects
Rapid industrialization and technology development have adverse side effects like soil contamination and degrading soil health. Due to the complexity involved in the conventional methods for remediation of soil, the use of microbes has arisen as a time-saver for bioremediation. However, bioremediation technology has limitations; several microorganisms cannot break toxic metals into harmless metabolites, and these have inhibitory effects on microbial activity. Modification in the outer membrane proteins of bacteria with potential bioremediation properties for improving metal binding abilities is the likely way to enhance their capacity for biotransformation of toxic metals. Future studies should focus on the factors involved in improving in situ bioremediation strategies using genetically engineered microorganisms (GEM) and also theapplicability and adaptability of these GEMs in all the possible adverse/stress conditions and multiple-heavy-metal-polluted conditions. The reluctance among the public to accept GEM for bioremediation also needs to be considered in future studies, and they must proved non-toxic to the environment.
Acknowledgments
The authors would like to thank all the three anonymous referees for their constructive comments and suggestions.
Author Contributions
Ruchita Dixit, Wasiullah, Deepti Malaviya and Kuppusamy Pandiyan mostly contributed equally at designing of manuscript. Renu Shukla, Asha Sahu, Bhanu P. Singh and Jai P. Rai carried out data collection and processing. Pawan Kumar Sharma was involved in the statistical analysis. Udai B. Singh, Harshad Lade and Diby Paul wrote and revise the paper. All authors read and approved the final manuscript.
Abbreviations
| Ag | silver |
| As | arsenic |
| As(V) | arsenate |
| As(III) | arsenite |
| Au | gold |
| Ba | barium |
| Bi | bismuth |
| Cd | cadmium |
| Co | cobalt |
| Cr | chromium |
| Cu | copper |
| Fe | Iron |
| Hg | mercury |
| Mn | manganese |
| Ni | nickel |
| Pb | lead |
| Se | selenium |
| Zn | zinc |
Conflicts of Interest
The authors declare no conflict of interest.
References
- Salem, H.M.; Eweida, E.A.; Farag, A. Heavy metals in drinking water and their environmental impact on human health. In ICEHM 2000; Cairo University: Giza, Egypt, 2000; pp. 542–556. [Google Scholar]
- Ahluwalia, S.S.; Goyal, D. Microbial and plant derived biomass for removal of heavy metals from wastewater. Bioresour. Technol. 2007, 98, 2243–2257. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.; Farag, S.; Moawad, H. Isolation and characterization of Pseudomonas resistant to heavy metals contaminants. Arab. J. Biotechnol. 2004, 7, 13–22. [Google Scholar]
- Kapoor, A.; Viraraghvan, T. Fungal biosorption—An alternative treatment option for heavy metal bearing wastewater: A review. Bioresour. Technol. 1995, 53, 195–206. [Google Scholar]
- Doelman, P.; Jansen, E.; Michels, M.; van Til, M. Effects of heavy metals in soil on microbial diversity and activity as shown by the sensitivity-resistance index, an ecologically relevant parameter. Biol. Fertil. Soils 1994, 17, 177–1784. [Google Scholar] [CrossRef]
- Wood, J.M.; Wang, H.K. Microbiol resistance to heavy metals. Environ. Sci. Technol. 1983, 17, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Tan, T.C. Monitoring BOD in the presence of heavy metal ions using a poly (4-vinylpyr-idine) coated microbial sensor. Biosen. Bioelectron. 1994, 9, 445–455. [Google Scholar] [CrossRef]
- Goblenz, A.; Wolf, K.; Bauda, P. The role of glutathione biosynthesis in heavy metal resistance in the fission yeast Schizosaccharomyces pombe. FEMS Microbiol. Rev. 1994, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- US EPA. Drinking Water Contaminants; United States Environmental Protection Agency (EPA): Washington, DC, USA, 2009. [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Silver; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 1990. [Google Scholar]
- Tripathi, R.D.; Srivastava, S.; Mishra, S.; Singh, N.; Tuli, R.; Gupta, D.K.; Maathuis, F.J.M. Arsenic hazards: Strategies for tolerance and remediation by plants. Trends Biotech. 2007, 25, 158–165. [Google Scholar] [CrossRef]
- Acobs, I.A.; Taddeo, J.; Kelly, K.; Valenziano, C. Poisoning as a result of barium styphnate explosion. Am. J. Ind. Med. 2002, 41, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Degraeve, N. Carcinogenic, teratogenic and mutagenic effects of cadmium. Mutat. Res. 1981, 86, 115–135. [Google Scholar] [CrossRef] [PubMed]
- Wuana, R.A.; Okieimen, F.E. Heavy metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. ISRN Ecol. 2011, 2011. Article 20. [Google Scholar]
- Neustadt, J.; Pieczenik, S. Toxic-metal contamination: Mercury. Integr. Med. 2007, 6, 36–37. [Google Scholar]
- Ainza, C.; Trevors, J.; Saier, M. Environmental mercury rising. Water Air Soil Poll. 2010, 205, 47–48. [Google Scholar] [CrossRef]
- Gulati, K.; Banerjee, B.; Bala Lall, S.; Ray, A. Effects of diesel exhaust, heavy metals and pesticides on various organ systems: Possible mechanisms and strategies for prevention and treatment. Indian J. Exp. Biol. 2010, 48, 710–721. [Google Scholar] [PubMed]
- Khan, M.A.; Ahmad, I.; Rahman, I. Effect of environmental pollution on heavy metals content of Withania somnifera. J. Chin. Chem. Soc. 2007, 54, 339–343. [Google Scholar]
- Das, N.; Vimala, R.; Karthika, P. Biosorption of heavy metals—An overview. Indian J. Biotechnol. 2008, 7, 159–169. [Google Scholar]
- Duda-Chodak, A.; Baszczyk, U. The impact of nickel on human health. J. Elementol. 2008, 13, 685–696. [Google Scholar]
- Padmavathiamma, P.K.; Li, L.Y. Phytoremediation technology: Hyperaccumulation metals in plants. Water Air Soil Poll. 2007, 184, 105–126. [Google Scholar] [CrossRef]
- Vinceti, M.; Wei, E.T.; Malagoli, C.; Bergomi, M.; Vivoli, G. Adverse health effects of selenium in humans. Rev. Environ. Health 2001, 16, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Hess, R.; Schmid, B. Zinc supplement overdose can have toxic effects. J. Pediat. Hematol. Onc. 2002, 24, 582–584. [Google Scholar] [CrossRef]
- Gadd, G.M. Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr. Opin. Biotechnol. 2000, 11, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.E.; Mak, K.Y.; Mohamed, N.; Noor, A.M. Removal and speciation of heavy metals along the treatment path of wastewater in subsurface-flow constructed wetlands. Water Sci. Technol. 2003, 48, 307–313. [Google Scholar] [PubMed]
- Malik, A. Metal bioremediation through growing cells. Environ. Int. 2004, 30, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Lin, H.L. Remediation of soil contaminated with the heavy metal (Cd2+). J. Hazard. Mater. 2005, 122, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Elekwachi, C.O.; Andresen, J.; Hodgman, T.C. Global use of bioremediation technologies for decontamination of ecosystems. J. Bioremed. Biodeg. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Gan, S.; Lau, E.V.; Ng, H.K. Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs). J. Hazard. Mater. 2009, 172, 532–549. [Google Scholar] [CrossRef] [PubMed]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Satinder, K.B.; Verma, M.; Surampalli, R.Y.; Misra, K.; Tyagi, R.D.; Meunier, N. Bioremediation of hazardous wastes-A review. Pract. Period. Hazard. Toxic Radioact. Waste Manag. 2006, 10, 59–72. [Google Scholar] [CrossRef]
- Onwubuya, K.; Cundy, A.; Puschenreiter, M.; Kumpiene, J.; Bone, B. Developing decision support tools for the selection of “gentle” remediation approaches. Sci. Total Environ. 2009, 407, 6132–6142. [Google Scholar] [CrossRef] [PubMed]
- Environmental Agency (EA). Reporting the Evidence: Dealing with Contaminated Land in England and Wales. A Review of Progress from 2000–2007 with Part 2A of the Environmental Protection Act. Available online: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/313964/geho0109bpha-e-e.pdf (accessed on 1 January 2015).
- US EPA. Treatment Technologies for Site Cleanup: Annual Status Report; United States Environmental Protection Agency (EPA): Washington, DC, USA, 2007. [Google Scholar]
- Hussein, H.; Krull, R.; Abou El-Ela, S.I.; Hempel, D.C. Interaction of the different heavy metal ions with immobilized bacterial culture degrading xenobiotic wastewater compounds. In Proceedings of the Second International Water Association World Water Conference, Berlin, Germany, 15–19 October 2001; pp. 15–19.
- Scott, J.A.; Karanjkar, A.M. Repeated cadmium biosorption by regenerated Enterobacter aerogenes biofilm attached to activated carbon. Biotechnol. Lett. 1992, 14, 737–740. [Google Scholar] [CrossRef]
- Ajmal, M.; Rafaqat, A.K.; Bilquees, A.S. Studies on removal and recovery of Cr (VI) from electroplating wastes. Water Res. 1996, 30, 1478–1482. [Google Scholar] [CrossRef]
- Dilek, F.B.; Gokcay, C.F.; Yetis, U. Combined effects of Ni(II) and Cr(VI) on activated sludge. Water Res. 1998, 32, 303–312. [Google Scholar] [CrossRef]
- Volesky, B.; Holan, Z.R. Biosorption of heavy metals. Biotechnol. Prog. 1995, 11, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, C.P. Application of Aspergillus oryzae and Rhizopus oryzae for Cu (II) removal. Water Res. 1996, 30, 1985–1990. [Google Scholar] [CrossRef]
- Xiao, X.; Luo, S.; Zeng, G.; Wei, W.; Wan, Y.; Chen, L.; Guo, H.; Cao, Z.; Yang, L.; Chen, J.; et al. Biosorpiton of cadmium by endophytic fungus (EF) Microsphaeropsis sp. LSE10 isolated from cadmium hyperaccumulator Solanum nigrum L. Bioresour. Technol. 2010, 101, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.N.; Zhang, Y.F.; He, L.Y.; Chen, Z.J.; Wang, Q.Y.; Qian, M.; Sheng, X.F. Genetic diversity and characterization of heavy metal-resistant-endophytic bacteria from two copper-tolerant plant species on copper mine wasteland. Bioresour. Technol. 2010, 101, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Modaihsh, A.; Al-Swailem, M.; Mahjoub, M. Heavy metal contents of commercial inorganic fertilizer used in the Kingdom of Saudi Arabia. Agric. Mar. Sci. 2004, 9, 21–25. [Google Scholar]
- Chehregani, A.; Malayeri, B.E. Removal of heavy metals by native accumulator plants. Int. J. Agric. Biol. 2007, 9, 462–465. [Google Scholar]
- Fulekar, M.; Singh, A.; Bhaduri, A.M. Genetic engineering strategies for enhancing phytoremediation of heavy metals. Afr. J. Biotechnol. 2009, 8, 529–535. [Google Scholar]
- Sabiha-Javied; Mehmood, T.; Chaudhry, M.M.; Tufai, M.; Irfan, N. Heavy metal pollution from phosphate rock used for the production of fertilizer in Pakistan. Microchem. J. 2009, 91, 94–99. [Google Scholar] [CrossRef]
- Sumner, M.E. Beneficial use of effluents, wastes, and biosolids. Commun. Soil Sci. Plant Anal. 2000, 31, 1701–1715. [Google Scholar] [CrossRef]
- D’Amore, J.J.; Al-Abed, S.R.; Scheckel, K.G.; Ryan, J.A. Methods for speciation of metals in soils: A review. J. Environ. Qual. 2005, 34, 1707–1745. [Google Scholar] [CrossRef] [PubMed]
- Lombi, E.; Gerzabek, M.H. Determination of mobile heavy metal fraction in soil: Results of a pot experiment with sewage sludge. Commun. Soil Sci. Plant Anal. 1998, 29, 2545–2556. [Google Scholar] [CrossRef]
- Sposito, G.; Page, A.L. Cycling of metal ions in the soil environment. In Metal Ions in Biological Systems, Circulation of Metals in the Environment; Sigel, H., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 1984; pp. 287–332. [Google Scholar]
- Ahemad, M. Remediation of metalliferous soils through the heavy metal resistant plant growth promoting bacteria: Paradigms and prospects. Arab. J. Chem. 2014. [Google Scholar] [CrossRef]
- Garbisu, C.; Alkorta, I. Phytoextraction: A cost-effective plant-based technology for the removal of metals from the environment. Bioresour. Technol. 2001, 77, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [PubMed]
- Roane, T.M.; Pepper, I.L. Microorganisms and metal pollution. In Environmental Microbiology; Maier, I.L., Pepper, C.B., Eds.; Gerba, Academic Press: London, UK, 2000; p. 55. [Google Scholar]
- Guiné, V.; Spadini, L.; Sarret, G.; Muris, M.; Delolme, C.; Gaudet, J.P.; Martins, J.M. Zinc sorption to three gram-negative bacteria: Combined titration, modeling and EXAFS study. Environ. Sci. Technol. 2006, 40, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Comte, S.; Guibaud, G.; Baudu, M. Biosorption properties of extracellular polymeric substances (EPS) towards Cd, Cu and Pb for different pH values. J. Hazard. Mater. 2008, 151, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.C.; Huang, Q.Y.; Wei, X.; Liang, W.; Rong, X.M.; Chen, W.L.; Cai, P. Microcalorimetric and potentiometric titration studies on the adsorption of copper by extracellular polymeric substances (EPS), minerals and their composites. Bioresour. Technol. 2010, 101, 5774–5779. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wei, X.; Cai, P.; Huang, Q.; Chen, H.; Liang, W.; Rong, X. Role of extracellular polymeric substances in Cu(II) adsorption on Bacillus subtilis and Pseudomonas putida. Bioresour. Technol. 2011, 102, 1137–1141. [Google Scholar] [CrossRef] [PubMed]
- Carter, P.; Cole, H.; Burton, J. Bioremediation: Successes and Shortfalls. In Proceedings of Key International Conference and Exhibition for Spill Prevention, Preparedness, Response and Restoration (Interspill), London, UK, 23 March 2006.
- Kinya, K.; Kimberly, L.D. Current use of bioremediation for TCE cleanup: Results of a survey. Remediat. J. 1996, 6, 1–14. [Google Scholar]
- Chen, C.; Wang, J.L. Characteristics of Zn2+ biosorption by Saccharomyces cerevisiae. Biomed. Environ. Sci. 2007, 20, 478–482. [Google Scholar] [PubMed]
- Talos, K.; Pager, C.; Tonk, S.; Majdik, C.; Kocsis, B.; Kilar, F.; Pernyeszi, T. Cadmium biosorption on native Saccharomyces cerevisiae cells in aqueous suspension. Acta Univ. Sapientiae Agric. Environ. 2009, 1, 20–30. [Google Scholar]
- Tigini, V.; Prigione, V.; Giansanti, P.; Mangiavillano, A.; Pannocchia, A.; Varese, G.C. Fungal biosorption, an innovative treatment for the decolourisation and detoxification of textile effluents. Water 2010, 2, 550–565. [Google Scholar] [CrossRef]
- Brierley, C.L. Bioremediation of metal-contaminated surface and groundwater. Geomicrobiol. J. 1990, 8, 201–223. [Google Scholar] [CrossRef]
- Pinedo-Rivilla, C.; Aleu, J.; Collado, I.G. Pollutants biodegradation by fungi. Curr. Org. Chem. 2009, 13, 1194–1214. [Google Scholar] [CrossRef]
- D’Annibale, A.; Leonardi, V.; Federici, E.; Baldi, F.; Zecchini, F.; Petruccioli, M. Leaching and microbial treatment of a soil contaminated by sulphide ore ashes and aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 2007, 74, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Tunali, S.; Akar, T.; Oezcan, A.S.; Kiran, I.; Oezcan, A. Equilibrium and kinetics of biosorption of lead(II) from aqueous solutions by Cephalosporium aphidicola. Sep. Purif. Technol. 2006, 47, 105–112. [Google Scholar] [CrossRef]
- Akar, T.; Tunali, S.; Cabuk, A. Study on the characterization of lead (II) biosorption by fungus Aspergillus parasiticus. Appl. Biochem. Biotech. 2007, 136, 389–406. [Google Scholar] [CrossRef]
- Kelly, D.J.A.; Budd, K.; Lefebvre, D.D. The biotransformation of mercury in pH-stat cultures of microfungi. Can. J. Bot. 2006, 84, 254–260. [Google Scholar] [CrossRef]
- Thavasi, R. Microbial biosurfactants: From an environment application point of view. J. Bioremed. Biodegrad. 2011, 2. Article 104e. [Google Scholar] [CrossRef]
- Evanko, C.R.; Dzombak, D.A. Remediation of metals-contaminated soil and groundwater. Environ. Sci. 1997, 412, 1–45. [Google Scholar]
- Silver, S. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Phillips, E.J.P. Novel mode of microbial energy metabolism: Organic carbon oxidation to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 1988, 54, 1472–1480. [Google Scholar]
- Lovley, D.R.; Coates, J.D.; Blunt-Harris, E.L.; Philips, E.J.P.; Woodward, J.C. Humic substances as electron acceptors for microbial respiration. Nature 1996, 382, 445–448. [Google Scholar] [CrossRef]
- Spormann, A.M.; Widdel, F. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 2000, 11, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Lovely, D.R. Dissimilatory metal rduction: From early life to bioremediation. ASM News 2002, 68, 231–237. [Google Scholar]
- Lovley, D.R.; Philips, E.J.; Gorby, Y.A.; Landa, E.R. Microbial reduction of uranium. Nature 1991, 350, 413–416. [Google Scholar] [CrossRef]
- Gómez Jiménez-T, R.; Moliternib, E.; Rodríguezb, L.; Fernándezc, F.J.; Villaseñorc, J. Feasibility of mixed enzymatic complexes to enhanced soil bioremediation processes. Procedia Environ. Sci. 2011, 9, 54–59. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Role in heavy metals detoxification and homeostatis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef] [PubMed]
- Kagi, J.H.R.; Schaffer, A. Biochemistry of metallothionein. Biochemistry 1988, 27, 8509–8515. [Google Scholar] [CrossRef] [PubMed]
- Huckle, J.W.; Morby, A.P.; Turner, J.S.; Robinson, N.J. Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol. Microbiol. 1993, 7, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Valls, M.; Atrian, S.; de Lorenzo, V.; La, F. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 2000, 18, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Mejare, M.; Bulow, L. Metal binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol. 2001, 19, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Singh, S.; Kim, J.Y.; Lee, W.; Mulchandani, A.; Chen, W. Bacteria metabolically engineered for enhanced phtochelatin production and cadmium accumulation. App. Environ. Microbiol. 2007, 73, 6317–6320. [Google Scholar] [CrossRef]
- Singh, S.; Kang, S.H.; Mulchandani, A.; Chen, W. Bioremediation: environmental cleanup through pathway engineering. Curr. Opin. Biotechnol. 2008, 19, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Brim, H.; Venkateshwaran, A.; Kostandarithes, H.M.; Fredrickson, J.K.; Daly, M.J. Engineering Deinococcus geothermalis for bioremediation of high temperature radioactive waste environments. App. Environ. Microbiol. 2003, 69, 4575–4582. [Google Scholar] [CrossRef]
- Rojas, L.A.; Yanez, C.; Gonzalez, M.; Lobos, S.; Smalla, K.; Seeger, M. Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 gener-ated for mercury bioremediation. PLoS One 20 2011, 6, e17555. [Google Scholar] [CrossRef]
- Sone, Y.; Mochizuki, Y.; Koizawa, K.; Nakamura, R.; Pan-Hou, H.; Itoh, T.; Kiyono, M. Mercurial resistance determinants in Pseudomonas strain K-62 plasmid pMR68. AMB Express 2013, 3. Article 41. [Google Scholar]
- Essa, A.M.M.; Macaskie, L.E.; Brown, N.L. Mechanisms of mercury bioremediation. Biochem. Soc. Trans. 2002, 30, 672–674. [Google Scholar] [CrossRef] [PubMed]
- Brim, H.; Osborne, J.P.; Kostandarithes, H.M.; Fredrickson, J.K.; Wackett, L.P.; Daly, M.J. Deinococcus radiodurans engineered for complete toluene degradation facilities Cr(IV) reduction. Microbiology 2006, 152, 2469–2477. [Google Scholar] [CrossRef] [PubMed]
- Penny, C.; Vuilleumier, S.; Bringel, F. Microbial degradation of tetrachloromethane: Mechanisms and perspectives for bioremediation. FEMS Microbiol. Ecol. 2010, 74, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals-Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.H.; Brooks, R.R.; Howes, A.W.; Kirkman, J.H.; Gregg, P.E.H. The potential of the high biomass nickel hyperaccumulator Berkheya coddii for phytoremediation and phytomining. J. Geochem. Explor. 1997, 60, 115–126. [Google Scholar] [CrossRef]
- Martinez, M.; Bernal, P.; Almela, C.; Vélez, D.; García-Agustín, P.; Serrano, R.; Navarro-Aviñó, J. An engineered plant that accumulates higher levels of heavy metals than Thlaspi caerulescens, with yields of 100 times more biomass in mine soils. Chemosphere 2006, 64, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Alkorta, I.; Hernández-Allica, J.; Becerril, J.M.; Amezaga, I.; Albizu, I.; Garbisu, C. Recent findings on the phytoremediation of soils contaminated with environmentally toxic heavy metals and metalloids such as Zinc, Cadmium, Lead, and Arsenic. Rev. Environ. Sci. Biotechnol. 2004, 3, 71–90. [Google Scholar] [CrossRef]
- Sekara, A.; Poniedzialeek, M.; Ciura, J.; Jedrszczyk, E. Cadmium and lead accumulation and distribution in the organs of nine crops: implications for phytoremediation. Pol. J. Environ. Stud. 2005, 14, 509–516. [Google Scholar]
- Mesjasz-Przybylowicz, J.; Nakonieczny, M.; Migula, P.; Augustyniak, M.; Tarnawska, M.; Reimold, W.U.; Koeberl, C.; Przybylowicz, W.; Glowacka, E. Uptake of cadmium, lead, nickel and zinc from soil and water solutions by the nickel hyperaccumulator Berkheya coddii. Acta Biol. Cracov. Bot. 2004, 46, 75–85. [Google Scholar]
- Erakhrumen, A.A. Phytoremediation: An environmentally sound technology for pollution prevention, control and remediation in developing countries. Educ. Res. Rev. 2007, 2, 151–156. [Google Scholar]
- Barceló, J.; Poschenrieder, C. Phytoremediation: principles and perspectives. Contrib. Sci. 2003, 2, 333–344. [Google Scholar]
- Vishnoi, S.R.; Srivastava, P.N. Phytoremediation-green for environmental clean. In Proceeding of Taal 2007: The 12th World Lake Conference, Jaipur, India, 29 October–2 November 2008.
- Karami, A.; Shamsuddin, Z.H. Phytoremediation of heavy metals with several efficiency enhancer methods. Afr. J. Biotechnol. 2010, 9, 3689–3698. [Google Scholar]
- Baker, A.J.M.; McGrath, S.P.; Sidoli, C.M.D.; Reeves, R.D. The possibility of in situ heavy metal decontamination of polluted soils using crops of metal accumulating plants. Resour. Conserv. Recycl. 1994, 11, 42–49. [Google Scholar]
- Prasad, M.N.V.; Freitas, H.M.D.O. Metal hyperaccumulation in plants—Biodiversity prospecting for phytoremediation technology. Electro. J. Biotechnol. 2003, 6, 285–321. [Google Scholar] [CrossRef]
- Dahmani-Muller, H.; Van-Oort, F.; Gelie, B.; Balabane, M. Strategies of heavy metal uptake by three plant species growing near a metal smelter. Environ. Pollut. 2000, 109, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.H.; Zhou, Q.X.; Wang, X. Cadmium-hyperaccumulator Solanum nigrum L. and its accumulating characteristics. Environ. Sci. 2005, 26, 167–171. [Google Scholar]
- Wei, S.; Zhou, Q.; Zhang, K.; Liang, J. Roles of rhizosphere in remediation of contaminated soils and its mechanisms. Ying Yong Sheng Tai Xue Bao 2003, 14, 143–147. [Google Scholar] [PubMed]
- Kuiper, I.; Lagendijk, E.L.; Bloemberg, G.V.; Lugtenberg, B.J.J. Rhizoremediation: A beneficial plant-microbe interaction. Mol. Plant-Microbe Interact. 2004, 17, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Burd, G.I.; Dixon, D.G.; Glick, B.R. A plant growth-promoting bacterium that decreases nickel toxicity in seedlings. Appl. Environ. Microbiol. 1998, 64, 3663–3668. [Google Scholar] [PubMed]
- Pulford, I.; Watson, C. Phytoremediation of heavy metal-contaminated land by trees-a review. Environ. Int. 2003, 29, 529–540. [Google Scholar]
- Volk, T.A.; Abrahamson, L.P.; Nowak, C.A.; Smart, L.B.; Tharakan, P.J.; White, E.H. The development of short-rotation willow in the northeastern United States for bioenergy and bioproducts, agroforestry and phytoremediation. Biomass Bioener. 2006, 30, 715–727. [Google Scholar] [CrossRef]
- Ruttens, A.; Boulet, J.; Weyens, N.; Smeets, K.; Adriaensen, K.; Meers, E.; van Slycken, S.; Tack, F.; Meiresonne, L.; Thewys, T.; et al. Short rotation coppice culture of willows and poplars as energy crops on metal contaminated agricultural soils. Int. J. Phytorem. 2011, 13, 194–207. [Google Scholar] [CrossRef]
- Huang, H.; Yu, N.; Wang, L.; Gupta, D.K.; He, Z.; Wang, K.; Zhu, Z.; Yan, X.; Li, T.; Yang, X.E. The phytoremediation potential of bioenergy crop Ricinus communis for DDTs and cadmium co-contaminated soil. Bioresour. Technol. 2011, 102, 11034–11038. [Google Scholar] [CrossRef] [PubMed]
- Meers, E.; van Slycken, S.; Adriaensen, K.; Ruttens, A.; Vangronsveld, J.; Du Laing, G.; Witters, N.; Thewys, T.; Tack, F.M. The use of bio-energy crops (Zea mays) for “phytoattenuation” of heavy metals on moderately contaminated soils: A field experiment. Chemosphere 2010, 78, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Abhilash, P.C.; Jamil, S.; Singh, N. Transgenic plants for enhanced biodegradation and phytoremediation of organic xenobiotics. Biotechnol. Adv. 2009, 27, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Jamil, S.; Abhilash, P.C.; Singh, N.; Sharma, P.N. Jatropha curcas: A potential crop for phytoremediation of coal fly ash. J. Hazard. Mater. 2009, 172, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Che, D.; Meagher, R.B.; Heaton, A.C.; Lima, A.; Rugh, C.L.; Merkle, S.A. Expression of mercuric ion reductase in Eastern cottonwood (Populus deltoides) confers mercuric ion reduction and resistance. Plant Biotechnol. J. 2003, 1, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Bitther, O.P.; Pilon-Smits, E.A.H.; Meagher, R.B.; Doty, S. Biotechnological approaches for phytoremediation. In Plant Biotechnology and Agriculture; Arie Altman, A., Hasegawa, P.M., Eds.; Academic Press: Oxford, UK, 2012; pp. 309–328. [Google Scholar]
- Bittsanszkya, A.; Kömives, T.; Gullner, G.; Gyulai, G.; Kiss, J.; Heszky, L.; Radimszky, L.; Rennenberg, H. Ability of transgenic poplars with elevated glutathione content to tolerate zinc(2+) stress. Environ. Int. 2005, 31, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Sayler, G.S.; Ripp, S. Field applications of genetically engineered microorganisms for bioremediation process. Curr. Opin. Biotechnol. 2000, 11, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Singh, M. Biosensors for heavy metals. J. Biometals 2005, 18, 121–129. [Google Scholar] [CrossRef]
- Bruschi, M.; Goulhen, F. New bioremediation technologies to remove heavy metals and radionuclides using Fe(III)-sulfate- and sulfur reducing bacteria. In Environmental Bioremediation Technologies; Singh, S.N., Tripathi, R.D., Eds.; Springer Publication: NY, USA, 2006; pp. 35–55. [Google Scholar]
- Divya, B.; Deepak Kumar, M. Plant-Microbe interaction with enhanced bioremediation. Res. J. BioTechnol. 2011, 6, 72–79. [Google Scholar]
- Bae, W.; Mehra, R.K.; Mulchandani, A.; Chen, W. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl. Environ. Microbiol. 2001, 67, 5335–5338. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.; Wu, C.H.; Kostal, J.; Mulchandani, A.; Chen, W. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. App. Environ. Microbiol. 2003, 69, 3176–3180. [Google Scholar] [CrossRef]
- Wu, C.H.; Wood, T.K.; Mulchandani, A.; Chen, W. Engineering plant-microbe symbiosis for rhizoremediation of heavy metals. Appl. Environ. Microbiol. 2006, 72, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Kostal, J.R.Y.; Wu, C.H.; Mulchandani, A.; Chen, W. Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Appl. Environ. Microbiol. 2004, 70, 4582–4587. [Google Scholar] [CrossRef] [PubMed]
- Hasin, A.A.; Gurman, S.J.; Murphy, L.M.; Perry, A.; Smith, T.J.; Gardiner, P.E. Remediation of chromium (VI) by a methane-oxidizing bacterium. Environ. Sci. Technol. 2010, 44, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Ackerley, D.F.; Gonzalez, C.F.; Keyhan, M.; Blake, R.; Matin, A. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 2004, 6, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Brim, H.; McFarlan, S.C.; Fredrickson, J.K.; Minton, K.W.; Zhai, M.; Wackett, L.P.; Daly, M.J. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 2000, 18, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, I.; Dutt, A.; Ali, A. Biomolecular engineering of Escherichia coli organomercurial lyase gene and its expression. Indian J. Biotech. 2002, 1, 117–120. [Google Scholar]
- Zhao, X.W.; Zhou, M.H.; Li, Q.B.; Lu, Y.H.; He, N.; Sun, D.H.; Deng, X. Simultaneous mercury bioaccumulation and cell propagation by genetically engineered Escherichia coli. Process Biochem. 2005, 40, 1611–1616. [Google Scholar] [CrossRef]
- Kiyono, M.; Pan-Hou, H. Genetic engineering of bacteria for environmental remediation of mercury. J. Health Sci. 2006, 52, 199–204. [Google Scholar] [CrossRef]
- Ng, S.P.; Davis, B.; Polombo, E.A.; Bhave, M. A Tn5051-like mer-containing transposon identified in a heavy metal tolerant strain Achromobacter sp. AO22. BMC Res. Notes 2009, 7, 2–38. [Google Scholar]
- Lopez, A.; Lazaro, N.; Morales, S.; Margues, A.M. Nickel biosorption by free and immobilized cells of Pseudomonas fluorescens 4F39: A comparative study. Water Air Soil Pollut. 2002, 135, 157–172. [Google Scholar] [CrossRef]
- Sriprang, R.; Hayashi, M.; Ono, H.; Takagi, M.; Hirata, K.; Murooka, Y. Enhanced accumulation of Cd2+ by a Mesorhizobium sp. transformed with a gene from Arabidopsis thaliana coding for phytochelatin synthase. Appl. Environ. Microbiol. 2003, 69, 79–796. [Google Scholar] [CrossRef]
- Van Aken, B.; Tehrani, R.; Schnoor, J. Endophyte-assisted phytoremediation of explosives in poplar trees by Methylobacterium populi BJ001T. In Endophytes of Forest Trees: Biology and Applications, Forestry Sciences; Pirttilä, A.M., Frank, A.C., Eds.; Springer: Heidelberg, The Netherlands, 2011; Volume 80, pp. 217–234. [Google Scholar]
- Singh, B.K. Emerging and genomic approaches in bioremediation. In Proceedings of the 4th International Contaminated Site Remediation Conference, Adelaide, Australia, 11–15 September 2011.
- Maestri, E.; Marmiroli, M. Genetic and molecular aspects of metal tolerance and hyperaccumualtion. In Metal Toxicity in Plants: Perception, Signalling and Remediation; Gupta, D.K., Sandalio, L.M., Eds.; Springer: Berlin, Germany, 2011; pp. 41–61. [Google Scholar]
- Ruis, O.N.; Daniell, H. Genetic engineering to enhance mercury phytoremediation. Curr. Opin. Biotechnol. 2009, 20, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Doty, S.L. Enhanced metabolism of halogenated hydrocarbons in transgenic plants contain mammalian P450 2E1. Proc. Natl. Acad. Sci. USA 2007, 97, 6287–6291. [Google Scholar] [CrossRef]
- Gullner, G. Enhanced tolerance of transgenic poplar plants overexpressing gamma-glutamylcysteine synthetase towards chloroacetanilide herbicides. J. Exp. Bot. 2001, 52, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Abhilash, P.C.; Powell, J.R.; Singh, H.B.; Singh, B.K. Plant–microbe interactions: Novel applications for exploitation in multipurpose remediation technologies. Trends Biotechnol. 2012, 30, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat. Rev. Microbiol. 2003, 1, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Prasad, M.N.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Massielo, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota—A review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.; Schulten, K.; Baneyx, F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Lennon, E.; McNeil, L.B.; Minton, K.W. Duplication insertion of drug resistance determinants in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 1998, 170, 2126–2135. [Google Scholar]
- Vishwanathan, B. Nanomaterials; Narosa Publishing House Pvt Ltd.: New Delhi, India, 2009. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
