Prospecting Araucaria-Associated Yeasts for Second-Generation Biorefineries
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Isolation
2.2. Micro-Scale Cultivation
2.3. Glycosyl Hydrolase Activities
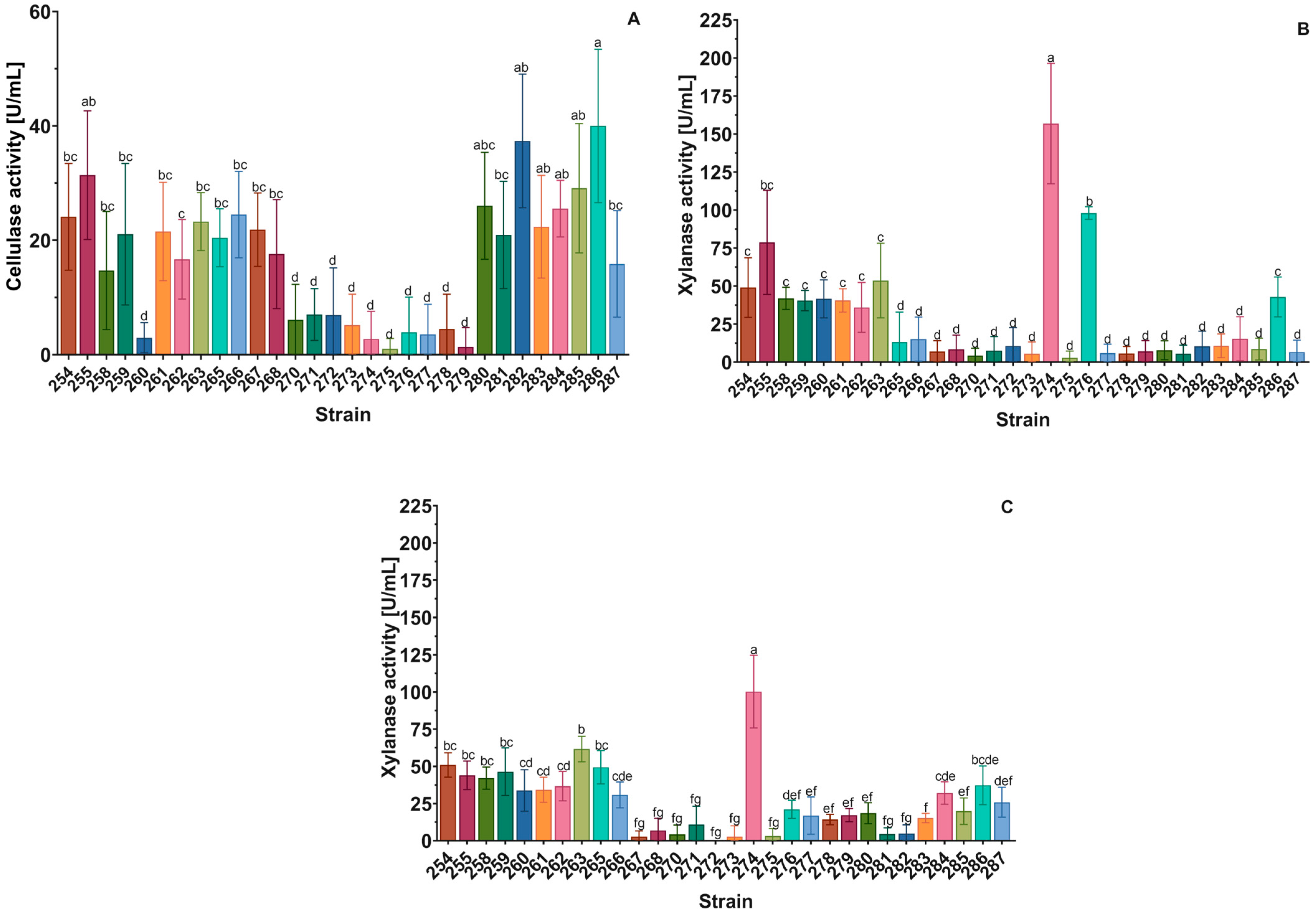
2.3.1. Cellulase and Xylanase Activity
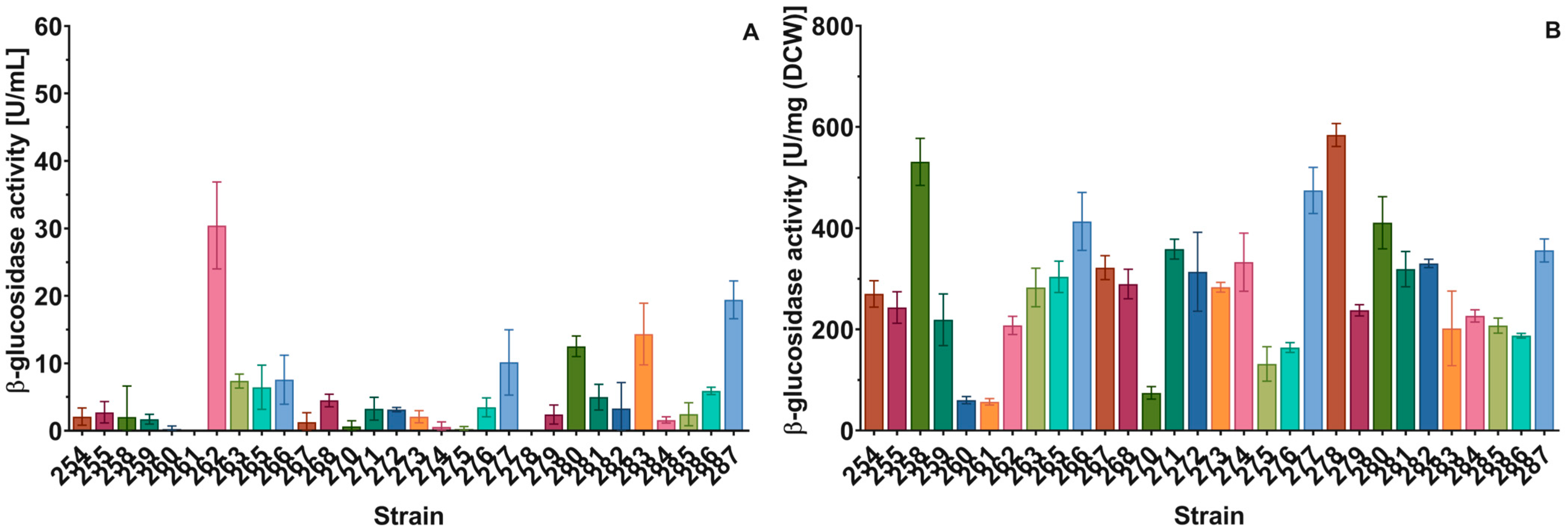
2.3.2. β-Glucosidase Activity in Permeabilized Yeast Cells
2.3.3. β-Glucosidase Activity in Culture Supernatants
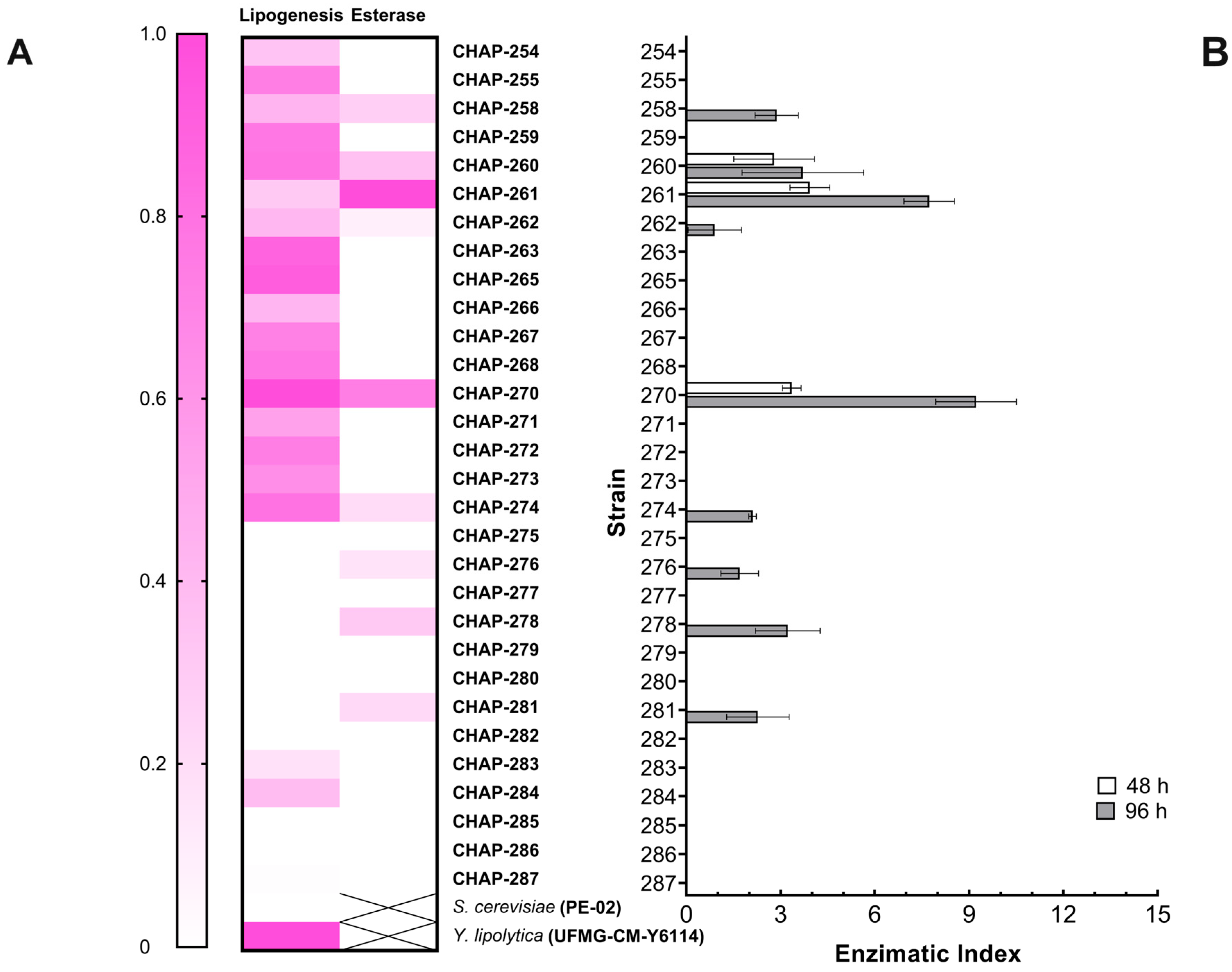
2.4. Screening of Oleaginous Yeasts
- Ya: Sample’s lipid index (0–1 scale);
- Y1: Value assigned to the negative control (0);
- Y2: Value assigned to the positive control (1);
- Xa: Mean color intensity of the sample;
- X1: Mean color intensity of the negative control;
- X2: Mean color intensity of the positive control.
2.5. Screening for Extracellular Esterase-Producing Yeasts
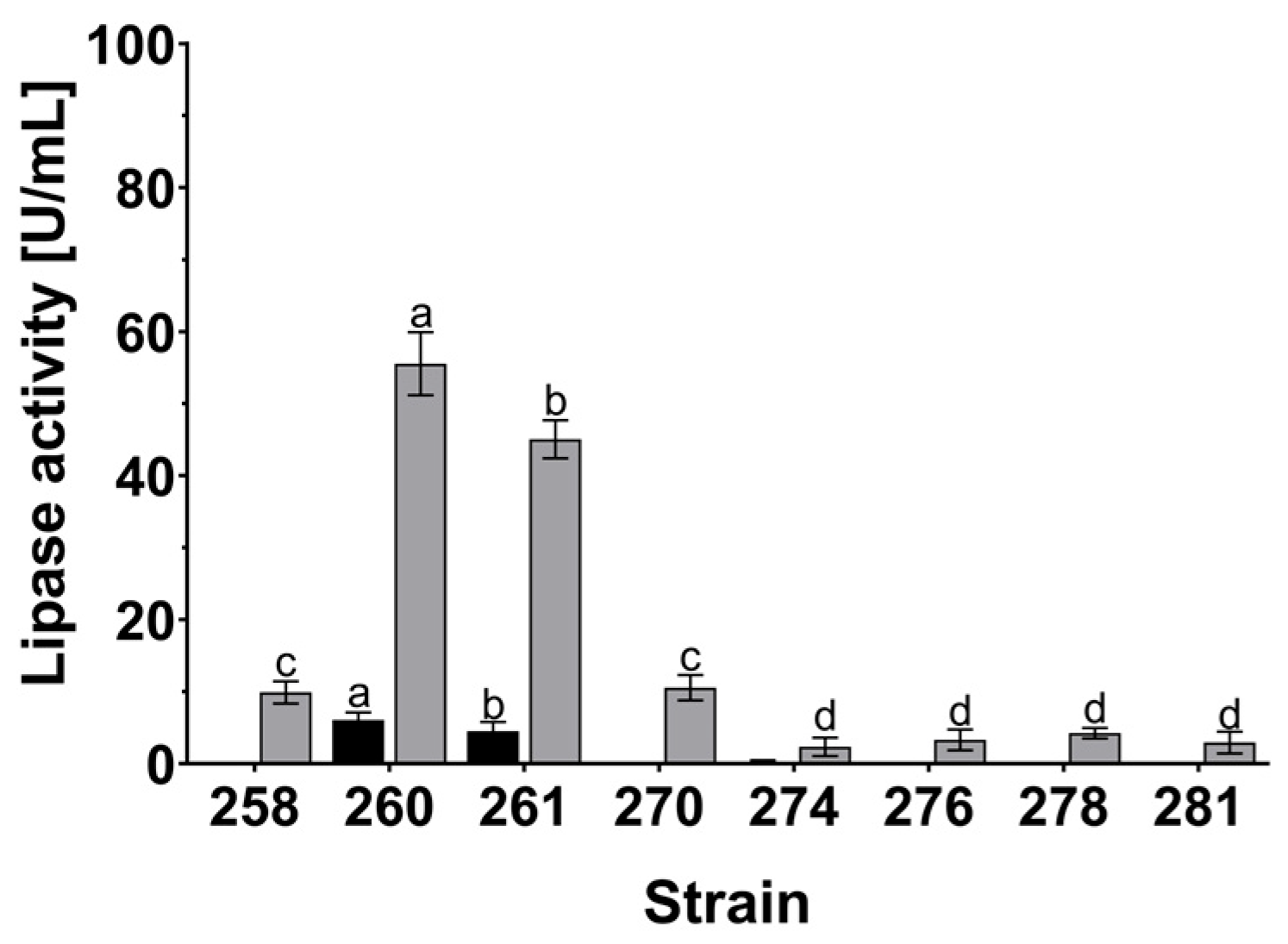
2.6. Screening for Extracellular Lipase Activity
2.7. Statistical Analysis
2.8. Genetic Fingerprinting of Yeast Isolates and UPGMA-Based Similarity Analysis
2.9. Taxonomic Analysis of Yeasts
3. Results
3.1. Specific Growth Rates
3.2. Prospecting Yeasts with High GH Activities
3.3. Oleaginous and Lipolytic Yeasts
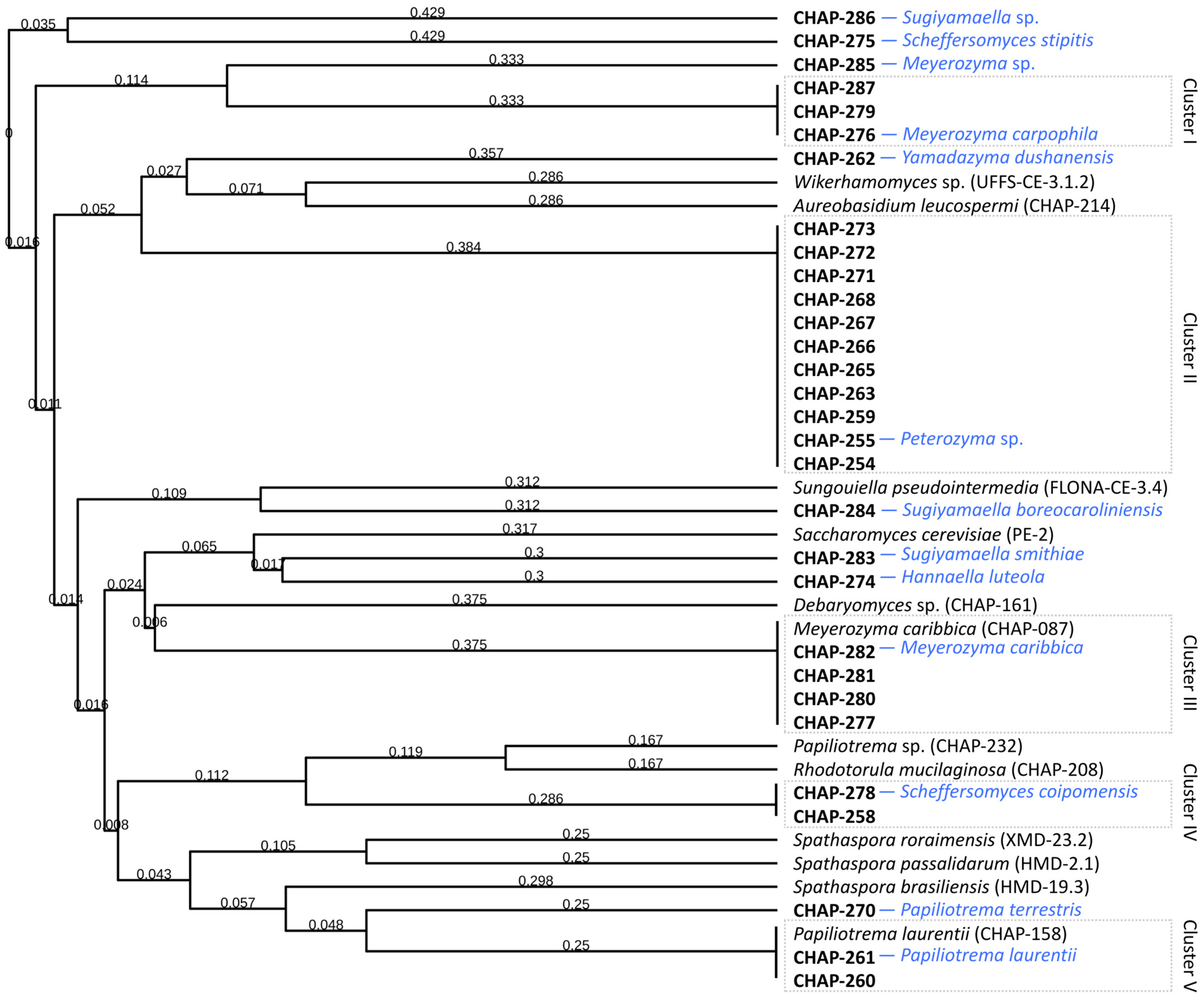
3.4. Taxonomic Identification of Yeasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alterthum, F. Biotecnologia Industrial, Volume 1—Fundamentos; Blucher: São Paulo, Brazil, 2020. [Google Scholar]
- Hasan, M.J.; Haque, P.; Rahman, M.M. Protease Enzyme Based Cleaner Leather Processing: A Review. J. Clean. Prod. 2022, 365, 132826. [Google Scholar] [CrossRef]
- Farinas, C.S.; Loyo, M.M.; Baraldo, A.; Tardioli, P.W.; Neto, V.B.; Couri, S. Finding Stable Cellulase and Xylanase: Evaluation of the Synergistic Effect of PH and Temperature. New Biotechnol. 2010, 27, 810–815. [Google Scholar] [CrossRef]
- Santos, A.A.; Kretzer, L.G.; Dourado, E.D.R.; Rosa, C.A.; Stambuk, B.U.; Alves, S.L. Expression of a Periplasmic β-Glucosidase from Yarrowia lipolytica Allows Efficient Cellobiose-Xylose Co-Fermentation by Industrial Xylose-Fermenting Saccharomyces cerevisiae Strains. Braz. J. Microbiol. 2025, 56, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Rathour, R.; Jha, S.; Pandey, K.; Srivastava, M.; Thakur, V.K.; Sengar, R.S.; Gupta, V.K.; Mazumder, P.B.; Khan, A.F.; et al. Microbial Beta Glucosidase Enzymes: Recent Advances in Biomass Conversation for Biofuels Application. Biomolecules 2019, 9, 220. [Google Scholar] [CrossRef]
- Baker, J.T.; Duarte, M.E.; Holanda, D.M.; Kim, S.W. Friend or Foe? Impacts of Dietary Xylans, Xylooligosaccharides, and Xylanases on Intestinal Health and Growth Performance of Monogastric Animals. Animals 2021, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Kaupert Neto, A.A.; Borin, G.P.; Goldman, G.H.; de Lima Damásio, A.R.; de Castro Oliveira, J.V. Insights into the Plant Polysaccharide Degradation Potential of the Xylanolytic Yeast Pseudozyma Brasiliensis. FEMS Yeast Res. 2016, 16, fov117. [Google Scholar] [CrossRef]
- Xiao, W.; Li, H.; Xia, W.; Yang, Y.; Hu, P.; Zhou, S.; Hu, Y.; Liu, X.; Dai, Y.; Jiang, Z. Co-Expression of Cellulase and Xylanase Genes in Sacchromyces cerevisiae toward Enhanced Bioethanol Production from Corn Stover. Bioengineered 2019, 10, 513–521. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Pei, Z.; Huang, J.; Wang, J.; Yang, S.; Li, H. Enhancing Lignocellulosic Biorefinery Sustainability: Mechanisms and Optimization of Microwave-Responsive Deep Eutectic Solvents for Rapid Delignification. Biofuel Res. J. 2025, 12, 2306–2318. [Google Scholar] [CrossRef]
- Huang, J.; Liu, T.; Wang, K.; Huang, Z.; Wang, J.; Rokhum, S.L.; Li, H. Room-Temperature and Carbon-Negative Production of Biodiesel via Synergy of Geminal-Atom and Photothermal Catalysis. Environ. Chem. Lett. 2024, 22, 1607–1613. [Google Scholar] [CrossRef]
- Boekhout, T.; Amend, A.S.; El Baidouri, F.; Gabaldón, T.; Geml, J.; Mittelbach, M.; Robert, V.; Tan, C.S.; Turchetti, B.; Vu, D.; et al. Trends in Yeast Diversity Discovery. Fungal Divers. 2022, 114, 491–537. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiol. Spectr. 2017, 5, FUNK-0052. [Google Scholar] [CrossRef]
- Giehl, A.; dos Santos, A.A.; Cadamuro, R.D.; Tadioto, V.; Guterres, I.Z.; Prá Zuchi, I.D.; do Amaral Minussi, G.; Fongaro, G.; Silva, I.T.; Alves, S.L. Biochemical and Biotechnological Insights into Fungus-Plant Interactions for Enhanced Sustainable Agricultural and Industrial Processes. Plants 2023, 12, 2688. [Google Scholar] [CrossRef]
- Lopes, M.R.; Lara, C.A.; Moura, M.E.F.; Uetanabaro, A.P.T.; Morais, P.B.; Vital, M.J.S.; Rosa, C.A. Characterisation of the Diversity and Physiology of Cellobiose-Fermenting Yeasts Isolated from Rotting Wood in Brazilian Ecosystems. Fungal Biol. 2018, 122, 668–676. [Google Scholar] [CrossRef]
- Sperandio, E.M.; Martins do Vale, H.M.; Moreira, G.A.M. Yeasts from Native Brazilian Cerrado Plants: Occurrence, Diversity and Use in the Biocontrol of Citrus Green Mould. Fungal Biol. 2015, 119, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Bazoti, S.F.; Golunski, S.; Pereira Siqueira, D.; Scapini, T.; Barrilli, É.T.; Alex Mayer, D.; Barros, K.O.; Rosa, C.A.; Stambuk, B.U.; Alves, S.L.; et al. Second-Generation Ethanol from Non-Detoxified Sugarcane Hydrolysate by a Rotting Wood Isolated Yeast Strain. Bioresour. Technol. 2017, 244, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.K.; Panatta, A.A.S.; Silveira, M.A.D.; Tav, C.; Johann, S.; Rodrigues, M.L.F.; Martins, C.V.B. Yeasts Isolated from a Lotic Continental Environment in Brazil Show Potential to Produce Amylase, Cellulase and Protease. Biotechnol. Rep. 2021, 30, e00630. [Google Scholar] [CrossRef]
- Abranches, J.; Valente, P.; Nóbrega, H.N.; Fernandez, F.A.S.; Mendonça-Hagler, L.C.; Hagler, A.N. Yeast Diversity and Killer Activity Dispersed in Fecal Pellets from Marsupials and Rodents in a Brazilian Tropical Habitat Mosaic. FEMS Microbiol. Ecol. 1998, 26, 27–33. [Google Scholar] [CrossRef]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How Much Is Left, and How Is the Remaining Forest Distributed? Implications for Conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- Kumar, K.K.; Deeba, F.; Pandey, A.K.; Islam, A.; Paul, D.; Gaur, N.A. Sustainable Lipid Production by Oleaginous Yeasts: Current Outlook and Challenges. Bioresour. Technol. 2025, 421, 132205. [Google Scholar] [CrossRef]
- Mitrea, L.; Călinoiu, L.-F.; Teleky, B.-E.; Szabo, K.; Martău, A.-G.; Ştefănescu, B.-E.; Dulf, F.-V.; Vodnar, D.-C. Waste Cooking Oil and Crude Glycerol as Efficient Renewable Biomass for the Production of Platform Organic Chemicals through Oleophilic Yeast Strain of Yarrowia lipolytica. Environ. Technol. Innov. 2022, 28, 102943. [Google Scholar] [CrossRef]
- Lei, Y.; Wang, X.; Sun, S.; He, B.; Sun, W.; Wang, K.; Chen, Z.; Guo, Z.; Li, Z. A Review of Lipid Accumulation by Oleaginous Yeasts: Culture Mode. Sci. Total Environ. 2024, 919, 170385. [Google Scholar] [CrossRef]
- Klug, L.; Daum, G. Yeast Lipid Metabolism at a Glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef]
- Takaku, H.; Matsuzawa, T.; Yaoi, K.; Yamazaki, H. Lipid Metabolism of the Oleaginous Yeast Lipomyces starkeyi. Appl. Microbiol. Biotechnol. 2020, 104, 6141–6148. [Google Scholar] [CrossRef] [PubMed]
- Bussamara, R.; Fuentefria, A.M.; de Oliveira, E.S.; Broetto, L.; Simcikova, M.; Valente, P.; Schrank, A.; Vainstein, M.H. Isolation of a Lipase-Secreting Yeast for Enzyme Production in a Pilot-Plant Scale Batch Fermentation. Bioresour. Technol. 2010, 101, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Meneses, D.P.; Paixão, L.M.N.; Fonteles, T.V.; Gudiña, E.J.; Rodrigues, L.R.; Fernandes, F.A.N.; Rodrigues, S. Esterase Production by Aureobasidium pullulans URM 7059 in Stirred Tank and Airlift Bioreactors Using Residual Biodiesel Glycerol as Substrate. Biochem. Eng. J. 2021, 168, 107954. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial Lipases and Their Industrial Applications: A Comprehensive Review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef]
- Ortiz, C.; Ferreira, M.L.; Barbosa, O.; Dos Santos, J.C.S.; Rodrigues, R.C.; Berenguer-Murcia, Á.; Briand, L.E.; Fernandez-Lafuente, R. Novozym 435: The “Perfect” Lipase Immobilized Biocatalyst? Catal. Sci. Technol. 2019, 9, 2380–2420. [Google Scholar] [CrossRef]
- Giraldo, L.; Gómez-Granados, F.; Moreno-Piraján, J.C. Biodiesel Production Using Palm Oil with a MOF-Lipase B Biocatalyst from Candida antarctica: A Kinetic and Thermodynamic Study. Int. J. Mol. Sci. 2023, 24, 10741. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, E.J.M.; Martins, P.M.M.; de Assis, J.G.R.; Batista, N.N.; de Oliveira Vilela, A.L.; da Rosa, S.D.V.F.; Dias, D.R.; Schwan, R.F. Self-Induced Anaerobiosis Fermentation in Coffees Inoculated with Yeast: Effect on Key Enzymes of the Germination Process and Its Relationship with the Decrease in Seed Germination. Food Res. Int. 2025, 199, 115376. [Google Scholar] [CrossRef]
- Lemes, A.C.; Silvério, S.C.; Rodrigues, S.; Rodrigues, L.R. Integrated Strategy for Purification of Esterase from Aureobasidium pullulans. Sep. Purif. Technol. 2019, 209, 409–418. [Google Scholar] [CrossRef]
- ICMBio, Chico Mendes Institute for Biodiversity Conservation. Chapecó National Forest. Available online: https://www.gov.br/icmbio/pt-br/assuntos/biodiversidade/unidade-de-conservacao/unidades-de-biomas/mata-atlantica/lista-de-ucs/flona-de-chapeco/flona-de-chapeco (accessed on 4 June 2025).
- IBGE–Instituto Brasileiro de Geografia e Estatística. Malhas Territoriais. Available online: https://www.ibge.gov.br/geociencias/organizacao-do-territorio/malhas-territoriais/15774-malhas.html (accessed on 4 June 2025).
- Tadioto, V.; Milani, L.M.; Barrilli, É.T.; Baptista, C.W.; Bohn, L.; Dresch, A.; Harakava, R.; Fogolari, O.; Mibielli, G.M.; Bender, J.P.; et al. Analysis of Glucose and Xylose Metabolism in New Indigenous Meyerozyma Caribbica Strains Isolated from Corn Residues. World J. Microbiol. Biotechnol. 2022, 38, 35. [Google Scholar] [CrossRef] [PubMed]
- Albarello, M.L.R.; Giehl, A.; Tadioto, V.; dos Santos, A.A.; Milani, L.M.; Bristot, J.C.S.; Tramontin, M.A.; Treichel, H.; Bernardi, O.; Stambuk, B.U.; et al. Analysis of the Holocellulolytic and Fermentative Potentials of Yeasts Isolated from the Gut of Spodoptera Frugiperda Larvae. Bioenergy Res. 2023, 16, 2046–2057. [Google Scholar] [CrossRef]
- dos Santos, A.A.; Deoti, J.R.; Müller, G.; Dário, M.G.; Stambuk, B.U.; Alves Junior, S.L. Dosagem de Açúcares Redutores Com o Reativo DNS Em Microplaca. Braz. J. Food Technol. 2017, 20, e2015113. [Google Scholar] [CrossRef]
- Barrilli, É.T.; Tadioto, V.; Milani, L.M.; Deoti, J.R.; Fogolari, O.; Müller, C.; Barros, K.O.; Rosa, C.A.; dos Santos, A.A.; Stambuk, B.U.; et al. Biochemical Analysis of Cellobiose Catabolism in Candida Pseudointermedia Strains Isolated from Rotten Wood. Arch. Microbiol. 2020, 202, 1729–1739. [Google Scholar] [CrossRef]
- Niehus, X.; Casas-Godoy, L.; Vargas-Sánchez, M.; Sandoval, G. A Fast and Simple Qualitative Method for Screening Oleaginous Yeasts on Agar. J. Lipids 2018, 2018, 5325804. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.M.D.; Ogaki, M.B.; Teixeira, E.A.A.; De Menezes, G.C.A.; Convey, P.; Rosa, C.A.; Rosa, L.H. Communities of Culturable Freshwater Fungi Present in Antarctic Lakes and Detection of Their Low-Temperature-Active Enzymes. Braz. J. Microbiol. 2023, 54, 1923–1933. [Google Scholar] [CrossRef]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of Genomic DNA from Yeasts for PCR-Based Applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Oliveira, V.A.; Vicente, M.A.; Fietto, L.G.; de Miranda Castro, I.; Coutrim, M.X.; Schüller, D.; Alves, H.; Casal, M.; de Oliveira Santos, J.; Araújo, L.D.; et al. Biochemical and Molecular Characterization of Saccharomyces cerevisiae Strains Obtained from Sugar-Cane Juice Fermentations and Their Impact in Cachaça Production. Appl. Environ. Microbiol. 2008, 74, 693–701. [Google Scholar] [CrossRef]
- Sipiczki, M.; Czentye, K.; Kállai, Z. High Intragenomic, Intergenomic, and Phenotypic Diversity in Pulcherrimin-Producing Metschnikowia Yeasts Indicates a Special Mode of Genome Evolution. Sci. Rep. 2024, 14, 10521. [Google Scholar] [CrossRef]
- Fenner, E.D.; Bressan, S.K.; dos Santos, A.A.; Giehl, A.; do Amaral Minussi, G.; Teixeira, E.A.A.; da Costa Diniz, M.; Werlang, L.; Fogolari, O.; Rosa, C.A.; et al. Ethanol and 2-Phenylethanol Production by Bee-Isolated Meyerozyma Caribbica Strains. Prep. Biochem. Biotechnol. 2025, 55, 359–369. [Google Scholar] [CrossRef]
- de Oliveira, C.G.; dos Santos, A.A.; Pritsch, E.J.P.; Bressan, S.K.; Giehl, A.; Fogolari, O.; Mossi, A.J.; Treichel, H.; Alves, S.L. Production of Indole-3-Acetic Acid and Degradation of 2,4-D by Yeasts Isolated from Pollinating Insects. Microorganisms 2025, 13, 1492. [Google Scholar] [CrossRef]
- Karnaouri, A.; Matsakas, L.; Krikigianni, E.; Rova, U.; Christakopoulos, P. Valorization of Waste Forest Biomass toward the Production of Cello-Oligosaccharides with Potential Prebiotic Activity by Utilizing Customized Enzyme Cocktails. Biotechnol. Biofuels 2019, 12, 285. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Cui, Q.; Feng, Y.; Xuan, J. Composition of Lignocellulose Hydrolysate in Different Biorefinery Strategies: Nutrients and Inhibitors. Molecules 2024, 29, 2275. [Google Scholar] [CrossRef] [PubMed]
- Kham, N.N.N.; Phovisay, S.; Unban, K.; Kanpiengjai, A.; Saenjum, C.; Lumyong, S.; Shetty, K.; Khanongnuch, C. A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production. J. Fungi 2024, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Duquesne, S.; Bozonnet, S.; Cioci, G.; Nicaud, J.M.; Marty, A.; O’Donohue, M.J. Development of Cellobiose-Degrading Ability in Yarrowia lipolytica Strain by Overexpression of Endogenous Genes. Biotechnol. Biofuels 2015, 8, 109. [Google Scholar] [CrossRef]
- Ramírez-Castrillón, M.; Mendes, S.D.C.; Inostroza-Ponta, M.; Valente, P. (GTG)5 MSP-PCR Fingerprinting as a Technique for Discrimination of Wine Associated Yeasts? PLoS ONE 2014, 9, e105870. [Google Scholar] [CrossRef]
- da Silva-Filho, E.A.; dos Santos, S.K.B.; do Monte Resende, A.; de Morais, J.O.F.; de Morais, M.A.; Simões, D.A. Yeast Population Dynamics of Industrial Fuel-Ethanol Fermentation Process Assessed by PCR-Fingerprinting. Antonie Leeuwenhoek 2005, 88, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Farias, D.; de Andrade, R.R.; Maugeri-Filho, F. Kinetic Modeling of Ethanol Production by Scheffersomyces stipitis from Xylose. Appl. Biochem. Biotechnol. 2014, 172, 361–379. [Google Scholar] [CrossRef]
- Adamczyk, P.A.; Coradetti, S.T.; Gladden, J.M. Non-Canonical d-Xylose and l-Arabinose Metabolism via d-Arabitol in the Oleaginous Yeast Rhodosporidium Toruloides. Microb. Cell Fact. 2023, 22, 145. [Google Scholar] [CrossRef]
- Dien, B.S.; Kurtzman, C.P.; Saha, B.C.; Bothast, R.J. Screening Forl-Arabinose Fermenting Yeasts. Appl. Biochem. Biotechnol. 1996, 57–58, 233–242. [Google Scholar] [CrossRef]
- Ko, H.; Park, Y.C. Mass Production and Characterization of an Endoglucanase from Coleoptera Insect (Monochamus saltuarius) in Yeast Kluyveromyces lactis. Protein Expr. Purif. 2024, 223, 106540. [Google Scholar] [CrossRef] [PubMed]
- Pang, A.P.; Luo, Y.; Hu, X.; Zhang, F.; Wang, H.; Gao, Y.; Durrani, S.; Li, C.; Shi, X.; Wu, F.G.; et al. Transmembrane Transport Process and Endoplasmic Reticulum Function Facilitate the Role of Gene Cel1b in Cellulase Production of Trichoderma reesei. Microb. Cell Fact. 2022, 21, 90. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, R.; Bishnoi, K.; Bishnoi, N.R. Optimization of Synergistic Parameters for Thermostable Cellulase Activity of Aspergillus Heteromorphus Using Response Surface Methodology. Biochem. Eng. J. 2009, 48, 28–35. [Google Scholar] [CrossRef]
- Tiwari, S.; Avchar, R.; Arora, R.; Lanjekar, V.; Dhakephalkar, P.K.; Dagar, S.S.; Baghela, A. Xylanolytic and Ethanologenic Potential of Gut Associated Yeasts from Different Species of Termites from India. Mycobiology 2020, 48, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.M.F.; Morais, C.G.; Lopes, M.R.; Santos, R.O.; Uetanabaro, A.P.T.; Morais, P.B.; Vital, M.J.S.; de Morais, M.A.; Lachance, M.A.; Rosa, C.A. D-Xylose Fermentation, Xylitol Production and Xylanase Activities by Seven New Species of Sugiyamaella. Antonie Leeuwenhoek 2017, 110, 53–67. [Google Scholar] [CrossRef]
- Rastogi, M.; Shrivastava, S.; Shukla, P. Bioprospecting of Xylanase Producing Fungal Strains: Multilocus Phylogenetic Analysis and Enzyme Activity Profiling. J. Basic Microbiol. 2022, 62, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Waghmare, P.R.; Dijkhuizen, L.; Meng, X.; Liu, W. Research Advances on the Consolidated Bioprocessing of Lignocellulosic Biomass. Eng. Microbiol. 2024, 4, 100139. [Google Scholar] [CrossRef]
- Cunha, J.T.; Romaní, A.; Inokuma, K.; Johansson, B.; Hasunuma, T.; Kondo, A.; Domingues, L. Consolidated Bioprocessing of Corn Cob-Derived Hemicellulose: Engineered Industrial Saccharomyces cerevisiae as Efficient Whole Cell Biocatalysts. Biotechnol. Biofuels 2020, 13, 138. [Google Scholar] [CrossRef]
- Kaur, G.; Kaur, P.; Kaur, J.; Singla, D.; Taggar, M.S. Xylanase, Xylooligosaccharide and Xylitol Production from Lignocellulosic Biomass: Exploring Biovalorization of Xylan from a Sustainable Biorefinery Perspective. Ind. Crops Prod. 2024, 215, 118610. [Google Scholar] [CrossRef]
- Bergmann, J.C.; Costa, O.Y.A.; Gladden, J.M.; Singer, S.; Heins, R.; D’haeseleer, P.; Simmons, B.A.; Quirino, B.F. Discovery of Two Novel β-Glucosidases from an Amazon Soil Metagenomic Library. FEMS Microbiol. Lett. 2014, 351, 147–155. [Google Scholar] [CrossRef]
- Lehman, R.M.; Osborne, S.L.; Ewing, P.M. When Are You Measuring Soil β-Glucosidase Activities in Cropping Systems? Agric. Environ. Lett. 2024, 9, e70002. [Google Scholar] [CrossRef]
- Miao, Y.; Zhong, Q. Isolation and Identification of β-Glucosidases-Producing Non-Saccharomyces Yeast Strains and Its Influence on the Aroma of Fermented Mango Juice. Molecules 2023, 28, 5890. [Google Scholar] [CrossRef]
- Knob, A.; Izidoro, S.C.; Lacerda, L.T.; Rodrigues, A.; de Lima, V.A. A Novel Lipolytic Yeast Meyerozyma guilliermondii: Efficient and Low-Cost Production of Acid and Promising Feed Lipase Using Cheese Whey. Biocatal. Agric. Biotechnol. 2020, 24, 101565. [Google Scholar] [CrossRef]
- Nimkande, V.D.; Bafana, A. A Review on the Utility of Microbial Lipases in Wastewater Treatment. J. Water Process Eng. 2022, 46, 102591. [Google Scholar] [CrossRef]
- Chaib, I.; Dakhmouche-Djekrif, S.; Bennamoun, L.; Nouadri, T. Extracellular Enzymes Producing Yeasts Study: Cost-Effective Production of α-Amylase by a Newly Isolated Thermophilic Yeast Geotrichum candidum PO27. AIMS Microbiol. 2024, 10, 83–106. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, S.; Aggelis, G. Lipids of Oleaginous Yeasts. Part II: Technology and Potential Applications. Eur. J. Lipid Sci. Technol. 2011, 113, 1052–1073. [Google Scholar] [CrossRef]
- Thevenieau, F.; Beopoulos, A.; Desfougeres, T.; Sabirova, J.; Albertin, K.; Zinjarde, S.; Nicaud, J.-M. Uptake and Assimilation of Hydrophobic Substrates by the Oleaginous Yeast Yarrowia lipolytica. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1513–1527. [Google Scholar]
- Dulermo, T.; Tréton, B.; Beopoulos, A.; Kabran Gnankon, A.P.; Haddouche, R.; Nicaud, J.-M. Characterization of the Two Intracellular Lipases of Y. lipolytica Encoded by TGL3 and TGL4 Genes: New Insights into the Role of Intracellular Lipases and Lipid Body Organisation. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Chhabra, M. Isolation, Identification and Characterization of Cystobasidium Oligophagum JRC1: A Cellulase and Lipase Producing Oleaginous Yeast. Bioresour. Technol. 2017, 223, 250–258. [Google Scholar] [CrossRef]
- Freitas, C.; Nobre, B.; Gouveia, L.; Roseiro, J.; Reis, A.; Silva, T.L. New at-line flow cytometric protocols for determining carotenoid content and cell viability during Rhodosporidium toruloides NCYC 921 batch growth. Process Biochem. 2014, 49, 554–562. [Google Scholar] [CrossRef]
- Bučková, M.; Puškárová, A.; Ženišová, K.; Kraková, L.; Piknová, Ľ.; Kuchta, T.; Pangallo, D. Novel Insights into Microbial Community Dynamics during the Fermentation of Central European Ice Wine. Int. J. Food Microbiol. 2018, 266, 42–51. [Google Scholar] [CrossRef]
- Lopes, M.; Araújo, C.; Aguedo, M.; Gomes, N.; Gonçalves, C.; Teixeira, J.A.; Belo, I. The Use of Olive Mill Wastewater by Wild Type Yarrowia lipolytica Strains: Medium Supplementation and Surfactant Presence Effect. J. Chem. Technol. Biotechnol. 2009, 84, 533–537. [Google Scholar] [CrossRef]
- Li, Y.; Liu, T.-J.; Zhao, M.-J.; Zhang, H.; Feng, F.Q. Screening, Purification, and Characterization of an Extracellular Lipase from Aureobasidium pullulans Isolated from Stuffed Buns Steamers. J. Zhejiang Univ. Sci. B 2019, 20, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Han, B.; Gui, X.; Wang, G.; Xu, L.; Yan, Y.; Madzak, C.; Pan, D.; Wang, Y.; Zha, G.; et al. Engineering Yarrowia lipolytica to Simultaneously Produce Lipase and Single Cell Protein from Agro-Industrial Wastes for Feed. Sci. Rep. 2018, 8, 758. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, V.M.; Borelli, B.M.; Lara, C.A.; Soares, M.A.; Pataro, C.; Bodevan, E.C.; Rosa, C.A. The Influence of Seasons and Ripening Time on Yeast Communities of a Traditional Brazilian Cheese. Food Res. Int. 2015, 69, 331–340. [Google Scholar] [CrossRef]
- Eom, G.T.; Lee, S.H.; Song, B.K.; Chung, K.-W.; Kim, Y.-W.; Song, J.K. High-Level Extracellular Production and Characterization of Candida antarctica Lipase B in Pichia pastoris. J. Biosci. Bioeng. 2013, 116, 165–170. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, X.-G.; Lu, Z.-P.; Xu, C.; Xu, H.-J.; Hu, Y. Enhancing the Catalytic Performance of Candida antarctica Lipase B by Chemical Modification with Alkylated Betaine Ionic Liquids. Front. Bioeng. Biotechnol. 2022, 10, 850890. [Google Scholar] [CrossRef]
- Surussawadee, J.; Khunnamwong, P.; Srisuk, N.; Limtong, S. Papiliotrema siamense f.a., Sp. Nov., a Yeast Species Isolated from Plant Leaves. Int. J. Syst. Evol. Microbiol. 2014, 64, 3058–3062. [Google Scholar] [CrossRef] [PubMed]
- Machado Pagani, D.; Brandão, L.R.; Santos, A.R.O.; Felix, C.R.; Pais Ramos, J.; Broetto, L.; Scorzetti, G.; Fell, J.W.; Augusto Rosa, C.; Valente, P.; et al. Papiliotrema leoncinii Sp. Nov. and Papiliotrema miconiae Sp. Nov., Two Tremellaceous Yeast Species from Brazil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1799–1806. [Google Scholar] [CrossRef]
- Maksimova, I.A.; Glushakova, A.M.; Thanh, V.N.; Kachalkin, A.V. Yamadazyma cocois f.a., Sp. Nov., an Ascomycetous Yeast Isolated from Coconuts. Int. J. Syst. Evol. Microbiol. 2020, 70, 3491–3496. [Google Scholar] [CrossRef]
- Khunnamwong, P.; Nualthaisong, P.; Sakolrak, B.; Nutaratat, P.; Limtong, S. Yamadazyma sisaketensis f.a., Sp. Nov. and Yamadazyma koratensis f.a., Sp. Nov., Two Novel Ascomycetous Yeast Species from Mushrooms and Cocoa Leaves in Thailand, and Reassignment of Candida Andamanensis, Candida jaroonii and Candida songkhlaensis to the Genus Yamadazyma. Int. J. Syst. Evol. Microbiol. 2023, 73, 006174. [Google Scholar] [CrossRef]
- Avesani, M.; Zapparoli, G.; Jindamorakot, S.; Limtong, S. Yamadazyma oleae f.a. Sp. Nov. and Yamadazyma molendinolei f.a. Sp. Nov., Two Novel Ascomycetous Yeast Species Isolated from Olive Oil Mills in Italy, and Reassignment of 11 Candida Species to the Genus Yamadazyma. Int. J. Syst. Evol. Microbiol. 2024, 74, 006592. [Google Scholar] [CrossRef]
- Seike, T.; Takekata, H.; Sakata, N.; Furusawa, C.; Matsuda, F. Yamadazyma thunbergiae Sp. Nov., a Novel Yeast Species Associated with Bengal Clock Vines and Soil in Okinawa, Japan. Int. J. Syst. Evol. Microbiol. 2024, 74, 006537. [Google Scholar] [CrossRef]
- Gao, W.-L.; Li, Y.; Chai, C.-Y.; Yan, Z.-L.; Hui, F.-L. New Species of Yamadazyma from Rotting Wood in China. MycoKeys 2021, 83, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Cadete, R.M.; Melo, M.A.; Lopes, M.R.; Pereira, G.M.D.; Zilli, J.E.; Vital, M.J.S.; Gomes, F.C.O.; Lachance, M.-A.; Rosa, C.A. Candida amazonensis Sp. Nov., an Ascomycetous Yeast Isolated from Rotting Wood in the Amazonian Forest. Int. J. Syst. Evol. Microbiol. 2012, 62, 1438–1440. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.R.; Batista, T.M.; Franco, G.R.; Ribeiro, L.R.; Santos, A.R.O.; Furtado, C.; Moreira, R.G.; Goes-Neto, A.; Vital, M.J.S.; Rosa, L.H.; et al. Scheffersomyces stambukii f.a., Sp. Nov., a d-Xylose-Fermenting Species Isolated from Rotting Wood. Int. J. Syst. Evol. Microbiol. 2018, 68, 2306–2312. [Google Scholar] [CrossRef]
- Suh, S.-O.; Houseknecht, J.L.; Gujjari, P.; Zhou, J.J. Scheffersomyces parashehatae f.a., Sp. Nov., Scheffersomyces xylosifermentans f.a., Sp. Nov., Candida broadrunensis Sp. Nov. and Candida manassasensis Sp. Nov., Novel Yeasts Associated with Wood-Ingesting Insects, and Their Ecological and Biofuel Implications. Int. J. Syst. Evol. Microbiol. 2013, 63, 4330–4339. [Google Scholar] [CrossRef]
- Lara, C.A.; Santos, R.O.; Cadete, R.M.; Ferreira, C.; Marques, S.; Gírio, F.; Oliveira, E.S.; Rosa, C.A.; Fonseca, C. Identification and Characterisation of Xylanolytic Yeasts Isolated from Decaying Wood and Sugarcane Bagasse in Brazil. Antonie Leeuwenhoek 2014, 105, 1107–1119. [Google Scholar] [CrossRef]
- Landell, M.F.; Brandão, L.R.; Barbosa, A.C.; Ramos, J.P.; Safar, S.V.B.; Gomes, F.C.O.; Sousa, F.M.P.; Morais, P.B.; Broetto, L.; Leoncini, O.; et al. Hannaella pagnoccae Sp. Nov., a Tremellaceous Yeast Species Isolated from Plants and Soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 1970–1977. [Google Scholar] [CrossRef]
- Aires, A.; Gonçalves, C.; Sampaio, J.P. Hannaella Floricola Sp. Nov., a Novel Basidiomycetous Yeast Species Isolated from a Flower of Lantana Camara in Portugal. Int. J. Syst. Evol. Microbiol. 2023, 73, 005740. [Google Scholar] [CrossRef] [PubMed]
- Vitanović, E.; Aldrich, J.R.; Boundy-Mills, K.; Čagalj, M.; Ebeler, S.E.; Burrack, H.; Zalom, F.G. Olive Fruit Fly, Bactrocera oleae (Diptera: Tephritidae), Attraction to Volatile Compounds Produced by Host and Insect-Associated Yeast Strains. J. Econ. Entomol. 2020, 113, 752–759. [Google Scholar] [CrossRef]
- Menkis, A.; Lynikienė, J.; Marčiulynas, A.; Gedminas, A.; Povilaitienė, A. The Great Spruce Bark Beetle (Dendroctonus micans Kug.) (Coleoptera: Scolytidae) in Lithuania: Occurrence, Phenology, Morphology and Communities of Associated Fungi. Bull. Entomol. Res. 2017, 107, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Martini, A. Extracellular Enzymatic Activity Profiles in Yeast and Yeast-like Strains Isolated from Tropical Environments. J. Appl. Microbiol. 2002, 93, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, E.L.M.; Ventorim, R.Z.; de Moura Ferreira, M.A.; da Silveira, W.B. Papiliotrema laurentii: General Features and Biotechnological Applications. Appl. Microbiol. Biotechnol. 2022, 106, 6963–6976. [Google Scholar] [CrossRef] [PubMed]
- Yalçın, H.T.; Fındık, B.; Terzi, Y.; Uyar, E.; Shatila, F. Isolation and Molecular Identification of Industrially Important Enzyme Producer Yeasts from Tree Barks and Fruits. Arch. Microbiol. 2021, 203, 1079–1088. [Google Scholar] [CrossRef]
- Sitepu, I.; Selby, T.; Lin, T.; Zhu, S.; Boundy-Mills, K. Carbon Source Utilization and Inhibitor Tolerance of 45 Oleaginous Yeast Species. J. Ind. Microbiol. Biotechnol. 2014, 41, 1061–1070. [Google Scholar] [CrossRef]
- Li, J.; Yang, T.; Yuan, F.; Lv, X.; Zhou, Y. Inhibitory Effect and Potential Antagonistic Mechanism of Isolated Epiphytic Yeasts against Botrytis Cinerea and Alternaria Alternata in Postharvest Blueberry Fruits. Foods 2024, 13, 1334. [Google Scholar] [CrossRef]
- Palmieri, D.; Ianiri, G.; Conte, T.; Castoria, R.; Lima, G.; De Curtis, F. Influence of Biocontrol and Integrated Strategies and Treatment Timing on Plum Brown Rot Incidence and Fungicide Residues in Fruits. Agriculture 2022, 12, 1656. [Google Scholar] [CrossRef]
- Jiang, Y.-L.; Bao, W.-J.; Liu, F.; Wang, G.-S.; Yurkov, A.M.; Ma, Q.; Hu, Z.-D.; Chen, X.-H.; Zhao, W.-N.; Li, A.-H.; et al. Proposal of One New Family, Seven New Genera and Seventy New Basidiomycetous Yeast Species Mostly Isolated from Tibet and Yunnan Provinces, China. Stud. Mycol. 2024, 109, 57–154. [Google Scholar] [CrossRef] [PubMed]


| Strain a | Substrate and Temperature of Isolation | Species | LSU GenBank Code b or Reference |
|---|---|---|---|
| CHAP-254 | Soil—11 °C | Peterozyma sp. | - |
| CHAP-255 | Soil—11 °C | Peterozyma sp. | PV994468 |
| CHAP-258 | Soil—11 °C | Scheffersomyces coipomensis | - |
| CHAP-259 | Soil—11 °C | Peterozyma sp. | - |
| CHAP-260 | Araucaria’s bark—11 °C | Papiliotrema laurentii | - |
| CHAP-261 | Araucaria’s bark—11 °C | Papiliotrema laurentii | PV994616 |
| CHAP-262 | Araucaria’s bark—11 °C | Yamadazyma dushanensis | PV994689 |
| CHAP-263 | Araucaria’s bark—11 °C | Peterozyma sp. | - |
| CHAP-265 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-266 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-267 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-268 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-270 | Litter—11 °C | Papiliotrema terrestris | PX022965 |
| CHAP-271 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-272 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-273 | Litter—11 °C | Peterozyma sp. | - |
| CHAP-274 | Litter—11 °C | Hannaella luteola | PV995078 |
| CHAP-275 | Litter—11 °C | Scheffersomyces stipitis | PV995079 |
| CHAP-276 | Soil—30 °C | Meyerozymacarpophila | PV995110 |
| CHAP-277 | Soil—30 °C | Meyerozyma caribbica | - |
| CHAP-278 | Soil—30 °C | Scheffersomyces coipomensis | PV995111 |
| CHAP-279 | Araucaria’s bark—30 °C | Meyerozyma carpophila | - |
| CHAP-280 | Araucaria’s bark—30 °C | Meyerozyma caribbica | - |
| CHAP-281 | Araucaria’s bark—30 °C | Meyerozyma caribbica | - |
| CHAP-282 | Litter—30 °C | Meyerozyma caribbica | PV995221 |
| CHAP-283 | Litter—30 °C | Sugiyamaella smithiae | PV995348 |
| CHAP-284 | Litter—30 °C | Sugiyamaella boreocaroliniensis | PV995352 |
| CHAP-285 | Litter—30 °C | Meyerozyma sp. | PV995365 |
| CHAP-286 | Litter—30 °C | Sugiyamaella sp. | PV995366 |
| CHAP-287 | Litter—30 °C | Meyerozyma carpophila | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giehl, A.; dos Santos, A.A.; Werlang, L.; Teixeira, E.A.A.; Lopes, J.C.; Treichel, H.; Duarte, R.T.D.; Rosa, C.A.; Stambuk, B.U.; Alves, S.L., Jr. Prospecting Araucaria-Associated Yeasts for Second-Generation Biorefineries. Sustainability 2025, 17, 8134. https://doi.org/10.3390/su17188134
Giehl A, dos Santos AA, Werlang L, Teixeira EAA, Lopes JC, Treichel H, Duarte RTD, Rosa CA, Stambuk BU, Alves SL Jr. Prospecting Araucaria-Associated Yeasts for Second-Generation Biorefineries. Sustainability. 2025; 17(18):8134. https://doi.org/10.3390/su17188134
Chicago/Turabian StyleGiehl, Anderson, Angela A. dos Santos, Larissa Werlang, Elisa A. A. Teixeira, Joana C. Lopes, Helen Treichel, Rubens T. D. Duarte, Carlos A. Rosa, Boris U. Stambuk, and Sérgio L. Alves, Jr. 2025. "Prospecting Araucaria-Associated Yeasts for Second-Generation Biorefineries" Sustainability 17, no. 18: 8134. https://doi.org/10.3390/su17188134
APA StyleGiehl, A., dos Santos, A. A., Werlang, L., Teixeira, E. A. A., Lopes, J. C., Treichel, H., Duarte, R. T. D., Rosa, C. A., Stambuk, B. U., & Alves, S. L., Jr. (2025). Prospecting Araucaria-Associated Yeasts for Second-Generation Biorefineries. Sustainability, 17(18), 8134. https://doi.org/10.3390/su17188134










