The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants
Abstract
:1. Introduction
2. Interactions between Plants and Microorganisms under Current and Climate Change Scenarios
2.1. Systemic Responses of Plants upon Phytopathogen Perception
2.2. Plant Defense Responses under Climate Change Scenarios
2.2.1. Population Dynamics of Native Phytopathogens
2.2.2. Phytopathogen Evolution
2.2.3. Systemic Acquired Resistance Signaling under Increased Temperature
2.3. Current Management Strategies Employing Plants’ ISR
2.3.1. Systemic Responses of Plants upon Beneficial Microorganisms’ Perception
2.3.2. Boosting Wheat Immunity: The Use of ISR-Inducing Microorganisms to Mitigate Phytopathogen Impact under Climate Change Scenarios
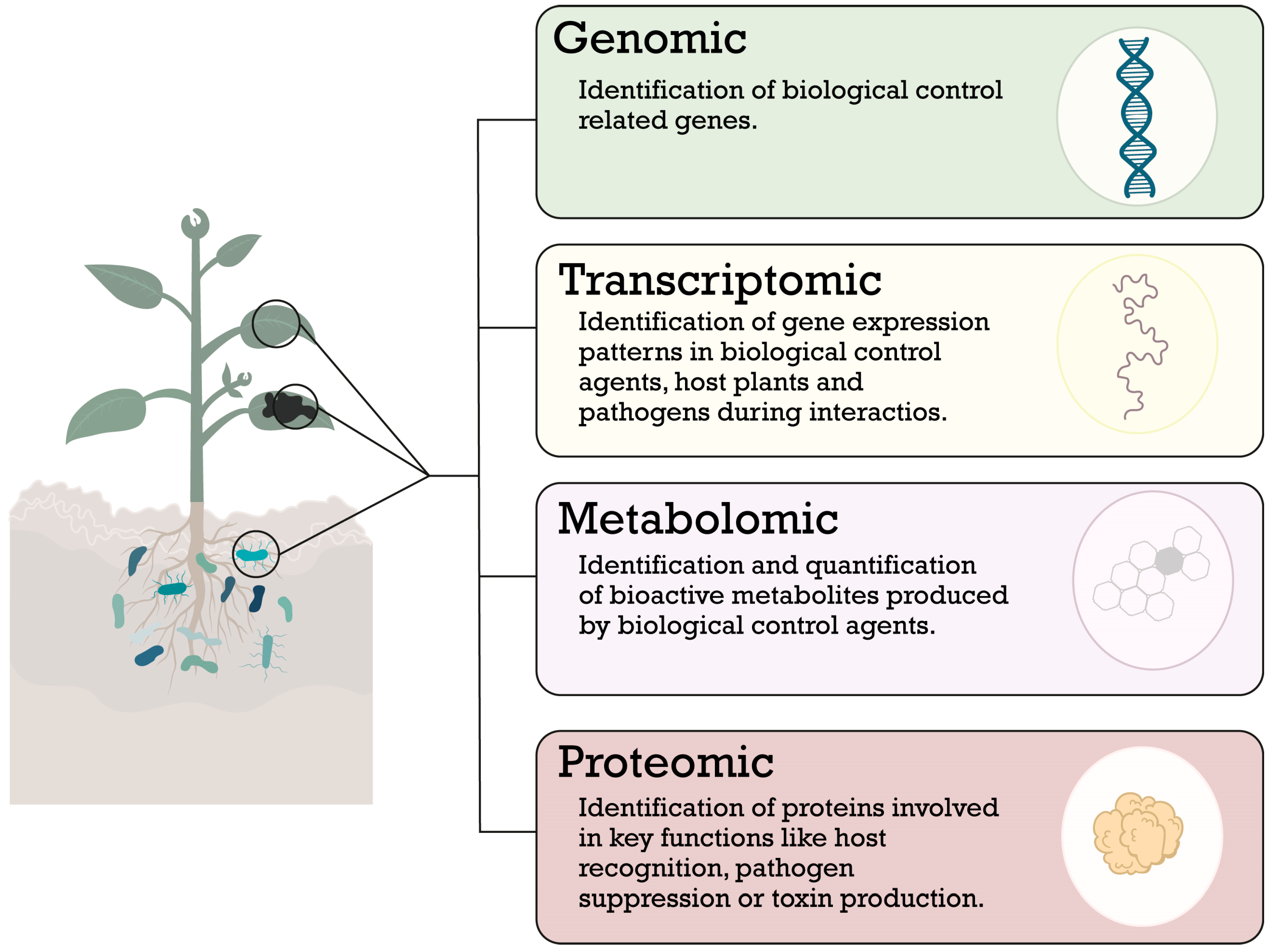
3. Elucidation of Plant and Microorganism Interactions for the Bioprospection of Beneficial Microorganisms
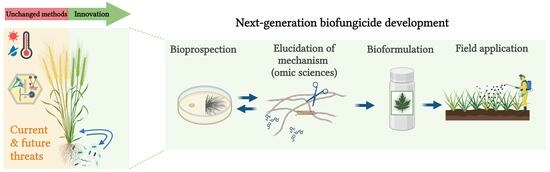
4. Development and On-Field Application of Beneficial Microorganisms
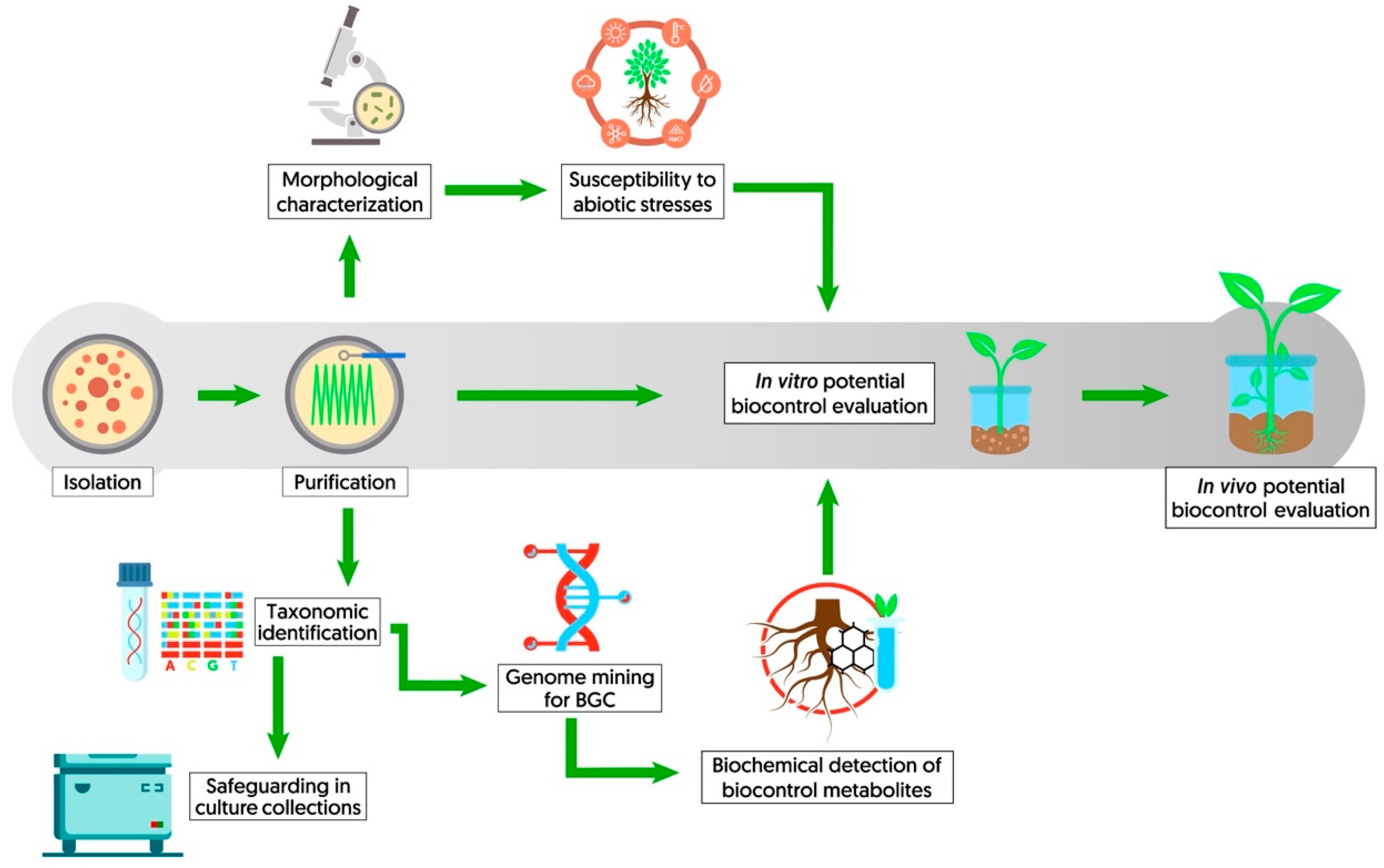
4.1. Bioprospection of a Biocontrol Microorganism
4.2. Formulation of Bioinoculants for Field Application
4.3. Field Application of Bioinoculants for Wheat: Existing Commercial Products and Guidelines
- The product must be of good quality (at least 1 × 107 viable cells g−1) and purchased from a reputed supplier, as well as applied according to the recommendations of the dose.
- The product must be used for the crop(s) specified on the product label.
- While inoculating, excess culture should be inoculated, or any remnants/residual culture should be immediately put in grooves of the field so that inoculum microorganisms start interacting with other microbiota in the rhizosphere and begin colonizing the rhizosphere.
- To achieve major/expected shelf life, the product should be stored in cool places and away from light sources (room temperature 25–28 °C or cooler, depending on the type of microbial product).
- Direct contact of the product with herbicides/weedicides/pesticides should be avoided.
- It is important to have detailed information about the strains and ingredients to know the ideal soil conditions for its application. For example, if the soil is highly acidic, the integration of soil amendments (lime or rock phosphate) is recommended.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCAs | Biological control agents |
| BGC | Biosynthetic gene clusters |
| CC | Coiled-coil |
| CFSs | Cell-free supernatants |
| CFU | Colony-forming units |
| ET | Ethylene |
| ETI | Effector-triggered immunity |
| ISR | Induced systemic resistance |
| JA | Jasmonic acid |
| MAMPs | Microbial-associated molecular patterns |
| MAPKs | Mitogen-activated protein kinases |
| MTI | Microbe-triggered immunity |
| NB-LRR | Nucleotide-binding and leucine-rich repeat |
| PAMPs | Pathogen-associated molecular patterns |
| PGPB | Plant growth-promoting bacterium |
| PR | Pathogenesis-related |
| PRRs | Pathogen recognition receptors |
| PTI | Pattern-triggered immunity |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
| SNP | Single-nucleotide polymorphism |
| TIR | Interleukin-1 receptor-like |
| UV | Ultraviolet |
| WDG | Water-dispersible granules |
| WGs | Wettable granules |
| WPs | Wettable powders |
References
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide research trends on wheat and barley: A bibliometric comparative analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef]
- USDA. FAS Wheat Explorer. Available online: https://ipad.fas.usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=0410000 (accessed on 3 April 2023).
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; Morley, T.; Rau, M.L.; Saghai, Y. A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat. Food 2021, 2, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Dalmau, J.; Berbel, J.; Ordóñez-Fernández, R. Nitrogen fertilization. A review of the risks associated with the inefficiency of its use and policy responses. Sustainability 2021, 13, 5625. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef]
- Beres, B.L.; Rahmani, E.; Clarke, J.M.; Grassini, P.; Pozniak, C.J.; Geddes, C.M.; Porker, K.D.; May, W.E.; Ransom, J.K. A systematic review of durum wheat: Enhancing production systems by exploring genotype, environment, and management (G × E × M) synergies. Front. Plant Sci. 2020, 11, 568657. [Google Scholar] [CrossRef]
- Dhakal, A.; Adhikari, C.; Manandhar, D.; Bhattarai, S.; Shrestha, S. Effect of abiotic stress in wheat: A review. Rev. Food Agric. 2021, 2, 69–72. [Google Scholar] [CrossRef]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef]
- Liu, B.; Martre, P.; Ewert, F.; Porter, J.R.; Challinor, A.J.; Müller, C.; Ruane, A.C.; Waha, K.; Thorburn, P.J.; Aggarwal, P.K.; et al. Global wheat production with 1.5 and 2.0 °c above pre-industrial warming. Glob. Chang. Biol. 2019, 25, 1428–1444. [Google Scholar] [CrossRef]
- Zhang, T.; He, Y.; DePauw, R.; Jin, Z.; Garvin, D.; Yue, X.; Anderson, W.; Li, T.; Dong, X.; Zhang, T.; et al. Climate change may outpace current wheat breeding yield improvements in north america. Nat. Commun. 2022, 13, 5591. [Google Scholar] [CrossRef]
- Bebber, D.P.; Holmes, T.; Gurr, S.J. The global spread of crop pests and pathogens. Glob. Ecol. Biogeogr. 2014, 23, 1398–1407. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Bebber, D.P.; Ramotowski, M.A.T.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Campos, M.; Góngora-Canul, C.; Das, S.; Kabir, M.R.; Valent, B.; Cruz, C.D. Epidemiological criteria to support breeding tactics against the emerging, high-consequence wheat blast disease. Plant Dis. 2020, 104, 2252–2261. [Google Scholar] [CrossRef]
- Villa-Rodríguez, E.; Parra-Cota, F.; Castro-Longoria, E.; López-Cervantes, J.; de los Santos-Villalobos, S. Bacillus subtilis TE3: A promising biological control agent against Bipolaris Sorokiniana, the causal agent of spot blotch in wheat (Triticum turgidum L. subsp. durum). Biol. Control 2019, 132, 135–143. [Google Scholar] [CrossRef]
- Halecki, W.; Bedla, D. Global wheat production and threats to supply chains in a volatile climate change and energy crisis. Resources 2022, 11, 118. [Google Scholar] [CrossRef]
- Ahmad, M.; Nadeem, S.M.; Zahir, Z.A. Plant-microbiome interactions in agroecosystem: An application. In Microbiome in Plant Health and Disease: Challenges and Opportunities; Springer: Singapore, 2019; pp. 251–291. ISBN 9789811384950. [Google Scholar]
- Xiong, C.; Lu, Y. Microbiomes in agroecosystem: Diversity, function and assembly mechanisms. Environ. Microbiol. Rep. 2022, 14, 833–849. [Google Scholar] [CrossRef]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Chen, Y.; Wenig, M.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250. [Google Scholar] [CrossRef]
- Zeier, J. Metabolic regulation of systemic acquired resistance. Curr. Opin. Plant Biol. 2021, 62, 102050. [Google Scholar] [CrossRef]
- Shine, M.B.; Xiao, X.; Kachroo, P.; Kachroo, A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 2019, 279, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-D.; BI, W.-S.; GAO, J.; YU, X.-M.; Wang, H.-Y.; Liu, D.-Q. Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 2018, 17, 2468–2477. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, L.; Liu, X.; Zhang, Q.; Zhuansun, X.; Fahima, T.; Krugman, T.; Sun, Q.; Xie, C. Glycerol-induced powdery mildew resistance in wheat by regulating plant fatty acid metabolism, plant hormones cross-talk, and pathogenesis-related genes. Int. J. Mol. Sci. 2020, 21, 673. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.O.; Friesen, T.L.; Xu, S.S.; Chen, M.S.; Giron, D.; Stuart, J.J. Pivoting from Arabidopsis to wheat to understand how agricultural plants integrate responses to biotic stress. J. Exp. Bot. 2015, 66, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.N.; Ren, M.; Zhan, J. Modeling plant diseases under climate change: Evolutionary perspectives. Trends Plant Sci. 2023, 28, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, E.J.; Miranda, B.R.; Dreaden, T.J.; Pinchot, C.C.; Jacobs, D.F. Beyond blight: Phytophthora root rot under climate change limits populations of reintroduced American chestnut. Ecosphere 2022, 13, e3917. [Google Scholar] [CrossRef]
- Shew, A.M.; Durand-Morat, A.; Putman, B.; Nalley, L.L.; Ghosh, A. Rice intensification in Bangladesh improves economic and environmental welfare. Environ. Sci. Policy 2019, 95, 46–57. [Google Scholar] [CrossRef]
- Parikka, P.; Hakala, K.; Tiilikkala, K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climatic change. Food Addit. Contam.—Part A 2012, 29, 1543–1555. [Google Scholar] [CrossRef]
- Manzotti, A.; Bergna, A.; Burow, M.; Jørgensen, H.J.L.; Cernava, T.; Berg, G.; Collinge, D.B.; Jensen, B. Insights into the community structure and lifestyle of the fungal root endophytes of tomato by combining amplicon sequencing and isolation approaches with phytohormone profiling. FEMS Microbiol. Ecol. 2020, 96, fiaa052. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Lugo-Enríquez, C.; de los Santos-Villalobos, S.; Parra-Cota, F.I.; Figueroa-López, P. First report of Cochliobolus sativus causing spot blotch on durum wheat (Triticum durum) in the Yaqui Valley, Mexico. Plant Dis. 2016, 100, 2329. [Google Scholar] [CrossRef]
- Raza, M.M.; Bebber, D.P. Climate change and plant pathogens. Curr. Opin. Microbiol. 2022, 70, 102233. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hilleary, R.; Seroka, A.; He, S.Y. Crops of the future: Building a climate-resilient plant immune system. Curr. Opin. Plant Biol. 2021, 60, 101997. [Google Scholar] [CrossRef] [PubMed]
- Frantzeskakis, L.; Di Pietro, A.; Rep, M.; Schirawski, J.; Wu, C.H.; Panstruga, R. Rapid evolution in plant–microbe interactions—A molecular genomics perspective. New Phytol. 2020, 225, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Buscaill, P.; van der Hoorn, R.A.L. Defeated by the nines: Nine extracellular strategies to avoid microbe-associated molecular patterns recognition in plants. Plant Cell 2021, 33, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Vy, T.T.P.; Yoshida, K.; Asano, H.; Mitsuoka, C.; Asuke, S.; Anh, V.L.; Cumagun, C.J.R.; Chuma, I.; Terauchi, R.; et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 2017, 357, 80–83. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Long, S.P. 30 years of free-air carbon dioxide enrichment (face): What have we learned about future crop productivity and its potential for adaptation? Glob. Chang. Biol. 2021, 27, 27–49. [Google Scholar] [CrossRef]
- Wu, E.J.; Wang, Y.P.; Yahuza, L.; He, M.H.; Sun, D.L.; Huang, Y.M.; Liu, Y.C.; Yang, L.N.; Zhu, W.; Zhan, J. Rapid adaptation of the irish potato famine pathogen Phytophthora infestans to changing temperature. Evol. Appl. 2020, 13, 768–780. [Google Scholar] [CrossRef]
- Moore, C.E.; Meacham-Hensold, K.; Lemonnier, P.; Slattery, R.A.; Benjamin, C.; Bernacchi, C.J.; Lawson, T.; Cavanagh, A.P. The effect of increasing temperature on crop photosynthesis: From enzymes to ecosystems. J. Exp. Bot. 2021, 72, 2822–2844. [Google Scholar] [CrossRef]
- Cohen, S.P.; Leach, J.E. High temperature-induced plant disease susceptibility: More than the sum of its parts. Curr. Opin. Plant Biol. 2020, 56, 235–241. [Google Scholar] [CrossRef]
- Onaga, G.; Wydra, K.D.; Koopmann, B.; Séré, Y.; Von Tiedemann, A. Elevated temperature increases in planta expression levels of virulence related genes in Magnaporthe oryzae and compromises resistance in Oryza sativa cv. nipponbare. Funct. Plant Biol. 2017, 44, 358–371. [Google Scholar] [CrossRef]
- Mikkelsen, B.L.; Jørgensen, R.B.; Lyngkjær, M.F. Complex interplay of future climate levels of CO2, ozone and temperature on susceptibility to fungal diseases in barley. Plant Pathol. 2015, 64, 319–327. [Google Scholar] [CrossRef]
- Janda, M.; Lamparová, L.; Zubíková, A.; Burketová, L.; Martinec, J.; Krčková, Z. Temporary heat stress suppresses pamp-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol. Plant Pathol. 2019, 20, 1005–1012. [Google Scholar] [CrossRef]
- Kim, J.H.; Castroverde, C.D.M.; Huang, S.; Li, C.; Hilleary, R.; Seroka, A.; Sohrabi, R.; Medina-Yerena, D.; Huot, B.; Wang, J.; et al. Increasing the resilience of plant immunity to a warming climate. Nature 2022, 607, 339–344. [Google Scholar] [CrossRef]
- Nnadi, N.E.; Carter, D.A. Climate change and the emergence of fungal pathogens. PLoS Pathog. 2021, 17, e1009503. [Google Scholar] [CrossRef]
- Romera, F.J.; García, M.J.; Lucena, C.; Martínez-Medina, A.; Aparicio, M.A.; Ramos, J.; Alcántara, E.; Angulo, M.; Pérez-Vicente, R. Induced systemic resistance (ISR) and Fe deficiency responses in dicot plants. Front. Plant Sci. 2019, 10, 444126. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Bryant, R. Effects of Temperature on Wheat-Pathogen Interactions. Ph.D. Thesis, University of East Anglia, Norwich, UK, 2013. [Google Scholar]
- Chaparro-Encinas, L.A.; Parra-Cota, F.I.; Cruz-Mendívil, A.; Santoyo, G.; Peña-Cabriales, J.J.; Castro-Espinoza, L.; de los Santos-Villalobos, S. Transcriptional regulation of cell growth and reprogramming of systemic response in wheat (Triticum turgidum subsp. durum) seedlings by Bacillus paralicheniformis TRQ65. Planta 2022, 255, 56. [Google Scholar] [CrossRef]
- Le Mire, G.; Siah, A.; Brisset, M.N.; Gaucher, M.; Deleu, M.; Jijakli, M.H. Surfactin protects wheat against Zymoseptoria tritici and activates both Salicylic Acid- and Jasmonic Acid-dependent defense responses. Agriculture 2018, 8, 11. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A.; Terefe, T.; Mhlongo, M.I. Metabolomic evaluation of PGPR defence priming in wheat (Triticum aestivum L.) cultivars infected with Puccinia Striiformis f. sp. tritici (stripe rust). Front. Plant Sci. 2023, 14, 1103413. [Google Scholar] [CrossRef]
- Mejri, S.; Siah, A.; Coutte, F.; Magnin-Robert, M.; Randoux, B.; Tisserant, B.; Krier, F.; Jacques, P.; Reignault, P.; Halama, P. Biocontrol of the wheat pathogen Zymoseptoria tritici using cyclic lipopeptides from Bacillus subtilis. Environ. Sci. Pollut. Res. 2018, 25, 29822–29833. [Google Scholar] [CrossRef]
- Moya-Elizondo, E.A.; Jacobsen, B.J. Integrated Management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of Systemic Acquired Resistance (SAR). Biol. Control 2016, 92, 153–163. [Google Scholar] [CrossRef]
- Samain, E.; Aussenac, T.; Selim, S. The effect of plant genotype, growth stage, and Mycosphaerella graminicola strains on the efficiency and durability of wheat-induced resistance by Paenibacillus sp. strain B2. Front. Plant Sci. 2019, 10, 587. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, T.; Li, Y.; Wang, X.; Chen, J. Effects of Trichoderma atroviride SG3403 and Bacillus subtilis 22 on the biocontrol of wheat head blight. J. Fungi 2022, 8, 1250. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.B.; Malviya, D.; Wasiullah; Singh, S.; Imran, M.; Pathak, N.; Alam, M.; Rai, J.P.; Singh, R.K.; Sarma, B.K.; et al. Compatible salt-tolerant rhizosphere microbe-mediated induction of phenylpropanoid cascade and induced systemic responses against Bipolaris sorokiniana (Sacc.) Shoemaker causing spot blotch disease in wheat (Triticum aestivum L.). Appl. Soil Ecol. 2016, 108, 300–306. [Google Scholar] [CrossRef]
- Tiwari, M.; Singh, R.; Jha, R.; Singh, P. Heritable priming by Trichoderma: A sustainable approach for wheat protection against Bipolaris sorokiniana. Front. Plant Sci. 2022, 13, 1050765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.T.; Zhang, H.; Tang, W. Transcriptomic analysis of wheat seedling responses to the systemic acquired resistance inducer n-hydroxypipecolic acid. Front. Microbiol. 2021, 12, 621336. [Google Scholar] [CrossRef]
- Hao, G.; Tiley, H.; McCormick, S. Chitin triggers tissue-specific immunity in wheat associated with Fusarium head blight. Front. Plant Sci. 2022, 13, 832502. [Google Scholar] [CrossRef]
- Jasem, A.M.; Sharifi, R.; Abbasi, S. Induced Systemic Resistance to wheat take-all disease by probiotic bacteria. J. Plant Prot. Res. 2018, 58, 304–310. [Google Scholar] [CrossRef]
- Li, L.; Guo, N.; Feng, Y.; Duan, M.; Li, C. Effect of Piriformospora indica-induced systemic resistance and basal immunity against Rhizoctonia cerealis and Fusarium graminearum in wheat. Front. Plant Sci. 2022, 13, 836940. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. The multifarious PGPR Serratia marcescens CDP-13 augments induced systemic resistance and enhanced salinity tolerance of wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155026. [Google Scholar] [CrossRef]
- Rumyantsev, S.D.; Alekseev, V.Y.; Sorokan, A.V.; Burkhanova, G.F.; Cherepanova, E.A.; Garafutdinov, R.R.; Maksimov, I.V.; Veselova, S.V. Additive effect of the composition of endophytic bacteria Bacillus subtilis on systemic resistance of wheat against greenbug aphid Schizaphis graminum due to lipopeptides. Life 2023, 13, 214. [Google Scholar] [CrossRef]
- Singh, U.B.; Malviya, D.; Singh, S.; Kumar, M.; Sahu, P.K.; Singh, H.V.; Kumar, S.; Roy, M.; Imran, M.; Rai, J.P.; et al. Trichoderma harzianum-and methyl jasmonate-induced resistance to Bipolaris sorokiniana through enhanced phenylpropanoid activities in bread wheat (Triticum aestivum L.). Front. Microbiol. 2019, 10, 453446. [Google Scholar] [CrossRef]
- González-Guzmán, A.; Rey, M.D.; Froussart, E.; Quesada-Moraga, E. Elucidating the effect of endophytic entomopathogenic fungi on bread wheat growth through signaling of immune response-related hormones. Appl. Environ. Microbiol. 2022, 88, e0088222. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Integrated omics approaches for deciphering antifungal metabolites produced by a novel Bacillus species, B. cabrialesii TE3T, against the spot blotch disease of wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, A.A.; Farooq, M.; Al-Sadi, A.M.; Nawaz, A.; Jabran, K.; Siddique, K.H.M. Impact of climate change on biology and management of wheat pests. Crop Prot. 2020, 137, 105304. [Google Scholar] [CrossRef]
- Sharma, P.; Mishra, S.; Singroha, G.; Kumar, R.S.; Singh, S.K.; Singh, G.P. Phylogeographic diversity analysis of Bipolaris sorokiniana (Sacc.) Shoemaker causing spot blotch disease in wheat and barley. Genes 2022, 13, 2206. [Google Scholar] [CrossRef]
- Gupta, P.K.; Chand, R.; Vasistha, N.K.; Pandey, S.P.; Kumar, U.; Mishra, V.K.; Joshi, A.K. Spot blotch disease of wheat: The current status of research on genetics and breeding. Plant Pathol. 2018, 67, 508–531. [Google Scholar] [CrossRef]
- Mehta, Y.R. Wheat diseases and their management; Springer International Publishing: Cham, Switzerland, 2014; Volume 256, ISBN 9783319064659. [Google Scholar]
- Chauhan, P.K.; Singh, D.P.; Karwasra, S.S. Morphological and pathogenic variability in Bipolaris sorokiniana causing spot blotch in wheat (Triticum aestivum, T. durum, T. dicoccum) in India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3499–3520. [Google Scholar] [CrossRef]
- Arya, S.; Kumar, R.; Prakash, O.; Rawat, A.; Pant, A.K. Impact of insecticides on soil and environment and their management strategies. In Agrochemicals in Soil and Environment: Impacts and Remediation; Springer Nature: Singapore, 2022; pp. 213–230. ISBN 9789811693106. [Google Scholar]
- He, D.C.; He, M.H.; Amalin, D.M.; Liu, W.; Alvindia, D.G.; Zhan, J. Biological control of plant diseases: An evolutionary and eco-economic consideration. Pathogens 2021, 10, 1311. [Google Scholar] [CrossRef]
- Rai, S.; Omar, A.F.; Rehan, M.; Al-Turki, A.; Sagar, A.; Ilyas, N.; Sayyed, R.Z.; Hasanuzzaman, M. Crop microbiome: Their role and advances in molecular and omic techniques for the sustenance of agriculture. Planta 2023, 257, 1–23. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 454982. [Google Scholar] [CrossRef]
- Sarethy, I.P.; Saharan, A. Genomics, proteomics and transcriptomics in the biological control of plant pathogens: A review. Indian Phytopathol. 2021, 74, 3–12. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as biological control agents of plant diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Malacrinò, A.; Wisniewski, M.; Cacciola, S.O.; Schena, L. Metabarcoding: A powerful tool to investigate microbial communities and shape future plant protection strategies. Biol. Control 2018, 120, 1–10. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Kumar, A.; Dayanandan, S.; Khan, M.L. Plants endophytes: Unveiling hidden agenda for bioprospecting toward sustainable agriculture. Crit. Rev. Biotechnol. 2020, 40, 1210–1231. [Google Scholar] [CrossRef] [PubMed]
- Diwan, D.; Rashid, M.M.; Vaishnav, A. Current Understanding of Plant-Microbe Interaction through the Lenses of Multi-Omics Approaches and Their Benefits in Sustainable Agriculture. Microbiol. Res. 2022, 265, 127180. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Santoyo, G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr. Plant Biol. 2021, 25, 100189. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Mohapatra, B.; Pattnaik, S.; Gupta, A. New-age genomic measures for uncovering plant-microbiome interactions: Tools, pipelines and guidance map for genomic data mining. In Advances in Agricultural and Industrial Microbiology: Volume-2: Applications of Microbes for Sustainable Agriculture and In-Silico Strategies; Springer Nature: Berlin/Heidelberg, Germany, 2022; pp. 207–232. ISBN 9789811696824. [Google Scholar] [CrossRef]
- Larena, I.; Espeso, E.A.; Carreras, M.; Villarino, M.; De Cal, A.; Melgarejo, P. Impact of genomic resources on improving the mode of action of biocontrol agents against plant pathogens. In How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases; Springer: Cham, Switzerland, 2020; pp. 203–229. [Google Scholar] [CrossRef]
- Jiang, C.H.; Yao, X.F.; Mi, D.D.; Li, Z.J.; Yang, B.Y.; Zheng, Y.; Qi, Y.J.; Guo, J.H. Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis F21 against Fusarium wilt on watermelon. Front. Microbiol. 2019, 10, 446178. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, identification, and complete genome assembly of an endophytic Bacillus velezensis YB-130, potential biocontrol agent against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef]
- Garkovenko, A.V.; Vasilyev, I.Y.; Ilnitskaya, E.V.; Radchenko, V.V.; Asaturova, A.M.; Kozitsyn, A.E.; Tomashevich, N.S.; Milovanov, A.V.; Grigoreva, T.V.; Shternshis, M.V. Draft genome sequence of Bacillus velezensis BZR 336g, a plant growth-promoting antifungal biocontrol agent isolated from winter wheat. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Luan, P.; Yi, Y.; Huang, Y.; Cui, L.; Hou, Z.; Zhu, L.; Ren, X.; Jia, S.; Liu, Y. Biocontrol potential and action mechanism of Bacillus amyloliquefaciens DB2 on Bipolaris sorokiniana. Front. Microbiol. 2023, 14, 1149363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liang, Q.; Li, Y.; Deng, Z.; Song, G.; Wang, H.; Yan, M.; Wang, X. Integrated transcriptome and metabolome analyses shed light on the defense mechanisms in tomato plants after (e)-2-hexenal fumigation. Genomics 2023, 115, 110592. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Charron, J.B.; Jabaji, S. Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis E68 against Fusarium graminearum DAOMC 180378, the causal agent of Fusarium head blight. PLoS ONE 2023, 18, e0277983. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Federspiel, J.D.; Sheng, X.; Cristea, I.M. Proteomics and integrative omic approaches for understanding host–pathogen interactions and infectious diseases. Mol. Syst. Biol. 2017, 13, 922. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, B.; Zhang, H.; Xu, A.; Qian, H. Integration of transcriptomic and proteomic analyses reveals new insights into the regulation of immune pathways in midgut of Samia ricini upon SariNPV Infection. Insects 2022, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Daim, I.A.; Bejai, S.; Meijer, J. Bacillus velezensis 5113 induced metabolic and molecular reprogramming during abiotic stress tolerance in wheat. Sci. Rep. 2019, 9, 16282. [Google Scholar] [CrossRef] [PubMed]
- Nehela, Y.; Atallah, O.; Xuan, T.D.; Elzaawely, A.A. Editorial: Exploring metabolic-based host-pathogen interactions. Front. Plant Sci. 2023, 14, 1247913. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.; Yasmin, H.; Hassan, M.N.; Naz, R.; Nosheen, A.; Sajjad, M.; Ilyas, N.; Keyani, R.; Jabeen, Z.; Mumtaz, S.; et al. Drought-tolerant Bacillus megaterium Isolated from semi-arid conditions induces systemic tolerance of wheat under drought conditions. Plant Cell Rep. 2022, 41, 549–569. [Google Scholar] [CrossRef]
- Cantoro, R.; Palazzini, J.M.; Yerkovich, N.; Miralles, D.J.; Chulze, S.N. Bacillus velezensis RC 218 as a biocontrol agent against Fusarium graminearum: Effect on penetration, growth and TRI5 expression in wheat spikes. BioControl 2021, 66, 259–270. [Google Scholar] [CrossRef]
- Kang, X.; Zhang, W.; Cai, X.; Zhu, T.; Xue, Y.; Liu, C. Bacillus velezensis CC09: A potential ‘vaccine’ for controlling wheat diseases. Mol. Plant-Microbe Interact. 2018, 31, 623–632. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, J.; Yang, K.; Wang, Y.; Tian, Y.; Liang, Z. Transcriptomic and metabonomic insights into the biocontrol mechanism of Trichoderma asperellum M45a against watermelon Fusarium wilt. PLoS ONE 2022, 17, e0272702. [Google Scholar] [CrossRef]
- Gokul, A.; Mabaso, J.; Henema, N.; Otomo, L.; Bakare, O.O.; Klein, A.; Daniel, A.I.; Omolola, A.; Niekerk, L.A.; Nkomo, M.; et al. Sustainable agriculture through the enhancement of microbial biocontrol agents: Current challenges and new perspectives. Appl. Sci. 2023, 13, 6507. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Thakur, N. Bioprospecting new-generation biocontrol strategies: A viable solution for attaining agricultural security and food safety. In Microbial Biocontrol: Sustainable Agriculture and Phytopathogen Management: Volume 1; Springer International Publishing: Berlin/Heidelberg, Germany, 2022; pp. 205–213. ISBN 9783030875121. [Google Scholar]
- Verma, M.L.; Rani, V.; Kumari, R.; Sharma, D.; Kumar, S.; Kushwaha, R. Bioprospecting: At the interface of plant and microbial communities. In Phytomicrobiome Interactions and Sustainable Agriculture; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2021; pp. 217–239. ISBN 9781119644798. [Google Scholar]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Parnell, J.J.; Berka, R.; Young, H.A.; Sturino, J.M.; Kang, Y.; Barnhart, D.M.; Dileo, M.V. From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 2016, 7, 209596. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef]
- Weber, T.; Rausch, C.; Lopez, P.; Hoof, I.; Gaykova, V.; Huson, D.H.; Wohlleben, W. CLUSEAN: A computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 2009, 140, 13–17. [Google Scholar] [CrossRef]
- Starcevic, A.; Zucko, J.; Simunkovic, J.; Long, P.F.; Cullum, J.; Hranueli, D. ClustScan: An integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008, 36, 6882–6892. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339. [Google Scholar] [CrossRef]
- Dutta, S.; Yu, S.M.; Lee, Y.H. Assessment of the contribution of antagonistic secondary metabolites to the antifungal and biocontrol activities of Pseudomonas fluorescens NBC275. Plant Pathol. J. 2020, 36, 491–496. [Google Scholar] [CrossRef]
- Tian, D.; Song, X.; Li, C.; Zhou, W.; Qin, L.; Wei, L.; Di, W.; Huang, S.; Li, B.; Huang, Q.; et al. Antifungal mechanism of Bacillus amyloliquefaciens Strain GKT04 against Fusarium wilt revealed using genomic and transcriptomic analyses. Microbiologyopen 2021, 10, e1192. [Google Scholar] [CrossRef]
- Santoyo, G.; Sánchez-Yáñez, J.M.; de los Santos-Villalobos, S. Methods for detecting biocontrol and plant growth-promoting traits in rhizobacteria. In Methods in Rhizosphere Biology Research; Reinhardt, D., Sharma, A., Eds.; Springer: Singapore, 2019; pp. 133–149. ISBN 9789811357671. [Google Scholar]
- Chaudhari, A.; Patel, J. Current status and future perspective on enzyme involving in biocontrol of plant pathogen. Int. J. Res. Appl. Sci. Biotechnol. 2021, 8, 49–55. [Google Scholar] [CrossRef]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Hernández-Salmerón, J.E.; Prieto-Barajas, C.M.; Valencia-Cantero, E.; Moreno-Hagelsieb, G.; Santoyo, G. Isolation and characterization of genetic variability in bacteria with β-hemolytic and antifungal activity isolated from the rhizosphere of Medicago truncatula Plants. Genet. Mol. Res. 2014, 13, 4967–4975. [Google Scholar] [CrossRef]
- Cossus, L.; Roux-Dalvai, F.; Kelly, I.; Nguyen, T.T.A.; Antoun, H.; Droit, A.; Tweddell, R.J. Interactions with plant pathogens influence lipopeptides production and antimicrobial activity of Bacillus subtilis Strain PTB185. Biol. Control 2021, 154, 104497. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-tolerant plant growth promoting rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 2019, 10, 469278. [Google Scholar] [CrossRef]
- Mukhtar, T.; ur Rehman, S.; Smith, D.; Sultan, T.; Seleiman, M.F.; Alsadon, A.A.; Amna; Ali, S.; Chaudhary, H.J.; Solieman, T.H.I.; et al. Mitigation of heat stress in Solanum lycopersicum L. by ACC-deaminase and exopolysaccharide producing Bacillus cereus: Effects on biochemical profiling. Sustainability 2020, 12, 2159. [Google Scholar] [CrossRef]
- Habib, S.H.; Kausar, H.; Saud, H.M.; Ismail, M.R.; Othman, R. Molecular characterization of stress tolerant plant growth promoting rhizobacteria (PGPR) for growth enhancement of rice. Int. J. Agric. Biol. 2016, 18, 184–191. [Google Scholar] [CrossRef]
- Kaur, A.; Devi, S.R.; Vyas, P. Stress-tolerant antagonistic plant growth-promoting rhizobacteria from Zea mays. J. Plant Prot. Res. 2018, 58, 115–123. [Google Scholar] [CrossRef]
- Dixit, V.K.; Misra, S.; Mishra, S.K.; Tewari, S.K.; Joshi, N.; Chauhan, P.S. Characterization of plant growth-promoting alkalotolerant alcaligenes and Bacillus strains for mitigating the alkaline stress in Zea mays. Antonie Van Leeuwenhoek 2020, 113, 889–905. [Google Scholar] [CrossRef] [PubMed]
- Nicot, P.C.; Blum, B.; Kohl, J.; Ruocco, M. Perspectives for future research-and-development projects on biological control of plant pests and diseases. In Classical and Augmentative Biological Control against Diseases and Pests: Critical Status Analysis and Review of Factors Influencing Their Success; IOBC/WPRS: Avignon, France, 2011; pp. 68–70. ISBN 9789290672432. [Google Scholar]
- Porter, S.S.; Sachs, J.L. Agriculture and the disruption of plant–microbial symbiosis. Trends Ecol. Evol. 2020, 35, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, T.; Dixit, M.; Gera, R.; Shukla, A.K.; Prakash, A.; Gupta, G.; Shukla, P. Techniques for improving formulations of bioinoculants. 3 Biotech 2020, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Ijaz, M.; Ali, Q.; Ashraf, S.; Kamran, M.; Rehman, A. Development of future bioformulations for sustainable agriculture. In Microbiome in Plant Health and Disease: Challenges and Opportunities; Springer: Singapore, 2019; pp. 421–446. ISBN 9789811384950. [Google Scholar]
- Bhattacharjee, A.; Dubey, S.; Sharma, S. “Next-generation bioformulations” for plant growth promotion and stress mitigation: A promising approach for sustainable agriculture. J. Plant Growth Regul. 2023, 42, 6741–6759. [Google Scholar] [CrossRef]
- Balla, A.; Silini, A.; Cherif-Silini, H.; Chenari Bouket, A.; Alenezi, F.N.; Belbahri, L. Recent advances in encapsulation techniques of plant growth-promoting microorganisms and their prospects in the sustainable agriculture. Appl. Sci. 2022, 12, 9020. [Google Scholar] [CrossRef]
- Yahya, M.; Rasul, M.; Sarwar, Y.; Suleman, M.; Tariq, M.; Hussain, S.Z.; Sajid, Z.I.; Imran, A.; Amin, I.; Reitz, T.; et al. Designing synergistic biostimulants formulation containing autochthonous phosphate-solubilizing bacteria for sustainable wheat production. Front. Microbiol. 2022, 13, 889073. [Google Scholar] [CrossRef]
- Azaroual, S.E.; El Mernissi, N.; Zeroual, Y.; Bouizgarne, B.; Meftah Kadmiri, I. Effect of Bacillus spp. strains on wheat nutrient assimilation and bioformulation by new spray drying approach using natural phosphate powder. Dry. Technol. 2022, 40, 2630–2644. [Google Scholar] [CrossRef]
- Comite, E.; El-Nakhel, C.; Rouphael, Y.; Ventorino, V.; Pepe, O.; Borzacchiello, A.; Vinale, F.; Rigano, D.; Staropoli, A.; Lorito, M.; et al. Bioformulations with beneficial microbial consortia a bioactive compound and plant biopolymers modulate sweet basil productivity photosynthetic activity and metabolites. Pathogens 2021, 10, 870. [Google Scholar] [CrossRef]
- Pellegrini, M.; Pagnani, G.; Bernardi, M.; Mattedi, A.; Spera, D.M.; Del Gallo, M. Cell-free supernatants of plant growth-promoting bacteria: A review of their use as biostimulant and microbial biocontrol agents in sustainable agriculture. Sustainability 2020, 12, 9917. [Google Scholar] [CrossRef]
- Shah, A.; Nazari, M.; Antar, M.; Msimbira, L.A.; Naamala, J.; Lyu, D.; Rabileh, M.; Zajonc, J.; Smith, D.L. PGPR in agriculture: A sustainable approach to increasing climate change resilience. Front. Sustain. Food Syst. 2021, 5, 667546. [Google Scholar] [CrossRef]
- Sunar, K.; Das, K.; Rai, A.K.; Gurung, S.A. Beneficial Microbial Consortia and Their Role in Sustainable Agriculture under Climate Change Conditions; Springer: Singapore, 2023; pp. 41–73. [Google Scholar] [CrossRef]
- Ortega-Urquieta, M.E.; Valenzuela-Ruíz, V.; Mitra, D.; Hyder, S.; Elsheery, N.I.; Kumar Das Mohapatra, P.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Draft genome sequence of Priestia sp. strain TSO9, a plant growth-promoting bacterium associated with wheat (Triticum turgidum subsp. durum) in the Yaqui Valley, Mexico. Plants 2022, 11, 2231. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rodriguez, E.; Lugo-Enríquez, C.; Ferguson, S.; Parra-Cota, F.I.; Cira-Chávez, L.A.; de los Santos-Villalobos, S. Trichoderma harzianum sensu lato TSM39: A wheat microbiome fungus that mitigates spot blotch disease of wheat (Triticum turgidum L. subsp. durum) caused by Bipolaris sorokiniana. Biol. Control 2022, 175, 105055. [Google Scholar] [CrossRef]
- García Montelongo, A.M.; Montoya-Martínez, A.C.; Morales-Sandoval, P.H.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Beneficial microorganisms as a sustainable alternative for mitigating biotic stresses in crops. Stresses 2023, 3, 210–228. [Google Scholar] [CrossRef]
- Lantmännen BioAgri. Organic Seed Treatment | LM Bioagri. Available online: https://www.lantmannenbioagri.com/biological-seed-treatment/ (accessed on 16 October 2023).
- XEDA ITALIA. XEDAVIR®—XEDA ITALIA. Available online: https://www.xeda.it/en/product/xedavir/ (accessed on 16 October 2023).
- Agrichem. Polyversum® Para El Control de Enfermedades—Agrichembio—Bioinnovación Para La Agricultura Integrada. Available online: https://agrichembio.com/productos/control-de-enfermedades/polyversum/ (accessed on 16 October 2023).
- Lallemand Plant Care. MYCOSTOP | Lallemand Plant Care. Available online: https://www.lallemandplantcare.com/en/canada/products/product-details/mycostop/ (accessed on 16 October 2023).
- Novozymes. Actinovate SP | Novozymes. Available online: https://www.novozymes.com/en/products/bioag/biocontrol/actinovate-sp (accessed on 16 October 2023).
- BAYER Serenade® ASO. Available online: https://www.cropscience.bayer.es/Productos/Fungicidas/Serenade-ASO (accessed on 16 October 2023).
- GreenCorp. RoyaOut—GreenCorp. Available online: https://greencorp.mx/producto/biocontrol/biofungicidas/royaout/ (accessed on 16 October 2023).
- GreenCorp. Best Ultra F—GreenCorp. Available online: https://greencorp.mx/producto/biocontrol/biofungicidas/best-ultra-f/ (accessed on 16 October 2023).
- Lallemand Plant Care. PRESTOP | Lallemand Plant Care. Available online: https://www.lallemandplantcare.com/en/canada/products/product-details/prestop/ (accessed on 16 October 2023).
- Mahajan, A.; Gupta, R.D. Bio-fertilizers: Their kinds and requirement in India. In Integrated Nutrient Management (INM) in a Sustainable Rice—Wheat Cropping System; Springer: Dordrecht, The Netherlands, 2009; pp. 75–100. [Google Scholar]
- Valenzuela Ruiz, V.; Gálvez Gamboa, G.T.; Villa Rodríguez, E.D.; Parra Cota, F.I.; Santoyo, G.; De los Santos Villalobos, S. Lipopéptidos producidos por agentes de control biológico del género Bacillus: Revisión de herramientas analíticas utilizadas para su estudio. Rev. Mex. Ciencias Agrícolas 2020, 11, 419–432. [Google Scholar] [CrossRef]
- Simón, M.R.; Börner, A.; Struik, P.C. Editorial: Fungal wheat diseases: Etiology, breeding, and integrated management. Front. Plant Sci. 2021, 12, 671060. [Google Scholar] [CrossRef]
- Reynoso, A.; Sautua, F.; Carmona, M.; Chulze, S.; Palazzini, J. Tan spot of wheat: Can biological control interact with actual management practices to counteract this global disease? Eur. J. Plant Pathol. 2023, 166, 27–38. [Google Scholar] [CrossRef]
- Piñeiro, V.; Arias, J.; Dürr, J.; Elverdin, P.; Ibáñez, A.M.; Kinengyere, A.; Opazo, C.M.; Owoo, N.; Page, J.R.; Prager, S.D.; et al. A scoping review on incentives for adoption of sustainable agricultural practices and their outcomes. Nat. Sustain. 2020, 3, 809–820. [Google Scholar] [CrossRef]
- Jacquet, F.; Jeuffroy, M.H.; Jouan, J.; Le Cadre, E.; Litrico, I.; Malausa, T.; Reboud, X.; Huyghe, C. Pesticide-free agriculture as a new paradigm for research. Agron. Sustain. Dev. 2022, 42, 8. [Google Scholar] [CrossRef]

| Wheat Disease | Phytopathogen/Pest | ISR-Inducing Agent | Reference |
|---|---|---|---|
| Septoria tritici blotch | Zymoseptoria tritici | Bacillus amyloliquefaciens | [53] |
| Septoria tritici blotch | Zymoseptoria tritici | Bacillus subtilis lipopeptide mixture (surfactin, fengycin, and mycosubtilin) | [51] |
| Spot blotch | Bipolaris sorokiniana | Trichoderma asperellum | [58] |
| Fusarium head blight | Fusarium graminearum | N-Hydroxypipecolic acid | [59] |
| Fusarium crown rot | Fusarium graminearum | Bacillus pumilis and Trichoderma harzianum | [54] |
| Fusarium head blight | Fusarium graminearum | Chitin | [60] |
| Spot blotch | Bipolaris sorokiniana | Bacillus amyloliquefaciens and Trichoderma harzianum | [57] |
| Take-all disease | Gaeumannomyces tritici | Bacillus subtilis | [61] |
| Sharp eyespot | Rhizoctonia cerealis | Piriformospora indica | [62] |
| Fusarium head blight | Fusarium graminearum | Piriformospora indica | [62] |
| Fusarium head blight | Fusarium moniliforme | Serratia marcescens | [63] |
| The greenbug aphid | Schizaphis graminum Rondani | Bacillus subtilis | [64] |
| Spot blotch | Bipolaris sorokiniana | Trichoderma harzianum | [65] |
| - | - | Bacillus paralicheniformis | [50] |
| - | - | Beauveria bassiana and Metarhizium brunneum | [66] |
| Fusarium head blight | Fusarium graminearum | Trichoderma atroviride | [56] |
| Stripe rust | Puccinia striiformis f. sp. tritici | Paenibacillus alvei | [52] |
| Septoria tritici blotch | Mycosphaerella graminicola | Paenibacillus sp. | [55] |
| Biological Control Agent | Commercial Product | Target Disease in Wheat | Company | Reference |
|---|---|---|---|---|
| Pseudomonas chlororaphis strain MA342 | Cerall® | Fusarium head blight, Tilletia tritici and Tilletia laevis wheat bunt | BioAgri AB, Uppsala, Sweden | [141] |
| Chitinolytic activities or by-products of microbial detoxification of mycotoxins. Trichoderma asperellum | Xedavir® | Fusarium verticillioides | Xeda | [142] |
| Mycophageous action. Pythium oligandrum strain M1 | Polyversum® | Fusarium graminearum head blight | AgrichemBio | [143] |
| Streptomyces spp. Antigerminative compounds | Mycostop® | Fusarium spp. and deoxynivalenol (DON) mycotoxins | Lallemand Specialties Inc. | [144] |
| Streptomyces spp. | Actinovate® | Fusarium spp. and deoxynivalenol (DON) mycotoxins | Actinovate SP | [145] |
| Bacillus subtilis strain QST713 | Serenade® ASO | Yellow rust (Puccinia striiformis) | Bayer | [146] |
| Clove extract + Bacillus subtilis + emulsifiers, conditioners, and diluents | Roya Out® | Leaf rust (Puccinia triticina) | Greencorp | [147] |
| Bacillus spp. + Azotobacter spp. + Pseudomonas spp. + plant extracts + conditioners and stabilizers | Best Ultra® F | Leaf rust (Puccinia triticina) | Greencorp | [148] |
| Gliocladium catenulatum strain J1446 | Prestop® (WG) | Damping off (Arthrinium sacchari), Root rot (Bipolaris sorokiniana, Fusarium spp.) | Lallemand Specialties Inc. | [149] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Avelar, I.; Montoya-Martínez, A.C.; Villa-Rodríguez, E.D.; Valenzuela-Ruiz, V.; Ayala Zepeda, M.; Parra-Cota, F.I.; de los Santos Villalobos, S. The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants. Sustainability 2023, 15, 15250. https://doi.org/10.3390/su152115250
Campos-Avelar I, Montoya-Martínez AC, Villa-Rodríguez ED, Valenzuela-Ruiz V, Ayala Zepeda M, Parra-Cota FI, de los Santos Villalobos S. The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants. Sustainability. 2023; 15(21):15250. https://doi.org/10.3390/su152115250
Chicago/Turabian StyleCampos-Avelar, Ixchel, Amelia C. Montoya-Martínez, Eber D. Villa-Rodríguez, Valeria Valenzuela-Ruiz, Marisol Ayala Zepeda, Fannie Isela Parra-Cota, and Sergio de los Santos Villalobos. 2023. "The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants" Sustainability 15, no. 21: 15250. https://doi.org/10.3390/su152115250
APA StyleCampos-Avelar, I., Montoya-Martínez, A. C., Villa-Rodríguez, E. D., Valenzuela-Ruiz, V., Ayala Zepeda, M., Parra-Cota, F. I., & de los Santos Villalobos, S. (2023). The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants. Sustainability, 15(21), 15250. https://doi.org/10.3390/su152115250