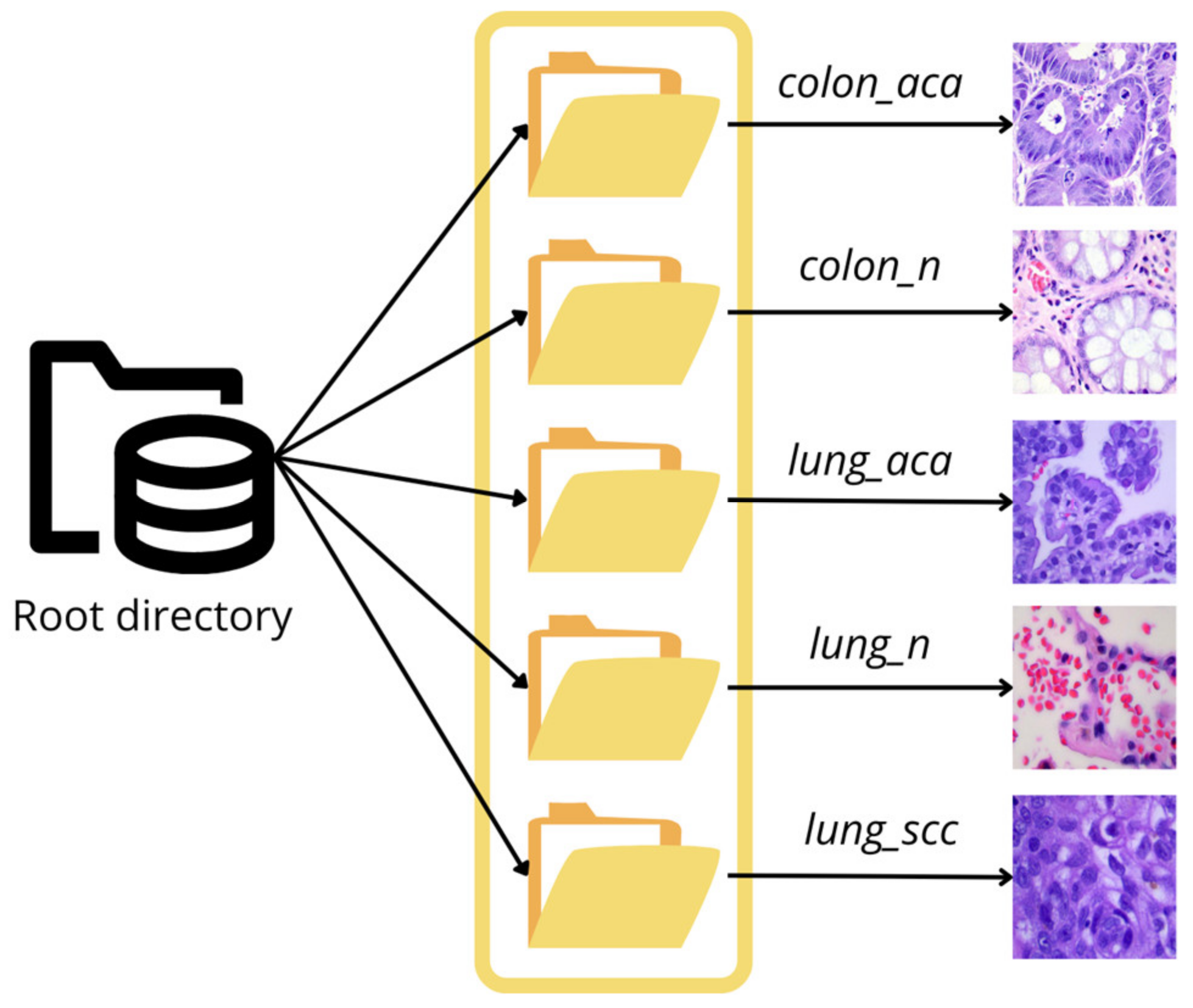
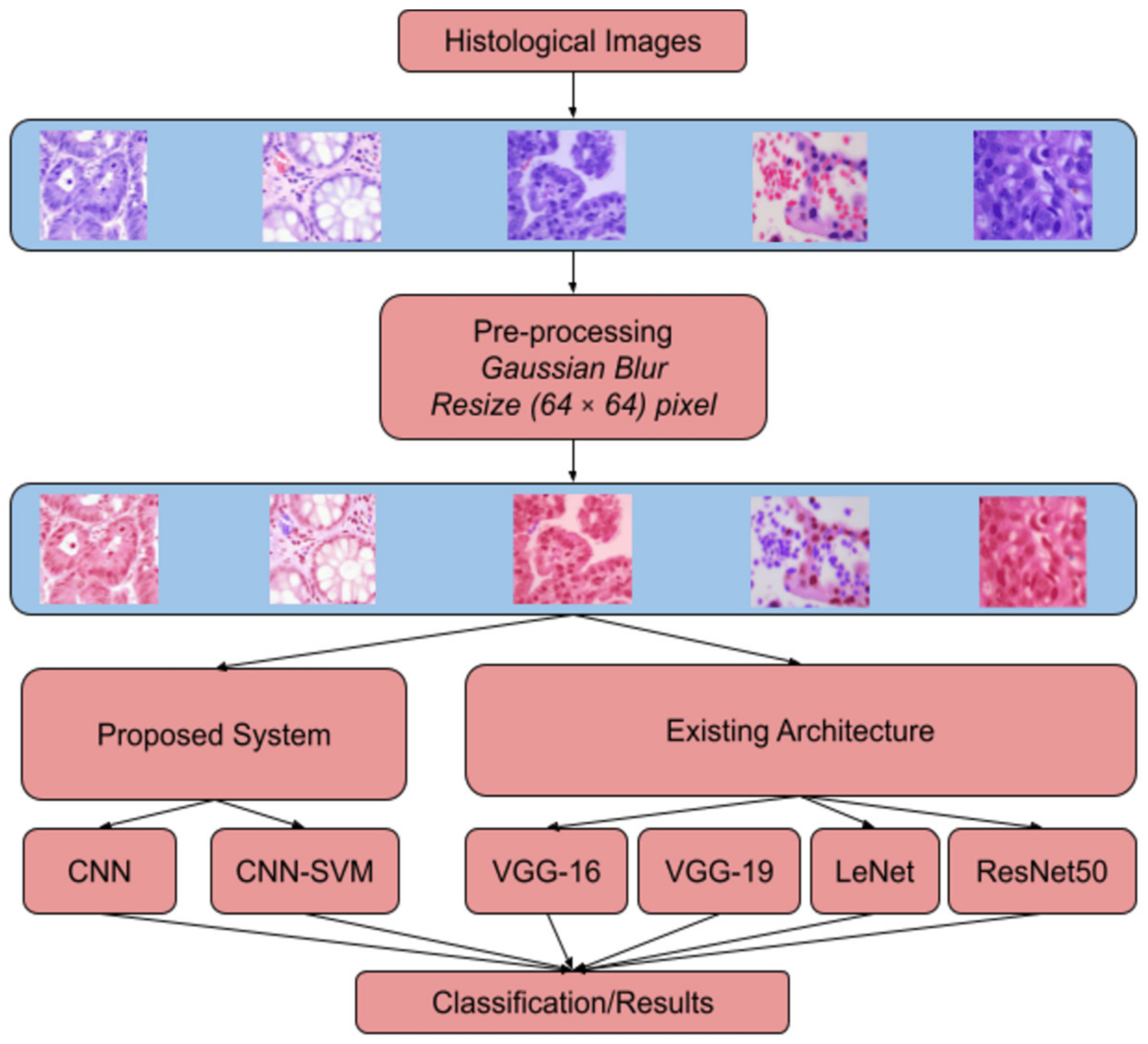
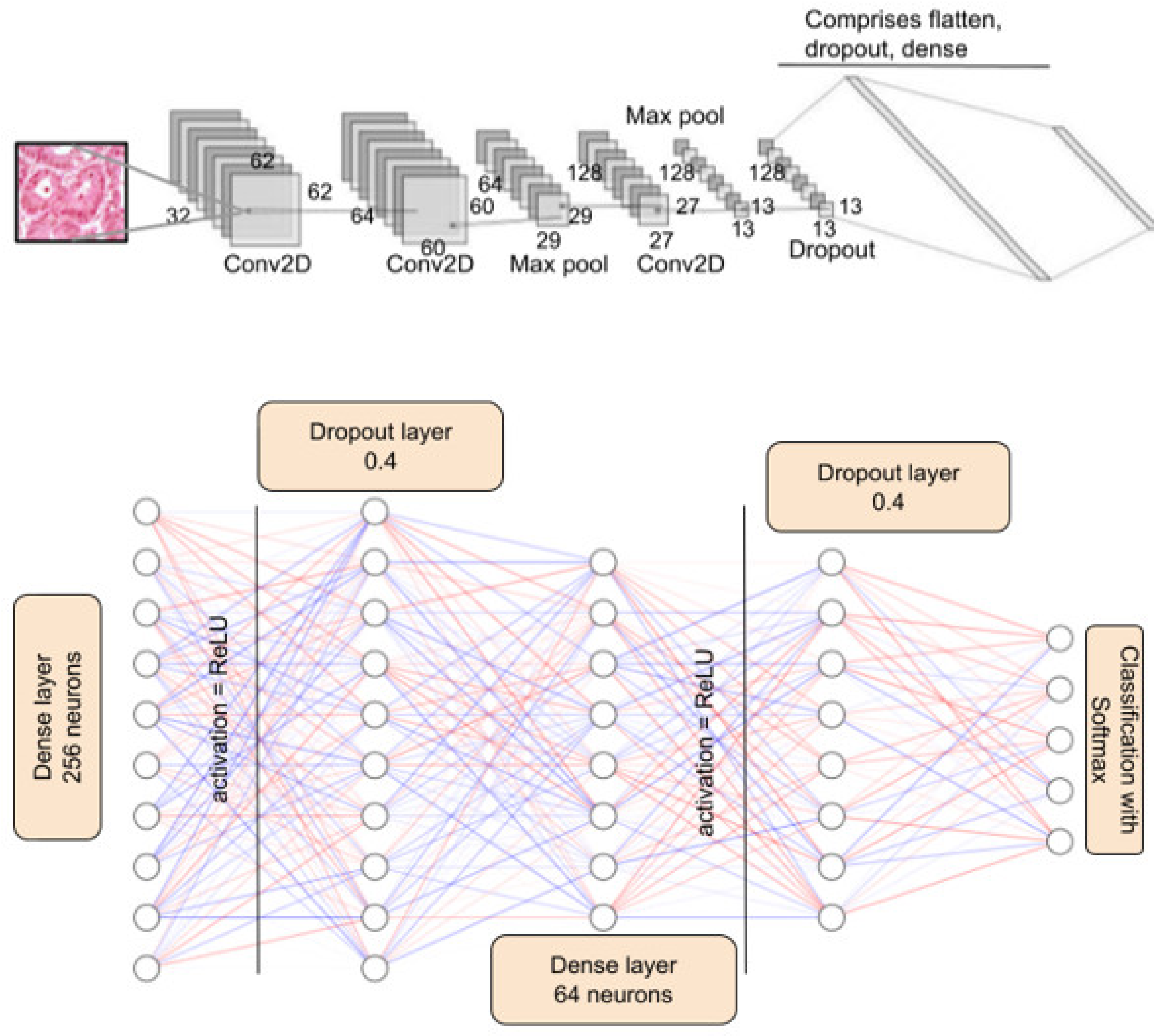
1. Introduction
The world today witnesses cancer as one of the most dreadful diseases impacting precious human life adversely. While cancer happens to be a generic term to refer to a large variety of diseases, it essentially involves enhanced formation of abnormal cells that propagate through the bloodstream and spread across the body, destroying normal cells, leading to the death of the affected individual. As reported by WHO [
1] (World Health Organization), this disease is estimated to have affected around 10 million people in the year 2020. The global cancer trend [
2] has been so concerning that an estimated 47% increase in the disease’s prevalence worldwide from 2020 to 2040 is predicted.
Adenocarcinoma [
3] relates to a common type of cancer found in glandular epithelial cells of human body. Lung, prostate, pancreas, liver, colorectal area, and breast form the primary sites for the adenocarcinoma. Unlike other carcinomas, these cancers do not exhibit any symptoms during their early stages and remain undetected. As data [
3] say, adenocarcinoma is responsible for around 40%, 95%, and 96% of non-small-cell lung cancers, pancreatic cancers, and colorectal cancers, respectively. It is also responsible for almost all prostate cancers and most breast cancers.
Histological images continue to be a standard method of cancer diagnosis. The advances made thus far in the area of health informatics have still proven to be unsuccessful in meeting the desired clinical requirements. Most diagnostics to date are still performed manually and rely heavily on the expertise and experience of histopathologists. These diagnostic techniques happen to be very time-consuming and difficult to grade in a reproducible manner.
Computer aided diagnosis using histopathological images has always been a topic of paramount interest in the field of cancer detection. Multiple works have been conducted in this domain using artificial intelligence (AI). Experimentation on AI-based cancer diagnosis using various machine learning and deep learning models has presently evolved as one of the prime areas of interest. Major studies undertaken in this domain include implementation of different AI-based models, improving existing AI-based models, or evaluating existing AI-based models to have an insight into their comparative efficacy. The development of methods to enhance image processing techniques to better extract features is provided as a solution to this problem statement. The development of filters to select the most efficient models among all the multiple machine learning models used as classifiers to improve accuracy is also one of the works conducted in this area. Existing works in this domain have recorded accuracies ranging from 70% to over 90%.
Compared to manual analysis, an AI-based system has the potential to provide rapid and consistent cancer detection and classification results. Therefore, the treatment and analysis of images using advanced machine learning and deep learning techniques need to be introduced to facilitate the increased rate of disease diagnosis in humans.
The primary objective of the present study is to develop an artificial intelligence-based tool that can assist in diagnosing adenocarcinoma from histopathology-based images. The study aims at detecting and classifying adenocarcinoma in the colon and lung regions of the human body using the LC25000 dataset [
4] procured from Kaggle.
Literature Study
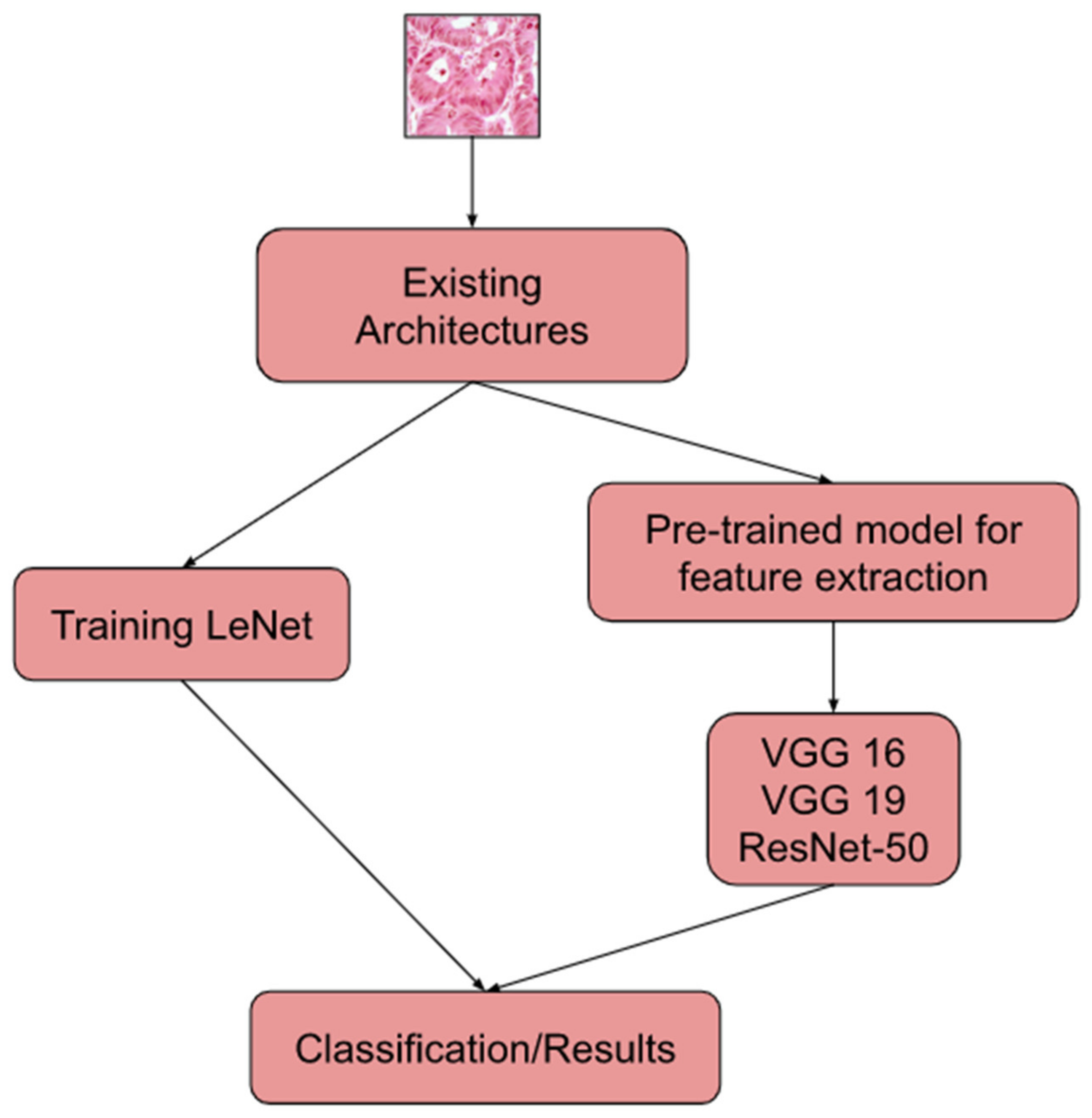
The use of machine learning (ML), deep learning (DL), and transfer learning (TL) to detect and classify has been the talk of the town for a while and there have been several approaches that performed successfully.
The research work [
5] used CNN architectures such as VGG16, VGG19, DenseNet169, and DenseNet201 to extract image characteristics from the LC25000 dataset. The extracted features were put into six widely used ML algorithms including Extreme Gradient Boosting (XGB), Random Forest (RF), Support Vector Machine (SVM), Light Gradient Boosting (LGB), Multi-Layer Perceptron (MLP), and Logistic Regression (LR) to evaluate the performance. Accuracy-based filtering of the findings allowed for the selection of the most effective algorithms. As classifiers, SVM, Logistic Regression, and MLP were chosen because of their superior performance. Using this method, cancers of the lung and colon were found. The authors of [
6] used the LC25000 dataset to automate the detection of lung and colon cancer. The pre-processing involved wavelet decomposition and application of 2D Fourier transform on channel-separated images. They used a CNN model with a Softmax classifier for feature extraction and classification tasks, achieving an accuracy of 96.37%. The research in [
7] used the LC25000 dataset for classifying histopathology images of lung cancer using CNN. Feature extraction was performed using ResNet50, VGG19, Inception_ResNet_V2, and DenseNet121. The Triplet loss function was used to enhance the performance. CNN having three hidden layers was used to classify the images. In this study, Inception-ResNetv2 performed well, having a test accuracy rate of 99.7%.
The authors in the research work [
8] classified adenocarcinoma of the lung region and adenocarcinoma of the colon region using a CNN model. They used the LC25000 dataset for this purpose. The images were first resized to 150 × 150 pixels, then some randomized shear and zoom transformations were applied to the images, followed by the normalization of images. A CNN model was applied separately for the lung dataset and the colon dataset, recording an accuracy of 97% and 96%, respectively. A research work was conducted to perform detection of lung cancer using CNN [
9]. The dataset used during the process was LC25000. The images of the dataset were first resized to 180 × 180, and the pixel values were then transformed to a range of (0, 1) to facilitate faster convergence. The CNN model had three hidden layers. The model recorded a training accuracy of 96.11% and validation accuracy of 97.2%. The work [
10] aimed at developing a model that classified lung cancer into adenocarcinoma, benign, and squamous cell carcinoma. They used the LC25000 dataset for this purpose. The model consists of a main path responsible for extracting small features and sub-paths which pass the medium- and high-level features to fully connected layers. The model recorded an accuracy of 98.53%.
The work carried out in [
11] designed a model for the diagnosis of lung and colon cancer using the LC25000 dataset. A pre-trained AlexNet model, after modifying four of its layers, was used. The model performed well for all classes except the “lung_ssc” class, achieving an accuracy of 89%. To improve the performance, image enhancement techniques such as image contrast enhancement were applied to the underperforming class, improving the accuracy to 98.4%. The research work [
12] used the AiCOLON dataset to train a CNN model. transfer learning was applied and results were compared with the built CNN model. The highest accuracy achieved was 96.98% with ResNet. CRC-5000, NCT-CRC-HE-100K, and merged (namely, the CRC-5000, the NCT-CRC-HE-100K) datasets were also used to test the ResNet model, recording an accuracy of 96.77%, 99.76%, and 99.98%, respectively. An accuracy of 98.66%, 99.12%, and 78.39% was achieved by SEGNET for the same datasets. SegNet was concluded to be an efficient model for cancer segmentation.
Both the LC25000 and Kather datasets were used in [
13] to develop a super-lightweight plug-and-play module (namely, Pyramidal Deep-Broad Learning (PDBL)), to equip CNN backbones, especially lightweight models, to increase tissue-level classification performance without a re-training burden. Experiments were performed to equip this module on ShuffLeNetV2, EfficientNetb0, and ResNet50, with accuracy of 74.61%, 79.87%, and 85.53% with no re-training. The work [
14] used the LC25000 dataset to develop a CNN model to categorize and classify the colon region for adenocarcinoma and benign cells. Lime and DeepLift were used as the optimization techniques to improve the understanding of results predicted via the model. The validation accuracy for diagnosis was found to be higher than 94% for distinguishing adenocarcinoma and benign colonic cells.
Extensive research work was carried out in [
15] to utilize the power of machine learning, feature engineering, and image processing for identifying classes of lung and colon cancer. They used the LC25000 dataset in their study. Machine learning models such as XGBoost, SVM, RF, LDA, and MLP were employed. Unsharp masking was used for image preprocessing. The Recursive Feature Elimination (RFE) method was used to eliminate the least important features. XGBoost recorded an accuracy of 99%. The SHAP method was used to show the contributions of each feature in the results predicted by models. The research work [
16] used CNN models with max pooling layers and average pooling layers along with MobileNetV2 for analyzing colon cancer using the LC25000 dataset. Using the ImageDataGenerator of the Keras library, images were augmented with flips, shear, zoom, rotation, and width and height range. Images were resized to 224 × 224 pixels and later converted to Numpy arrays for further use. The models were trained with various epochs and reported an accuracy of 97.49%, 95.48%, and 99.67%, respectively, for CNN models with max pooling and average pooling and MobileNetV2.
The research work in [
17] developed four different CNN models, varying from two pairs of convolutional layers and max pooling layers to four pairs of convolutional layers and max pooling layers with different number of filters and kernel sizes, using lung images procured from the LC25000 dataset. Three different sizes of input images were taken into consideration. The best result accuracy on the test dataset was 96.6% with input size 768 × 768 pixels with the CNN model having four convolutional layers and max pooling layers. It was observed that, with an increase in input image size and convolutional layers, the accuracy increases. The research carried out in [
18] used ResNet18, ResNet30, and ResNet50 architectures for colonic adenocarcinoma diagnosis using histopathological images. Two image datasets were used, LC25000 and CRAG. A total of 10,000 images from the first dataset were used to train and validate the CNN architectures in an 80:20 train and test set ratio. Images from the latter dataset were split into 40% and 60% for training and testing, respectively. This was performed to check how the model behaves using a self-supervised learning step, where networks were trained with the LC25000 dataset. Validation accuracy of 93.91%, 93.04%, and 93.04% was recorded for each of the three CNN architectures, respectively. The authors of [
19] proposed the development of homology-based image processing techniques along with conventional texture analysis that gives better results for classification when fed into machine learning models such as Perceptron, Logistic Regression, KNN, SVM with Linear Kernel, SVM with Radial-Basis Function Kernel, Decision Tree, Random Forest, and Gradient Tree Boosting. They primarily used two datasets, private and public. The public dataset was the LC25000. Images were processed by converting them into grayscale, applying binarization, and then finally converting them into betti numbers. The accuracy was 78.33% and 99.43%, respectively, for the private and public dataset.
In the research work [
20], a CNN model with three convolutional layers, having 32, 64, and 128 filters, respectively, was designed. The study aimed at the detection of breast cancer using histopathological images from the BreakHis dataset. The images were resized to 350 × 230 pixels and reshaped, maintaining the aspect ratio, achieved using OpenCV library in Python. It achieved an accuracy of 99.86%. The research work [
21] used the ACDC@LUNGHP dataset to compare the performance of two prominent CNN architectures, VGG16 and ResNet50. In this case, VGG16 was seen to slightly outperform ResNet50, with an accuracy of 75.41%. The research work [
22] used the PatchCamelyon benchmark dataset to compare the performance of 14 existing architectures of CNN. They concluded that DenseNet161 outperformed other architectures, with an AUC score of 0.9924. Three approaches were carried out during the process. The first approach correspondeded to utilizing the weights from the pre-trained network. The second approach related to training only fully connected layers. The last approach corresponded to training the entire model. The work [
23] used the BreakHis dataset to detect Breast Cancer using histopathological images using CNN. Image augmentation was applied to improve the performance of the model and it was finally concluded with training accuracy of 96.7% and testing accuracy of 90.4%.
3. Results and Discussion
The models were built and run using Google Colab. The Colab environment was used to train the models on a large number of data using GPU acceleration.
For the building of the models, several Python libraries were utilized to make the process more efficient and improve the performance. Tensorflow and Keras libraries were essential in building the models, compiling the models, and training the models as per the used dataset. Numpy was used to handle arrays, and OpenCV was used for reading and pre-processing the images. ImageGenerator was used for data augumentation.
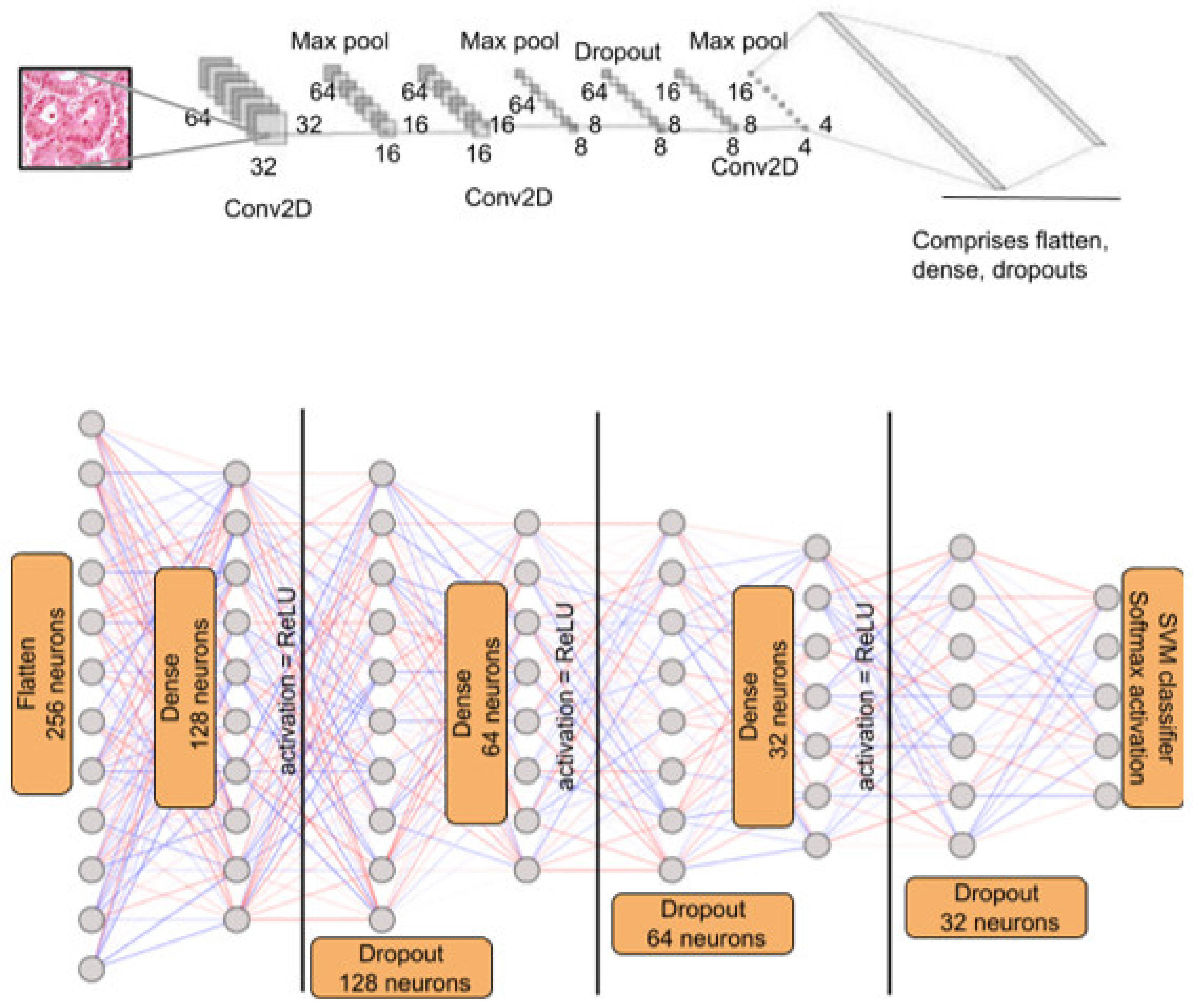
In the pre-processing segment, Gaussian Blur with a kernel size of 3 × 3 was used, and SigmaX and SigmaY values were 90 in both cases. The images were initially set to 128 by 128. However, the image size was large for smoother processing in the Colab environment. Hence, the images were resized to 64 by 64.
The model initially designed severely overfit the training set. Hence, the hyperparameters were altered, and the total number of model parameters was reduced. As a result of the above process, we could finally arrive at the proposed model.
In this section, we discuss the results obtained by the CNN models the classification of lung and colon adenocarcinoma from histopathological images. The models were applied to three cases. In the first case, the models trained only on lung classes, the second case is of only colon classes, and the third case has lung and colon classes combined. In each case, 80% of the images are for training the model, and 20% are for testing.
Accuracy metrics were used to compare the results. Accuracy is a metric generally used when all the classes are equally important. Since the dataset used in this study is balanced, and all the classes have the same significance, accuracy metrics seemed reliable. Accuracy is the ratio of correct predictions to the total number of predictions. Equation (2) corresponds to the mathematical formula for the accuracy metric.
Table 2 displays the training and testing accuracies obtained for all the discussed models on a subset of the data, each class containing 500 images.
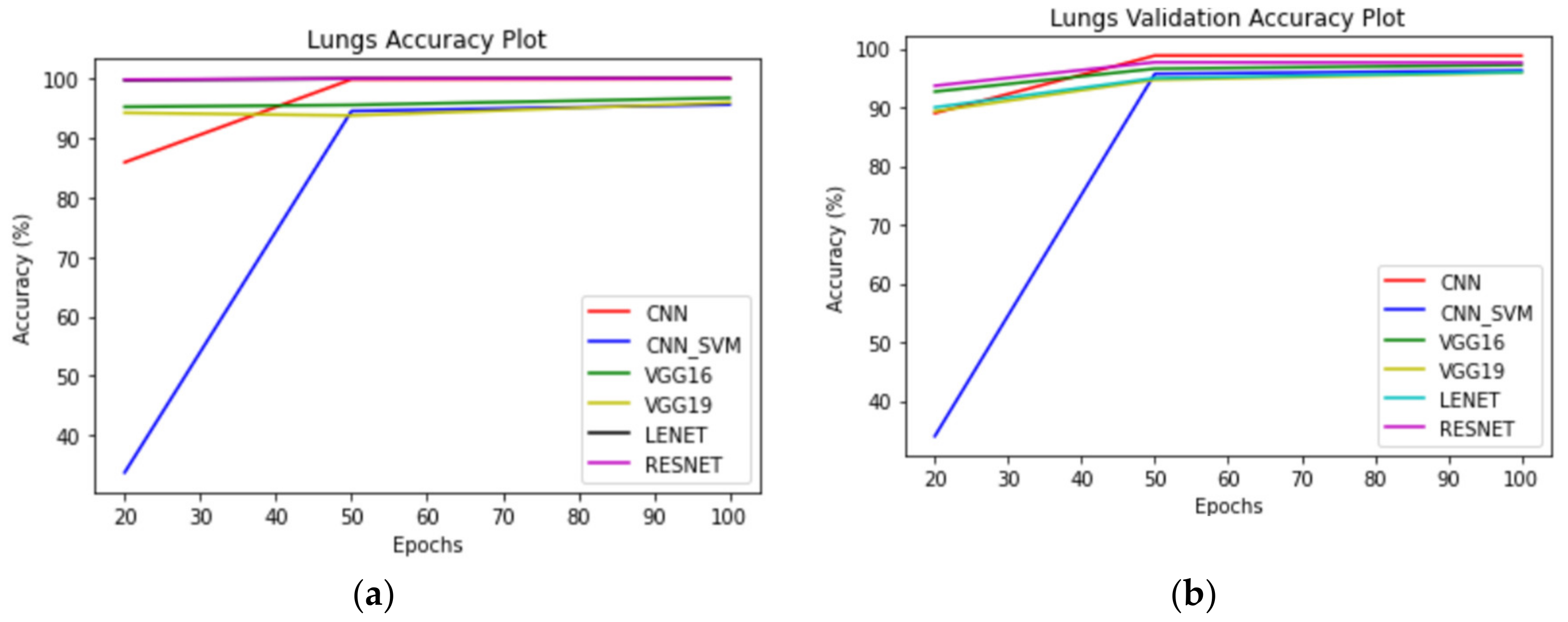
For subsets, each model was trained for 20 epochs. For the lung classes of the dataset, best performance was recorded for the ResNet50 model with the training and testing accuracies noted as 99.83% and 93.67%.
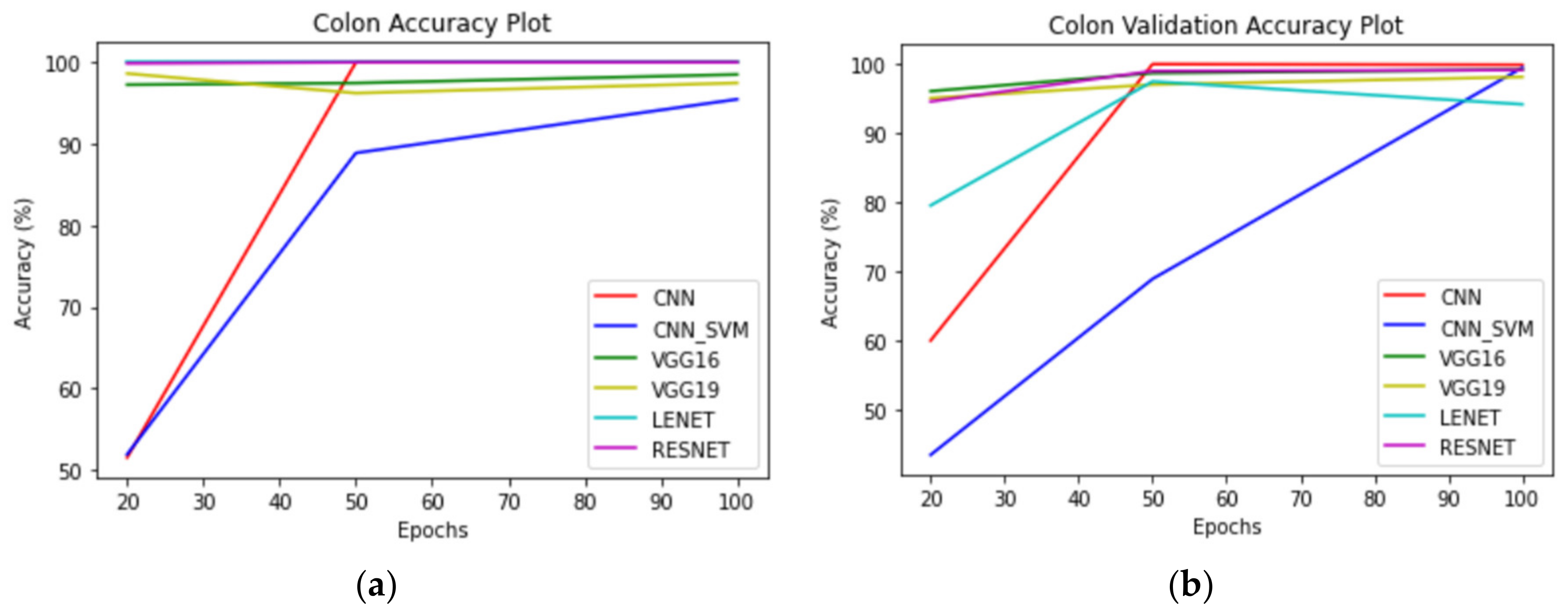
For the colon classes of the subset taken alone, LeNet-5 gave us 100% training accuracy. However, it is pertinent to note that the testing accuracy was only 79.50%, which is a case of overfitting. On the other hand, ResNet50 had a training accuracy of 99.87% and a testing accuracy of 94.50%. It is pertinent to note that VGG16 performed better than the rest of the models on the unseen data, recording the highest validation accuracy of 96.00%.
When histopathological images obtained from both lung and colon regions are analyzed, ResNet50 performed the best, with the highest training accuracy and testing accuracy of 100% and 95.40%.
The pre-existing architectures were found to perform better than the newly proposed system when the number of data used for training the model was drastically reduced.
It is pertinent to note that the concept of data augmentation was not implemented in any model run for 20 epochs.
All the models were then trained on the entire dataset i.e., 25,000 images, evenly distributed across five classes. The same train–test split of 80:20 was used. The models were trained for 50 as well as 100 epochs.
Table 3 displays the results obtained when the models were trained for 50 epochs.
For 50 epochs, in the case of histopathological images of only lungs, both ResNet50 and LeNet were found to record the highest training accuracy of 100%, and the validation accuracy of ResNet50 and LeNet was 97.67% and 94.97%. It is pertinent to note that the training and testing accuracies of the AdenoCanNet were 99.87% and 98.80%. The validation accuracy reported by AdenoCanNet architecture was the highest among the other models, indicating its better performance on unseen data. Hence, it can be concluded that AdenoCanNet performs better in the case of histopathological images in the lung region.
For colon images, as visible from
Table 3, ResNet50 and LeNet performed the best on the training dataset with 100% training accuracy. The validation accuracy of ResNet and LeNet was 98.90% and 97.40%. It is pertinent to note that the training and validation accuracy achieved by
AdenoCanNet were 99.99% and 99.90%. With the training and testing accuracy of
AdenoCanNet being almost 100%, it can be concluded that
AdenoCanNet performs better in the case of histopathological images obtained from the colon region.
When histopathological images obtained from both lung and colon regions are taken into consideration, ResNet performed well, the training and testing accuracy being 100%. However, the proposed architecture, AdenoCanNet, also recorded a training accuracy of 99.88% and validation accuracy of 98.90%, comparable with the performance of ResNet attained on the complete dataset.
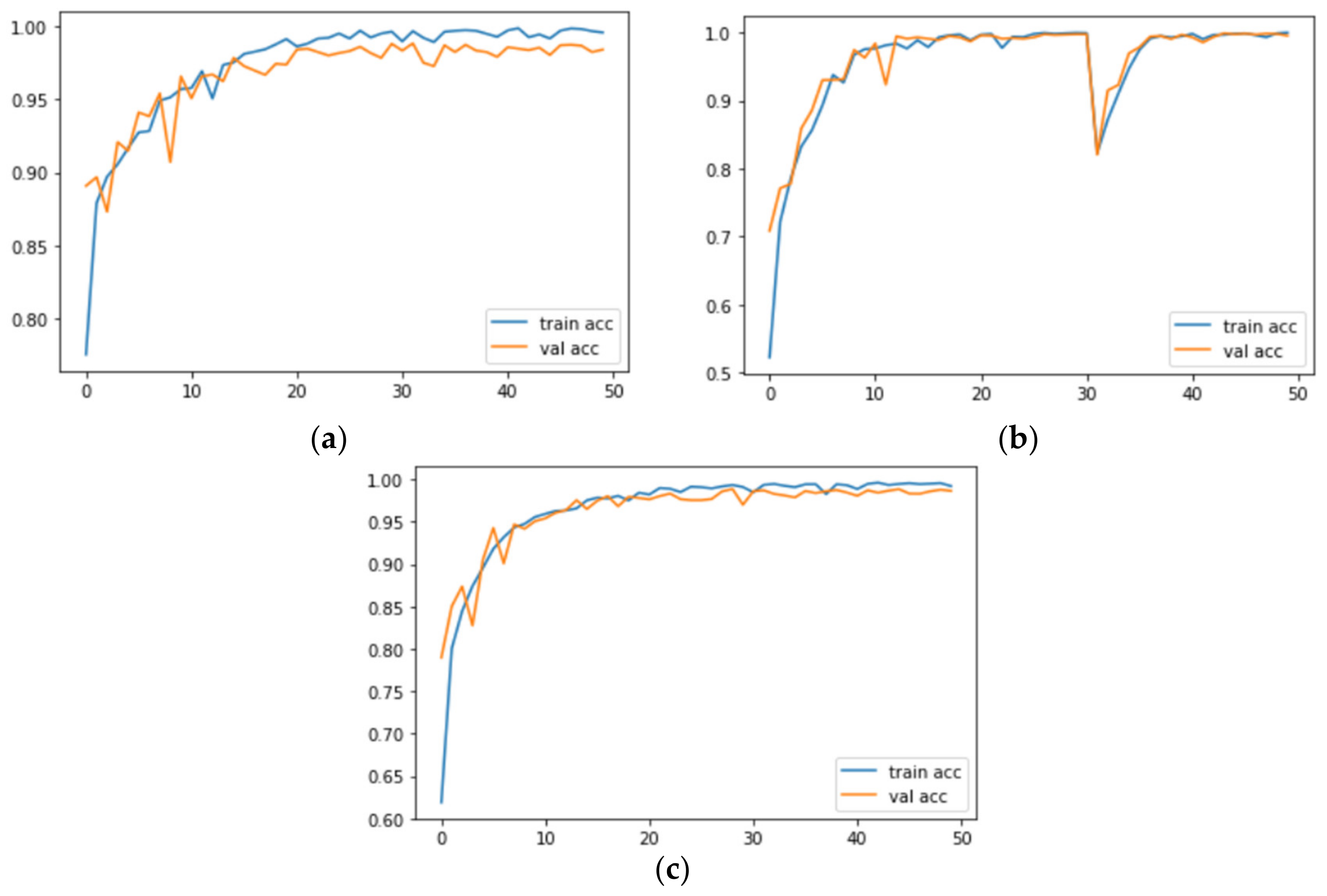
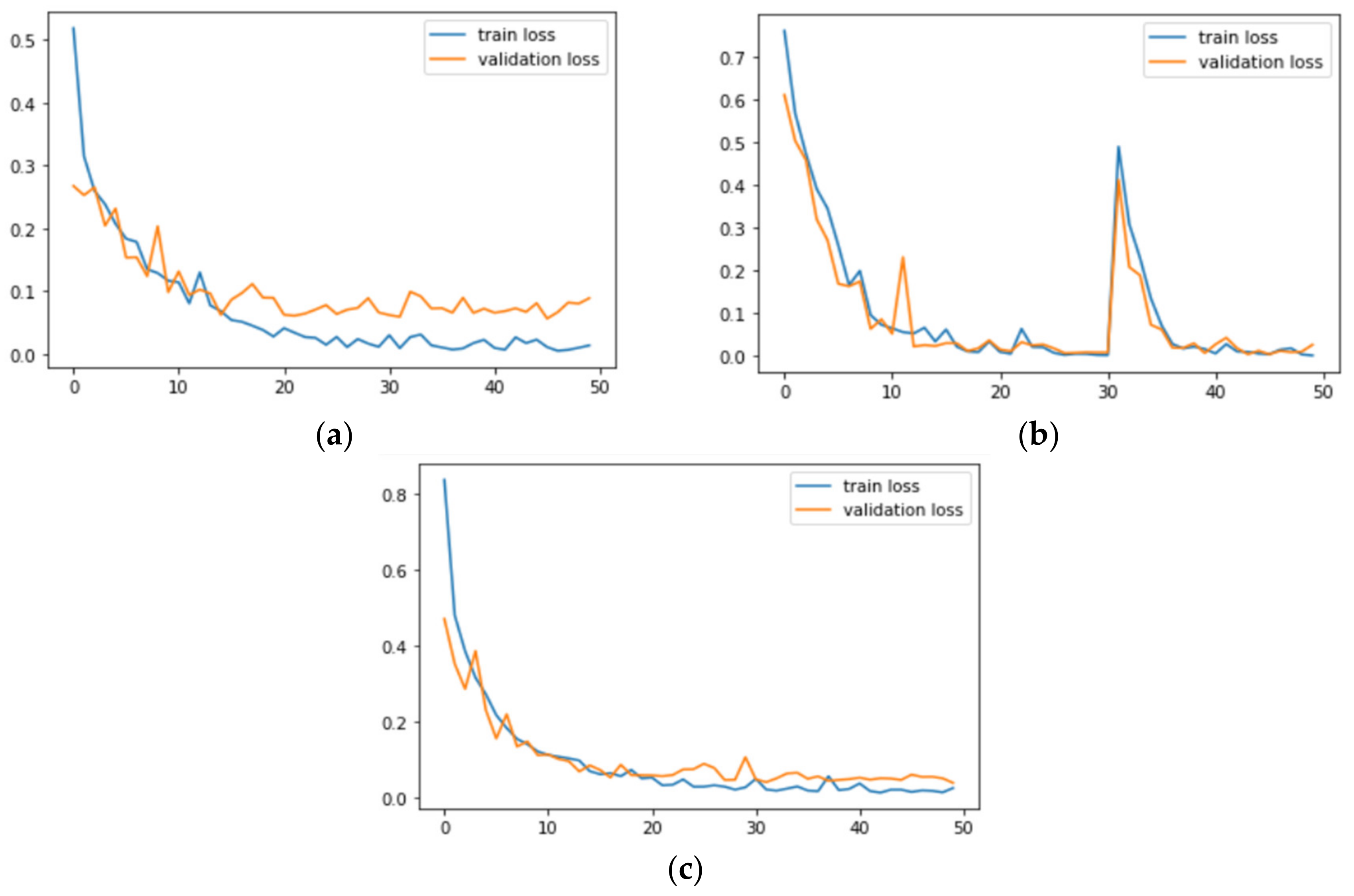
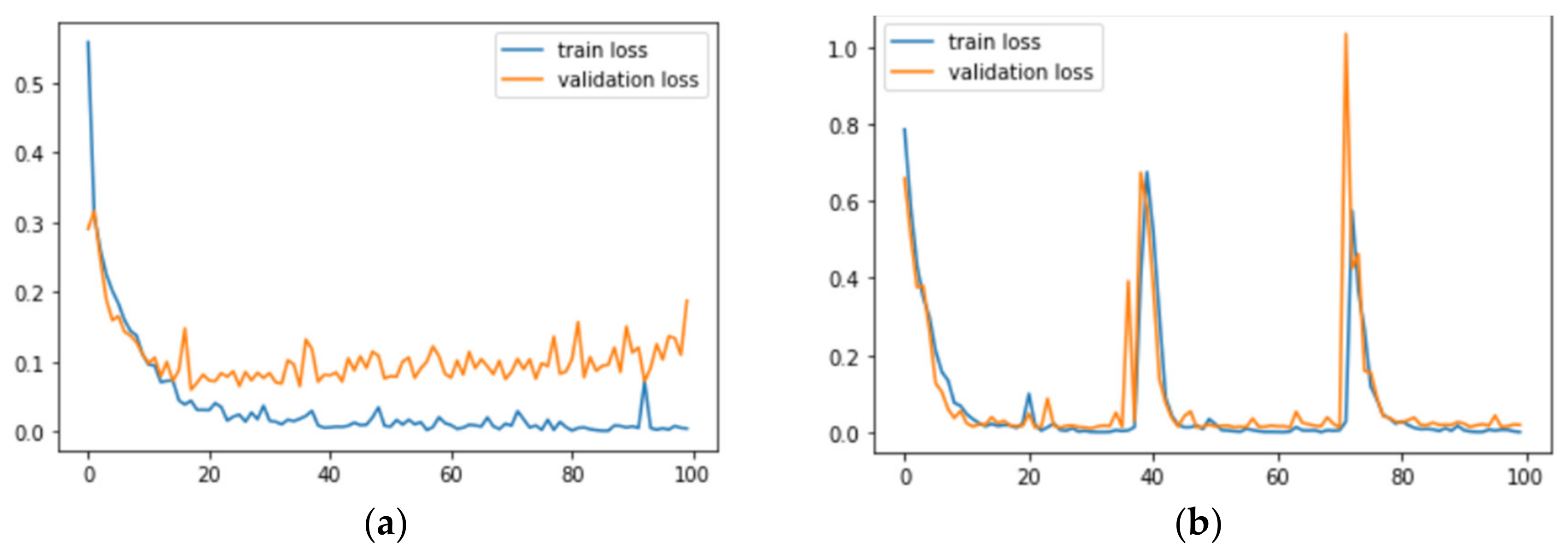
Figure 7 and
Figure 8 correspond to the accuracy and loss graphs of
AdenoCanNet trained for 50 epochs on different sets of the LC25000.
Table 4 discusses the results obtained when the models were trained for 100 epochs.
The results obtained for the models trained on histopathological images in the lung dataset run for 100 epochs show that ResNet50 and LeNet models achieved the highest training accuracy of 100%. The validation accuracy of ResNet and LeNet models correspond to 97.60% and 96.00%. It is worth noting that, among all the pre-existing architectures, VGG16 displayed a better performance on the validation dataset, achieving an accuracy of 97.20%. It is pertinent to note that the accuracy achieved by AdenoCanNet achieved a training accuracy of 99.96% and a validation accuracy of 98.77%. Among all the models, the suggested AdenoCanNet performs better on the unseen validation dataset. The training accuracy achieved by the CNN model is comparable with the performance of the ResNet and the LeNet models.
For the colon subset taken alone, the suggested AdenoCanNet with LeNet and ResNet recorded the highest accuracy of 100%. The validation accuracy obtained using the mentioned AdenoCanNet was the highest among all the other models, recording a value of 99.80%. It is worth noting that the proposed model of AdenoCanSVM also recorded a good training and validation accuracy of 95.46% and 99.40%. An accuracy of 98.50% and 99.10% reported for training and testing sets for the VGG16 model makes it another good model.
When histopathological images obtained from both lung and colon regions are taken into consideration, ResNet performed well, with both training and testing accuracy being 100%. However,
AdenoCanNet attained a training accuracy of 99.88% and validation accuracy of 99.00%, which is comparable with the performance of ResNet on the complete dataset.
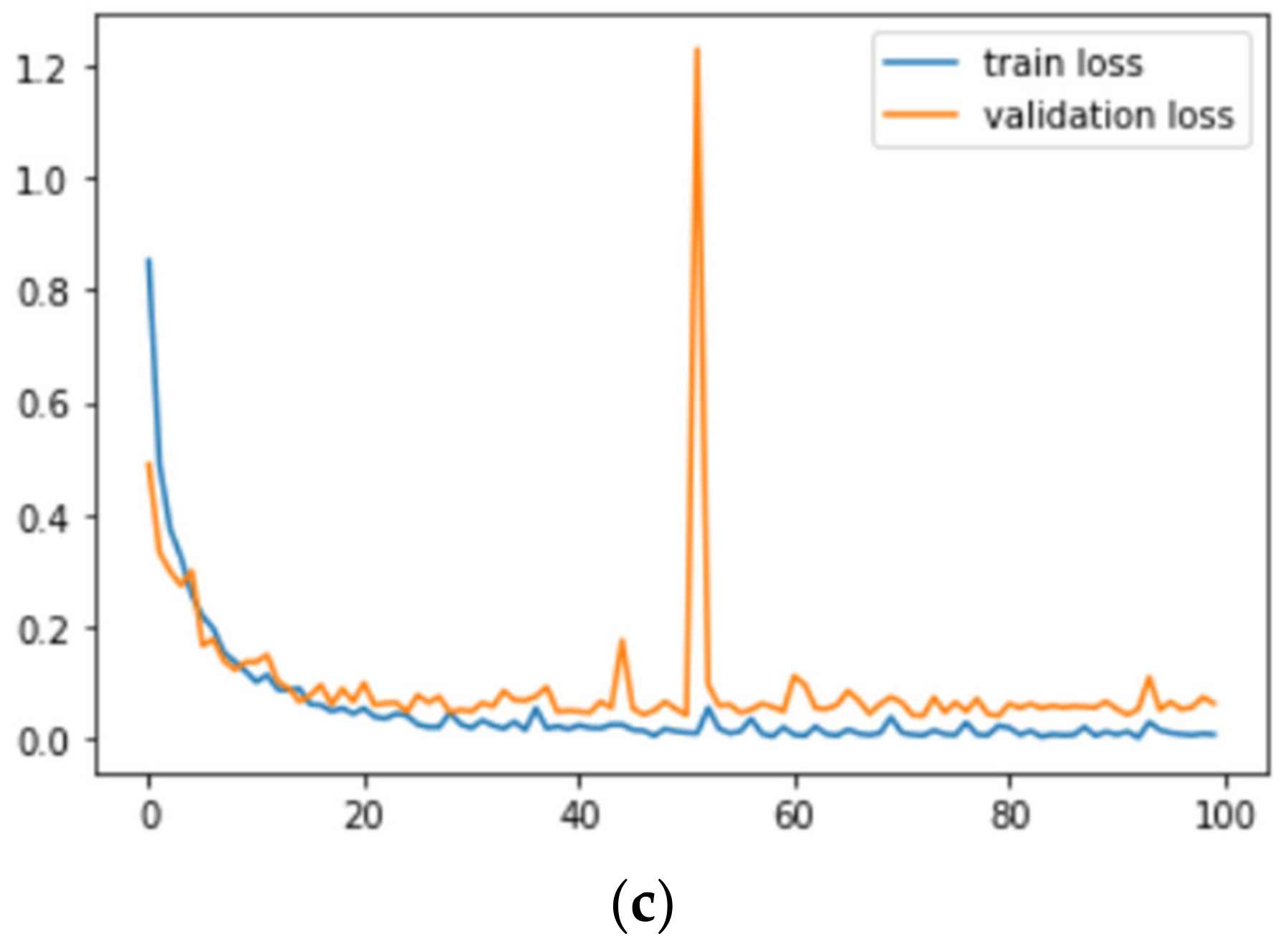
Figure 9 and
Figure 10 correspond to the accuracy and loss graphs of
AdenoCanNet trained for 100 epochs on different sets of the LC25000.
With the entire dataset into consideration, ResNet50 performs better in many cases. However, it is pertinent to note that ResNet50 is a 50-layer deep neural network, and training this model requires intense computation and time. The proposed AdenoCanNet consists of three convolution layers, two max pooling layers, and one dropout layer. The accuracies of this model and ResNet turn out to be comparable, and in some cases, AdenoCanNet outperforms the ResNet50 model.
When the models were trained on the entire dataset, the testing accuracy of AdenoCanNet outperformed the ResNet50 model when only lung and colon classes were considered.
LeNet-5 also displayed substantial results, however the AdenoCanNet outperformed the model in terms of testing accuracy for all cases when the entire dataset is considered.
For VGG16 and VGG19, the performance recorded was greatly comparable to the other architectures mentioned; however, AdenoCanNet outperformed both the pre-existing models in terms of training and testing accuracy in the case of the entire dataset.
Table 5 compares the results of the existing work on the detection and classification of lung and colon adenocarcinoma. Most works have opted for the LC25000 dataset and opted for CNN models; hence, these works are directly comparable.
For lung classes taken alone, Sanidhya Mangal et al. achieved an accuracy of 97%, and for colon classes alone Zarrin Tasnim et al. achieved a 96.6% accuracy rate. For CNN models experimented on the LC25000 dataset, the highest recorded accuracy turned out to be 99.8% achieved by Ben Hamida et al. With the obtained results and in comparison to the works discussed in the earlier section of this study, it can be concluded that, when considering the entire dataset of 25,000 histopathological images, AdenoCanNet introduced in this study performs with high accuracy and reliability, outperforming most of the existing models.
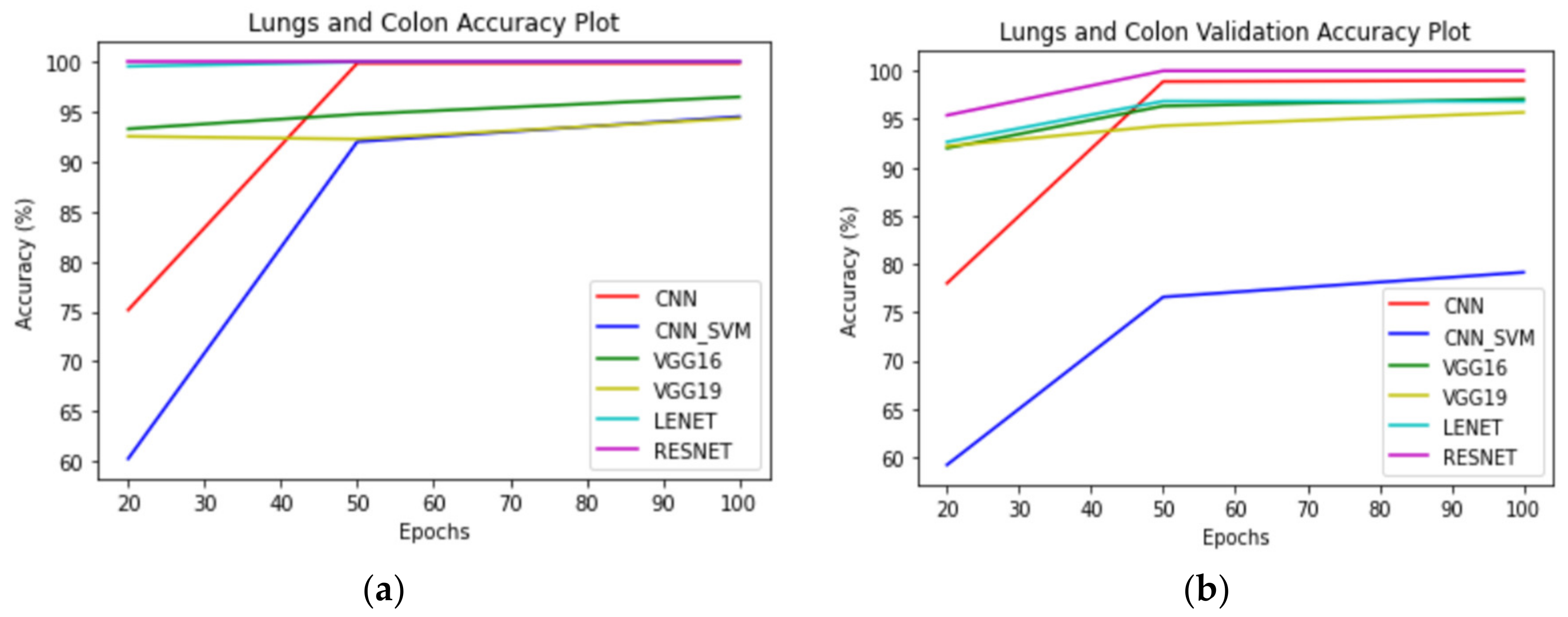
Figure 11,
Figure 12 and
Figure 13 correspond to performance comparison plots for lung, colon, and lung and colon regions of the LC25000 dataset. As observed from the above comparisons and explanation, the
AdenoCanNet model exhibited a better performance for the entire dataset for each category considered. Some recent works have used relatively deep neural networks or filtering multiple machine learning models based on accuracy obtained from them. The proposed model, having just three convolution layers, two max pooling layers, and one dropout layer, with minimum image preprocessing, achieved about 99% validation accuracy and 99.88% accuracy for detection and classification on the entire dataset of both lung and colon tissues. A 99.96% accuracy and 98.77% validation accuracy on the entire dataset of lung tissues were achieved, as well as 100% accuracy and 99.80% validation accuracy on the entire dataset of colon tissues with 100 epochs.