Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing
Abstract
1. Introduction
2. Materials and Methods
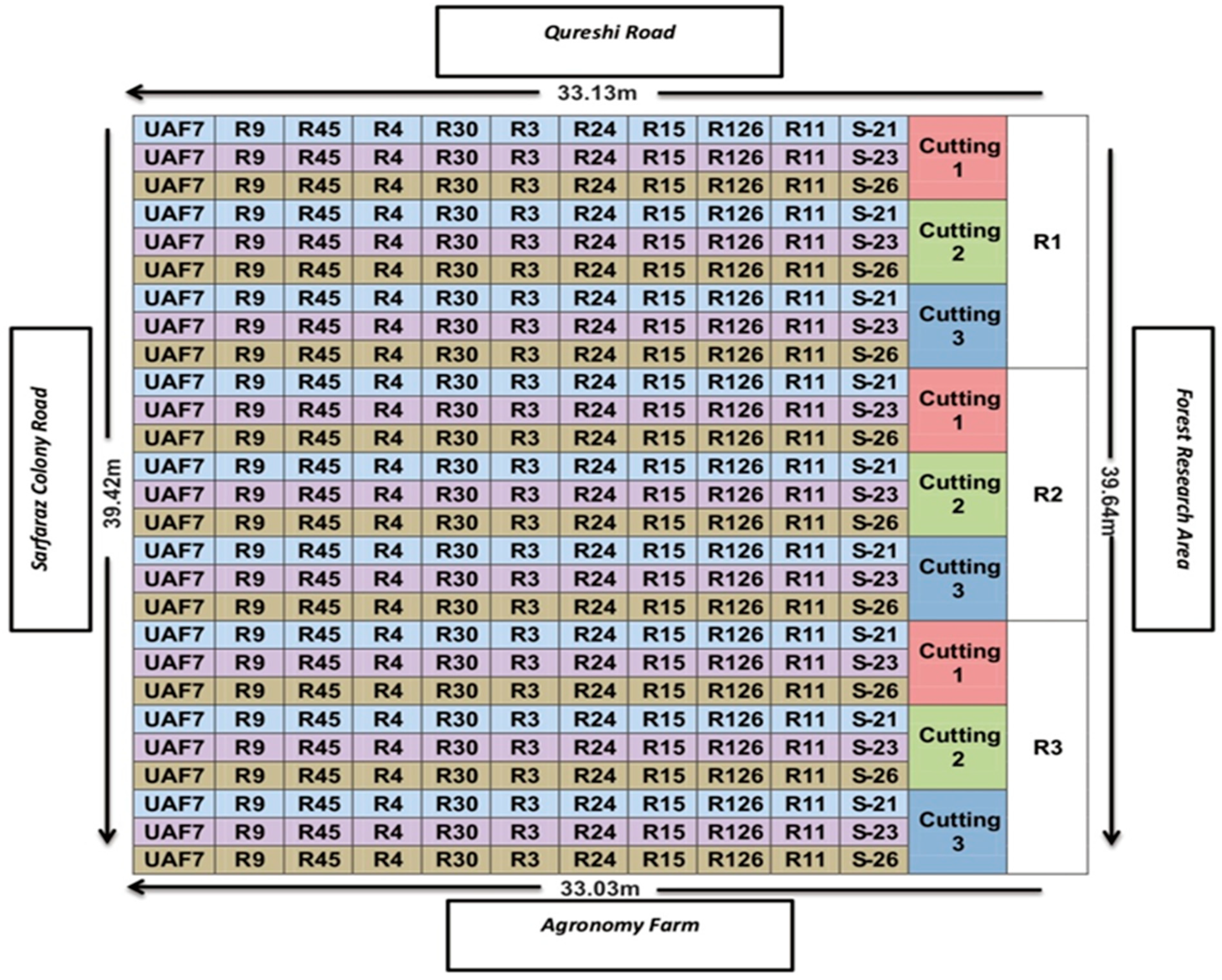
2.1. Experimental Site, Design, and Agronomic Managements
2.2. Measurements of Morphological and Quality Parameters
2.3. Statistical Analysis
3. Results
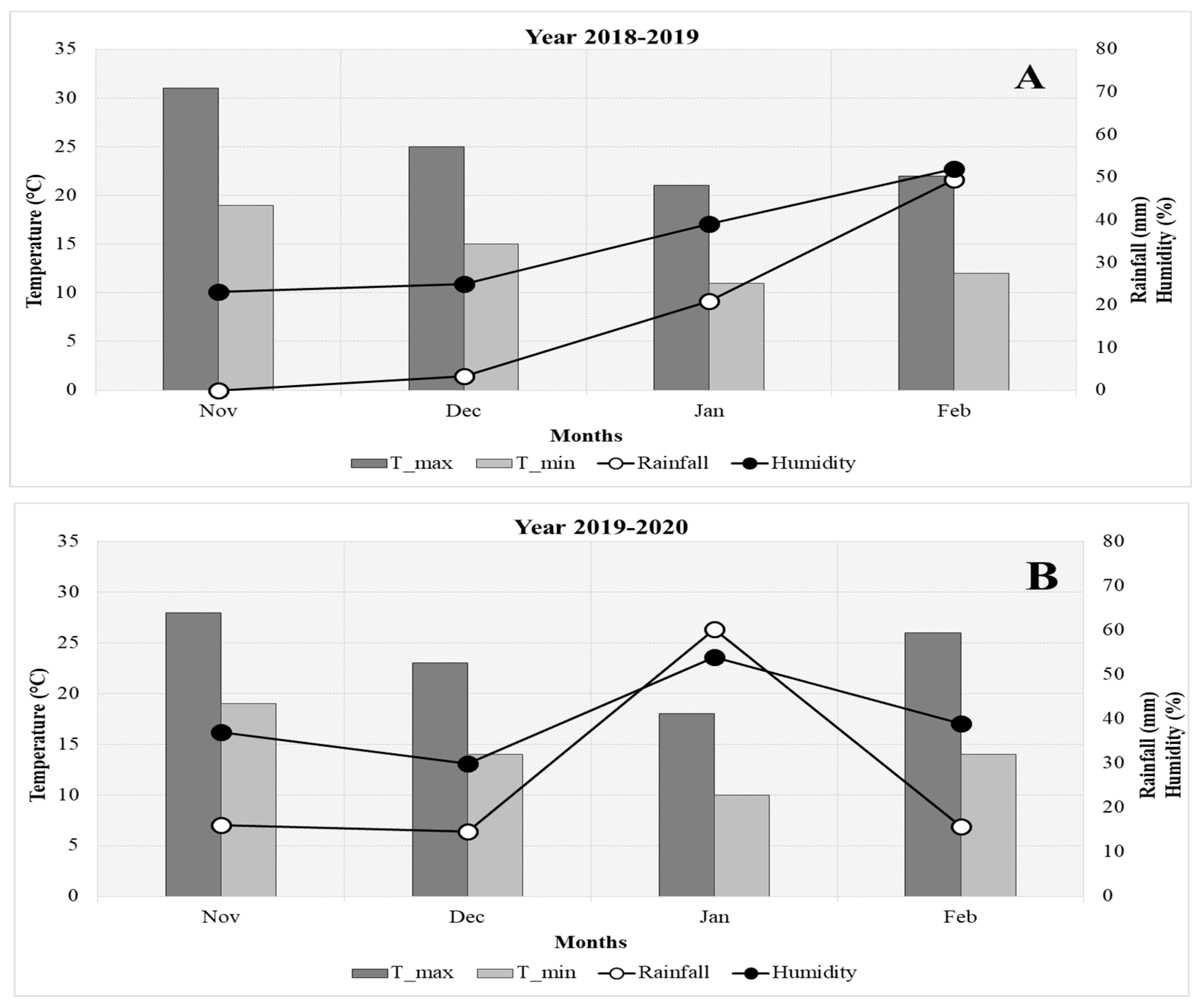
3.1. Climatic Conditions
3.2. Morphological and Forage Quality Traits of Quinoa Genotypes
3.3. MGIDI Index Selection
3.3.1. Selection of Best Application combination (Cut × Space) to Obtain the Maximum Desired Gains
3.3.2. Selection of Best Performing Genotypes
3.4. Correlation Analysis
3.4.1. Correlation Analysis of Cutting Level 1 in Both Years
3.4.2. Correlation Analysis of Cutting Level 2 in Both Years
3.4.3. Correlation Analysis of Cutting Level 3 in Both Years
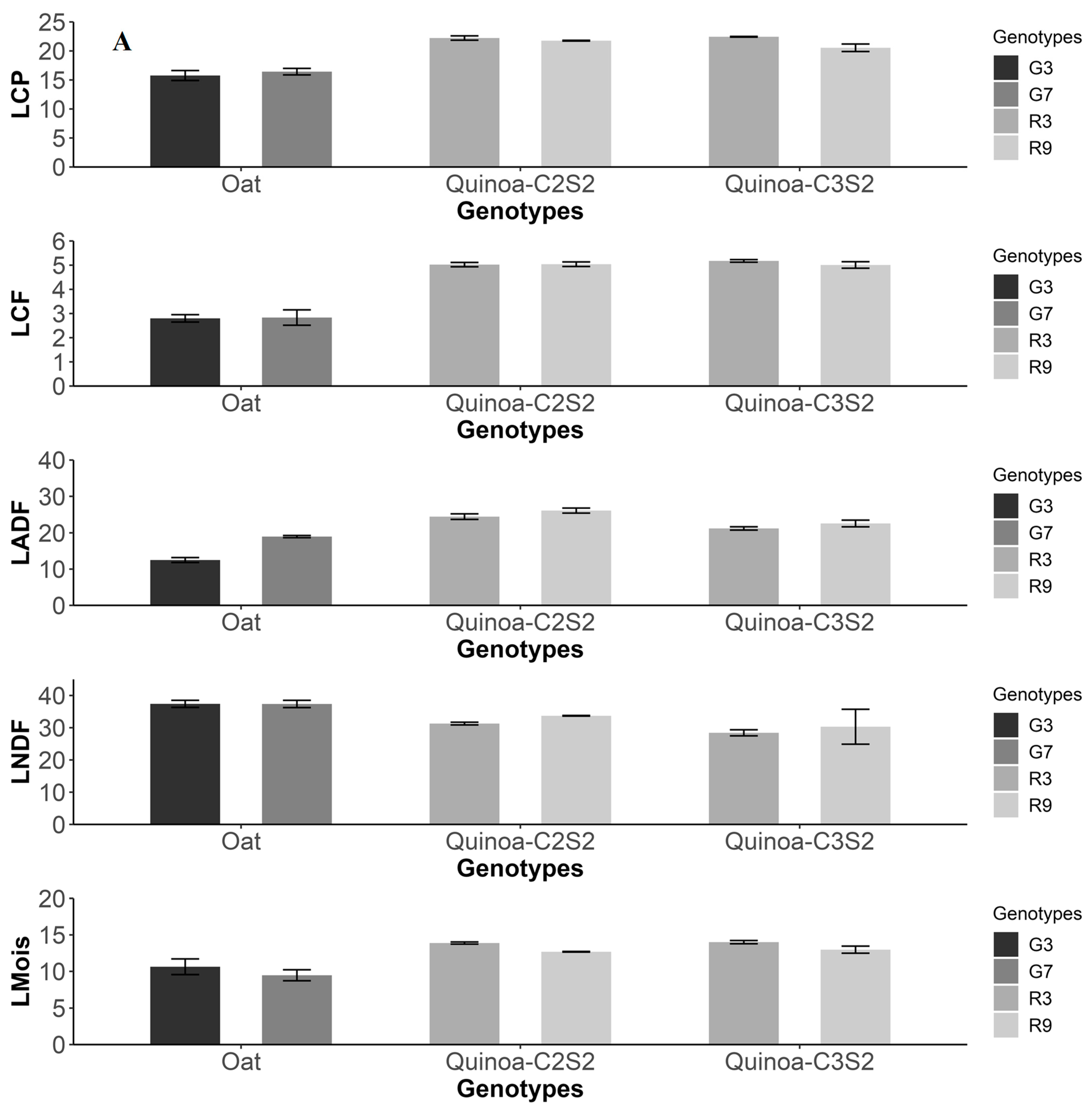
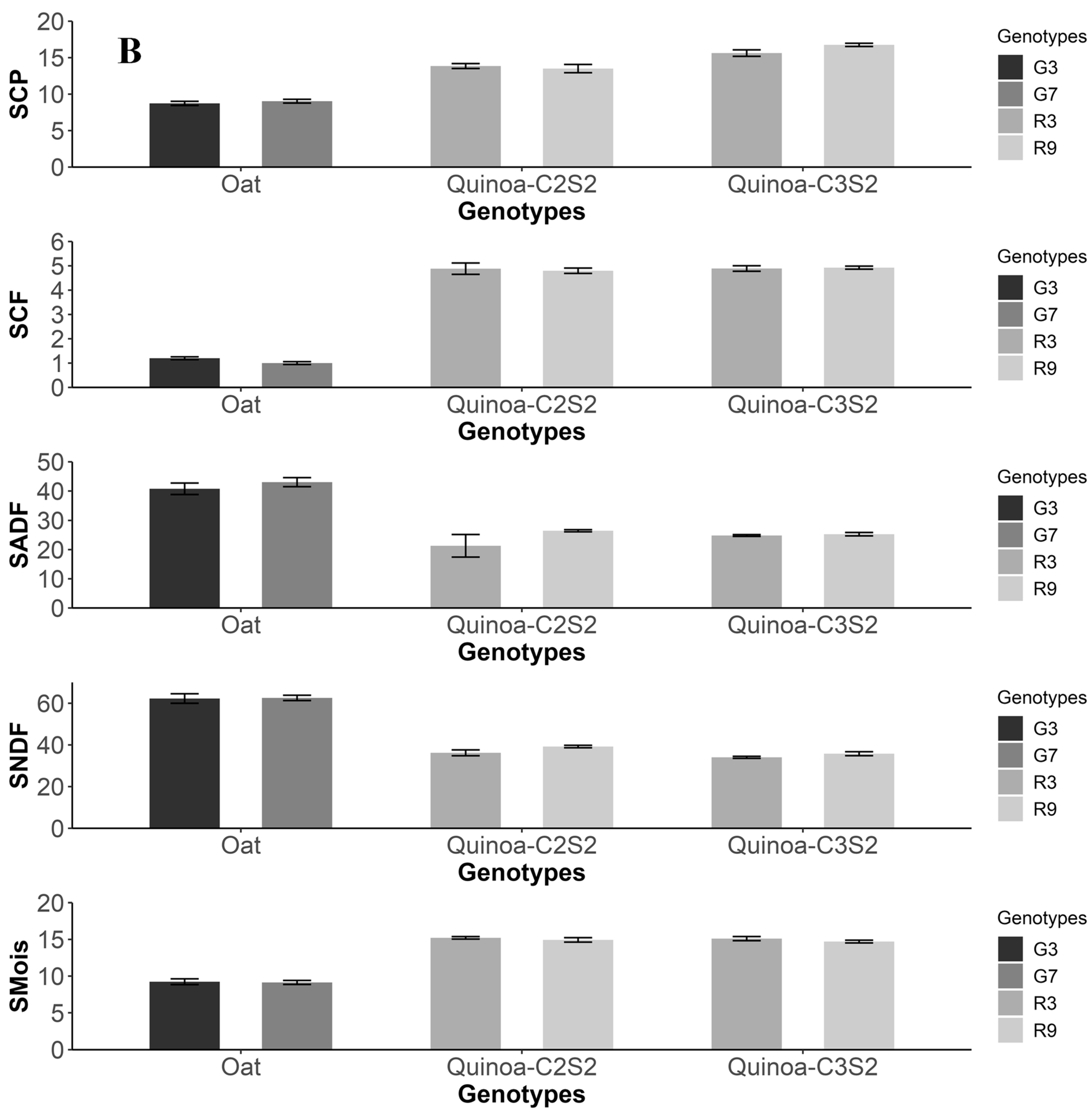
3.5. Comparison with Oat
4. Discussion
4.1. Environmental Factors Markedly Influenced Morphological and Forage Quality Traits of Quinoa in Both Years
4.2. Effect of Different Treatments on Morphological and Quality Traits of Quinoa
4.3. Treatment Combination and Genotypes Selection Using the MGIDI
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz-Bobea, A.; Ault, T.R.; Carrillo, C.M.; Chambers, R.G.; Lobell, D.B. Anthropogenic climate change has slowed global agricultural productivity growth. Nat. Clim. Chang. 2021, 11, 306–312. [Google Scholar] [CrossRef]
- Steensland, A. 2019 Global Agricultural Productivity Report: Productivity Growth for Sustainable Diets and More (Virginia Tech, 2019). Available online: http://hdl.handle.net/10919/96429 (accessed on 16 October 2019).
- Chambers, R.G.; Pieralli, S.; Sheng, Y. The millennium droughts and Australian agricultural productivity performance: A nonparametric analysis. Am. J. Agric. Econ. 2020, 102, 1383–1403. [Google Scholar] [CrossRef]
- Gil, J. Global climate sensitivity of cropping frequency and yields. Nat. Food 2023, 4, 114–123. [Google Scholar] [CrossRef]
- Navarro-Racines, C.; Tarapues, J.; Thornton, P.; Jarvis, A.; Ramirez-Villegas, J. High-resolution and bias-corrected CMIP5 projections for climate change impact assessments. Sci. Data 2020, 7, 7–15. [Google Scholar] [CrossRef]
- Fu, G.; Wang, J.; Li, S. Response of forage nutritional quality to climate change and human activities in alpine grasslands. Sci. Total Environ. 2022, 845, 157552. [Google Scholar] [CrossRef]
- Wijerathna-Yapa, A.; Pathirana, R. Sustainable Agro-Food Systems for Addressing Climate Change and Food Security. Agriculture 2022, 12, 1554. [Google Scholar] [CrossRef]
- Hirel, B.; Tétu, T.; Lea, P.J.; Dubois, F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 2011, 9, 1452–1485. [Google Scholar] [CrossRef]
- Donald, C.T. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Hazel, L.N. The genetic basis for constructing selection indexes. Genetics 1943, 28, 476–490. [Google Scholar] [CrossRef]
- Smith, H.F. A discriminant function for plant selection. Ann. Eugen. 1936, 7, 240–250. [Google Scholar] [CrossRef]
- Olivoto, T.; Nardino, M. MGIDI: Toward an effective multivariate selection in biological experiments. Bioinformatics 2021, 37, 1383–1389. [Google Scholar] [CrossRef]
- Farhad, M.; Tripathi, S.B.; Singh, R.P.; Joshi, A.K.; Bhati, P.K.; Vishwakarma, M.K.; Kumar, U. Multi-trait selection of bread wheat ideotypes for adaptation to early sown condition. Crop Sci. 2022, 62, 67–82. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Sanjani, S.; Nikkhah-Chamanabad, H.; Mehrvar, M.R.; Asadi, A.; Amini, A. Identification of salt-tolerant barley genotypes using multiple-traits index and yield performance at the early growth and maturity stages. Bull. Natl. Res. Cent. 2021, 45, 117. [Google Scholar] [CrossRef]
- León, R.; Rosero, A.; García, J.L.; Morelo, J.; Orozco, A.; Silva, G.; Ceballos, H. Multi-trait selection indices for identifying new cassava varieties adapted to the Caribbean region of Colombia. Agronomy 2021, 11, 1694. [Google Scholar] [CrossRef]
- Uddin, M.S.; Billah, M.; Afroz, R.; Rahman, S.; Jahan, N.; Hossain, M.G.; Hossain, A. Evaluation of 130 Eggplant (Solanum melongena L.) genotypes for future breeding program based on qualitative and quantitative traits, and various genetic parameters. Horticulturae 2021, 7, 376. [Google Scholar] [CrossRef]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.; Tester, M. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Risi, J.C. The Chenopodium grains of the Andes: Inca crops for modern agriculture. Adv. Appl. Biol. 1984, 10, 145–216. [Google Scholar]
- Bastidas, E.G.; Roura, R.; Rizzolo, D.A.D.; Massanés, T.; Gomis, R. Quinoa (Chenopodium quinoa Willd), from nutritional value to potential health benefits: An integrative review. J. Nutr. Food Sci. 2016, 6, 3–14. [Google Scholar] [CrossRef]
- Alamri, E.; Amany, B.; Bayomy, H. Quinoa seeds (Chenopodium Quinoa): Nutritional value and potential biological effects on hyperglycemic rats. J. King Saud Univ. Sci. 2023, 35, 102427. [Google Scholar] [CrossRef]
- Guo, Y. Heat-induced yield loss of quinoa. Nat. Food 2020, 1, 101–103. [Google Scholar] [CrossRef]
- Nalbandian, E.; Pietrysiak, E.; Murphy, K.M.; Ganjyal, G.M. Different breeding lines of quinoa significantly influence the quality of baked cookies and cooked grains. J. Food Sci. 2022, 87, 5225–5239. [Google Scholar] [CrossRef]
- Dangi, S. Oat as green fodder and its intercropping benefits: A review. Agric. Rev. 2021, 42, 66–72. [Google Scholar] [CrossRef]
- Pant, S.R.; Ghimire, R.P.; Prenil, K.C.; Upreti, S. Growth and yield of different oat (Avena sativa) varieties in Lalitpur district of Nepal. J. Agric. Nat. Resour. 2022, 5, 34–39. [Google Scholar] [CrossRef]
- Chapter VI—Fodder Oats in Pakistan. Available online: https://www.fao.org/3/y5765e/y5765e0a.htm#:~:text=Oats%20as%20fodder,land%20under%20forages%20in%20Pakistan (accessed on 1 January 2004).
- Uusitalo, V.; Leino, M. Neutralizing global warming impacts of crop production using biochar from side flows and buffer zones: A case study of oat production in the boreal climate zone. J. Clean. Prod. 2019, 227, 48–57. [Google Scholar] [CrossRef]
- Ugrenović, V.; Popović, V.; Ugrinović, M.; Filipović, V.; Mačkić, K.; Ljubičić, N.; Lakić, Ž. Black Oat (Avena strigosa Schreb.) Ontogenesis and Agronomic Performance in Organic Cropping System and Pannonian Environments. Agriculture 2021, 11, 55. [Google Scholar] [CrossRef]
- Demir, B.; Bilgiçli, N. Changes in chemical and anti-nutritional properties of pasta enriched with raw and germinated quinoa (Chenopodium quinoa Willd.) flours. J. Food Sci. Technol. 2020, 57, 3884–3892. [Google Scholar] [CrossRef]
- Ebeid, H.M.; Kholif, A.E.; El-Bordeny, N.; Chrenkova, M.; Mlynekova, Z.; Hansen, H.H. Nutritive value of quinoa (Chenopodium quinoa) as a feed for ruminants: In sacco degradability and in vitro gas production. Environ. Sci. Pollut. Res. 2022, 29, 35241–35252. [Google Scholar] [CrossRef]
- Kamal, H.; Habib, H.M.; Ali, A.; Show, P.L.; Koyande, A.K.; Kheadr, E.; Ibrahim, W.H. Food waste valorization potential: Fiber, sugar, and color profiles of 18 date seed varieties (Phoenix dactylifera, L.). J. Saudi Soc. Agric. Sci. 2023, 22, 133–138. [Google Scholar] [CrossRef]
- Farhadi, A.; Paknejad, F.; Golzardi, F.; Ilkaee, M.N.; Aghayari, F. Effects of limited irrigation and nitrogen rate on the herbage yield, water productivity, and nutritive value of sorghum silage. Commun. Soil Sci. Plant Anal. 2022, 53, 576–589. [Google Scholar] [CrossRef]
- Staniak, M.; Czopek, K.; Stępień-Warda, A.; Kocira, A.; Przybyś, M. Cold stress during flowering alters plant structure, yield and seed quality of different soybean genotypes. Agronomy 2021, 11, 2059. [Google Scholar] [CrossRef]
- Ayyat, M.S.; A-Abdel-Rahman, G.; Ayyat, A.M.N.; Abdel-Rahman, M.S.; Al-Sagheer, A.A. Evaluation of leaf protein concentrate from Beta vulgaris and Daucus carota as a substitute for soybean meal in Oreochromis niloticus fingerlings diets. Aquac. Res. 2021, 52, 3256–3269. [Google Scholar] [CrossRef]
- Jahanzaib-Saddiq, M.S.; Basra, S.M.; Nawaz, M.F.; Iqbal, S.; Khan, S.; Hafeez, M.B. Inorganic NPK supplementation improves biomass and quality of multicut moringa fodder. J. Plant Nutr. 2023, 46, 1519–1532. [Google Scholar] [CrossRef]
- Asher, A.; Dagan, R.; Galili, S.; Rubinovich, L. Effect of Row Spacing on Quinoa (Chenopodium quinoa) Growth, Yield, and Grain Quality under a Mediterranean Climate. Agriculture 2022, 12, 1298. [Google Scholar] [CrossRef]
- Zulkadir, G. The agroecological impact of different sowing dates and row spacing applications in quinoa (Chenopodium quinoa willd.). Appl. Ecol. Environ. Sci. 2021, 19, 751–762. [Google Scholar] [CrossRef]
- Menšík, L.; Kincl, D.; Nerušil, P.; Srbek, J.; Hlisnikovský, L.; Smutný, V. Water erosion reduction using different soil tillage approaches for maize (Zea mays L.) in the Czech Republic. Land 2020, 9, 358. [Google Scholar] [CrossRef]
- Zicarelli, F.; Sarubbi, F.; Iommelli, P.; Grossi, M.; Lotito, D.; Lombardi, P.; Musco, N. Nutritional Characterization of Hay Produced in Campania Region: Analysis by the near Infrared Spectroscopy (NIRS) Technology. Animals 2022, 12, 3035. [Google Scholar] [CrossRef]
- Hand, D.J.; Taylor, C.C. Multivariate Analysis of Variance and Repeated Measures: A Practical Approach for Behavioural Scientists; CRC Press: Boca Raton, FL, USA, 1987; Volume 5. [Google Scholar]
- Olivoto, T.; Lúcio, A.D.C. metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Maamri, K.; Zidane, O.D.; Chaabena, A.; Fiene, G.; Bazile, D. Adaptation of some quinoa genotypes (Chenopodium quinoa Willd.), grown in a saharan climate in Algeria. Life 2022, 12, 1854. [Google Scholar] [CrossRef]
- Alvar-Beltrán, J.; Verdi, L.; Dalla Marta, A.; Dao, A.; Vivoli, R.; Sanou, J.; Orlandini, S. The effect of heat stress on quinoa (cv. Titicaca) under controlled climatic conditions. J. Agric. Sci. 2020, 158, 255–261. [Google Scholar] [CrossRef]
- Bakhtavar, M.A.; Afzal, I. Climate smart Dry Chain Technology for safe storage of quinoa seeds. Sci. Rep. 2020, 10, 12554. [Google Scholar] [CrossRef]
- Behrouzi, D.; Diyanat, M.; Majidi, E.; Mirhadi, M.J.; Shirkhani, A. Yield and quality of forage maize as a function of diverse irrigation regimes and biofertilizer in the west of Iran. J. Plant Nutr. 2022, 46, 2246–2256. [Google Scholar] [CrossRef]
- Noor, M.A.; Fiaz, S.; Nawaz, A.; Nawaz, M.M. The effects of cutting interval on agro-qualitative traits of different millet (Pennisetum americanum L.) cultivars. J. Saudi Soc. Agric. Sci. 2018, 17, 317–322. [Google Scholar] [CrossRef]
- İbrahim, A.T.I.S.; Celiktas, N.; Ersin, C.A.N.; Yilmaz, S. The effects of cutting intervals and seeding rates on forage yield and quality of alfalfa. Turk. J. Field Crops 2019, 24, 12–20. [Google Scholar] [CrossRef]
- Zaheer, S.; Arif, M.; Akhtar, K.; Khan, A.; Khan, A.; Bibi, S.; Wei, F. Grazing and cutting under different nitrogen rates, application methods and planting density strongly influence qualitative traits and yield of canola crop. Agronomy 2020, 10, 404. [Google Scholar] [CrossRef]
- Ma, J.; Dai, H.; Liu, H.; Du, W. Effects of Cutting Stages and Additives on the Fermentation Quality of Triticale, Rye and Oat Silage in Qinghai-Tibet Plateau. Agronomy 2022, 12, 3113. [Google Scholar] [CrossRef]
- Yilmaz, Ş.; Ertekin, I.; İbrahim, A.T.I.Ş. Forage yield and quality of quinoa (Chenopodium quinoa Willd.) genotypes harvested at different cutting stages under Mediterranean conditions. Turk. J. Field Crops 2021, 26, 202–209. [Google Scholar] [CrossRef]
- Liu, M.; Yang, M.; Yang, H. Biomass production and nutritional characteristics of quinoa subjected to cutting and sowing date in the midwestern China. Grassl. Sci. 2021, 67, 215–224. [Google Scholar] [CrossRef]
- Ciftci, S.; Zulkadir, G.; Gökçe, M.S.; Karaburu, E.; Bozdağ, E.; Idikut, L. The effect of row distances on quinoa yield and yield components in the late planting period. Int. J. Res. Publ. Rev. 2020, 1, 37–42. [Google Scholar]
- Reddy, K.R.; Singh, R.; Singh, E. Effect of spacing and nitrogen management on yield and economics of quinoa (Chenopodium quinoa). J. Pharm. Innov. 2021, 10, 543–545. [Google Scholar]
- Temel, S.; Yolcu, S. The effect of different sowing time and harvesting stages on the herbage yield and quality of quinoa (Chenopodium quinoa Willd.). Turk. J. Field Crops 2020, 25, 41–49. [Google Scholar] [CrossRef]
- Temel, İ.; Keskin, B. Farklı sıra arası ve sıra üzeri mesafelerinin kinoa (Chenopodium quinoa Willd.)’nın besin içeriğine etkisi. Turk. J. Field Crops 2019, 5, 110–116. [Google Scholar] [CrossRef]
- Sief, A.S.; El-Deepah, H.R.A.; Kamel, A.S.M.; Ibrahim, J.F. Effect of various inter and intra spaces on the yield and quality of quinoa (Chenopodium quinoa Willd.). J. Plant Prod. 2015, 6, 371–383. [Google Scholar] [CrossRef]
- Alwala, S.; Kwolek, T.; McPherson, M.; Pellow, J.; Meyer, D. A comprehensive comparison between Eberhart and Russell joint regression and GGE biplot analyses to identify stable and high yielding maize hybrids. Field Crops Res. 2010, 119, 225–230. [Google Scholar] [CrossRef]
- Benakanahalli, N.K.; Sridhara, S.; Ramesh, N.; Olivoto, T.; Sreekantappa, G.; Tamam, N.; Abdelmohsen, S.A. A framework for identification of stable genotypes basedon MTSI and MGDII indexes: An example in guar (Cymopsis tetragonoloba L.). Agronomy 2021, 11, 1221. [Google Scholar] [CrossRef]
- Loro, M.V.; Carvalho, I.R.; Cargnelutti Filho, A.; Hoffmann, J.F.; Kehl, K. Wheat grain biofortification for essential amino acids. Pesq. Agropec. Bras. 2023, 58, e02860. [Google Scholar] [CrossRef]
- Osuna-Caballero, S.; Rispail, N.; Barilli, E.; Rubiales, D. Identification and Characterization of Novel Sources of Resistance to Rust Caused by Uromyces pisi in Pisum spp. Plants 2022, 11, 2268. [Google Scholar] [CrossRef]
- Adewumi, A.S.; Asare, P.A.; Adejumobi, I.I.; Adu, M.O.; Taah, K.J.; Adewale, S.; Agre, P.A. Multi-Trait Selection Index for Superior Agronomic and Tuber Quality Traits in Bush Yam (Dioscorea praehensilis Benth.). Agronomy 2023, 13, 682. [Google Scholar] [CrossRef]
- Hoogendoorn, J.; Rickson, J.M.; Gale, M.D. Differences in leaf and stem anatomy related to plant height of tall and dwarf wheat (Triticum aestivum L.). J. Plant Physiol. 1990, 136, 72–77. [Google Scholar] [CrossRef]
- Díaz-Pérez, J.C. Bell pepper (Capsicum annum L.) crop as affected by shade level: Microenvironment, plant growth, leaf gas exchange, and leaf mineral nutrient concentration. J. Am. Soc. Hortic. Sci. 2013, 48, 175–182. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Hyles, J.; Joaquim, P.; Azanza, F.; Bonnett, D.; Ellis, M.E.; Richards, R.A. A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor. Appl. Genet. 2007, 115, 59–66. [Google Scholar] [CrossRef]
- Park, Y.; Runkle, E.S. Far-red radiation promotes growth of seedlings by increasing leaf expansion and whole-plant net assimilation. Environ. Exp. Bot. 2017, 136, 41–49. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.L.; Kamran, M.; Hamayun, M.; Kang, S.M.; Kim, Y.H.; Lee, I.J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 2012, 17, 10754–10773. [Google Scholar] [CrossRef]
- Salesse-Smith, C.E.; Sharwood, R.E.; Busch, F.A.; Kromdijk, J.; Bardal, V.; Stern, D.B. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 2018, 4, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Egli, D.B. Alterations in plant growth and dry matter distribution in soybean. Agron. J. 1988, 80, 86–90. [Google Scholar] [CrossRef]
- Kamran, M.; Cui, W.; Ahmad, I.; Meng, X.; Zhang, X.; Su, W.; Liu, T. Effect of paclobutrazol, a potential growth regulator on stalk mechanical strength, lignin accumulation and its relation with lodging resistance of maize. Plant Growth Regul. 2018, 84, 317–332. [Google Scholar] [CrossRef]
- Chen, H.; Wang, C.; Huasai, S.; Chen, A. Effects of dietary forage to concentrate ratio on nutrient digestibility, ruminal fermentation and rumen bacterial composition in Angus cows. Sci. Rep. 2021, 11, 17023. [Google Scholar] [CrossRef]
- Pavlů, K.; Kassahun, T.; Pavlů, V.V.; Pavlů, L.; Blažek, P.; Homolka, P. The effects of first defoliation and previous management intensity on forage quality of a semi-natural species-rich grassland. PLoS ONE 2021, 16, e0248804. [Google Scholar] [CrossRef]
- Wang, H.; Coulman, B.; Bai, Y.; Tar’an, B.; Biligetu, B. Genetic diversity and local adaption of alfalfa populations (Medicago sativa L.) under long-term grazing. Sci. Rep. 2023, 13, 1632–1640. [Google Scholar] [CrossRef]
- Muir, J.P. Effect of dairy compost application and plant maturity on forage kenaf cultivar fiber concentration and in sacco disappearance. Crop Sci. 2002, 42, 248–254. [Google Scholar] [CrossRef]
- Beauchemin, K.A. Using ADF and NDF in dairy cattle diet formulation—A western Canadian perspective. Anim. Feed Sci. Technol. 1996, 58, 101–111. [Google Scholar] [CrossRef]
- Javanmard, A.; Nasab, A.D.M.; Javanshir, A.; Moghaddam, M.; Janmohammadi, H. Forage yield and quality in intercropping of maize with different legumes as double-cropped. J. Food Agric. Environ. 2009, 7, 163–166. [Google Scholar]
- Kering, M.K.; Guretzky, J.; Funderburg, E.; Mosali, J. Effect of nitrogen fertilizer rate and harvest season on forage yield, quality, and macronutrient concentrations in midland Bermuda grass. Commun. Soil Sci. Plant Anal. 2011, 42, 1958–1971. [Google Scholar] [CrossRef]
- Muhammad, U.H.; Gurmani, Z.A.; Khan, S.; Bakhsh, A. PARC Oat: A new Lodging-resistant, Late-maturing and High-yielding Variety of Oats (Avena sativa L.) for Pakistan. Pak. J. Agric. Res. 2020, 33, 601. [Google Scholar] [CrossRef]
- Peng, Y.; Ren, C. Genome origin and evolution of common oat. Nat. Genet. 2022, 54, 1074–1075. [Google Scholar] [CrossRef]
 ) represent plant residues after harvesting. Note: For leaves’ quality traits, the prefix “L” was used with each trait, in other words, LCP, LCF, LADF, and LNDF. Similarly, for stems, the prefix “S” was used with each trait, in other words, SCP, SCF, SADF and SNDF.
) represent plant residues after harvesting. Note: For leaves’ quality traits, the prefix “L” was used with each trait, in other words, LCP, LCF, LADF, and LNDF. Similarly, for stems, the prefix “S” was used with each trait, in other words, SCP, SCF, SADF and SNDF.
 ) represent plant residues after harvesting. Note: For leaves’ quality traits, the prefix “L” was used with each trait, in other words, LCP, LCF, LADF, and LNDF. Similarly, for stems, the prefix “S” was used with each trait, in other words, SCP, SCF, SADF and SNDF.
) represent plant residues after harvesting. Note: For leaves’ quality traits, the prefix “L” was used with each trait, in other words, LCP, LCF, LADF, and LNDF. Similarly, for stems, the prefix “S” was used with each trait, in other words, SCP, SCF, SADF and SNDF.
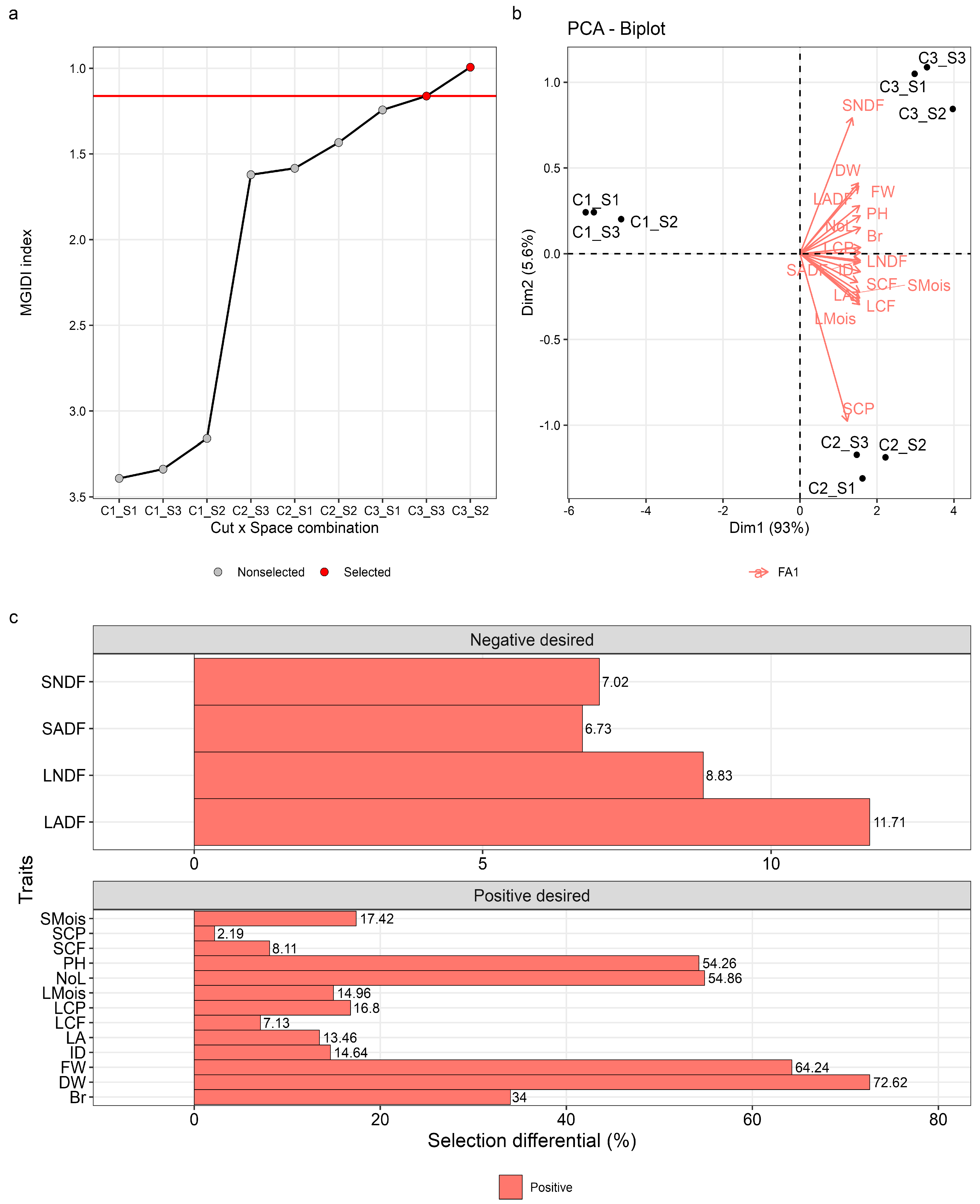

| Factors | Traits | Sense | Xo | Xs | SD | SD% | Goal |
|---|---|---|---|---|---|---|---|
| FA1 | PH | increase | 59.35 | 91.55 | 32.20 | 54.26 | 100 |
| FA1 | NoL | increase | 148.94 | 230.66 | 81.71 | 54.86 | 100 |
| FA1 | LA | increase | 34.96 | 39.67 | 4.71 | 13.46 | 100 |
| FA1 | Br | increase | 25.85 | 34.65 | 8.79 | 34.00 | 100 |
| FA1 | ID | increase | 4.61 | 5.29 | 0.68 | 14.64 | 100 |
| FA1 | FW | increase | 91.32 | 150.00 | 58.67 | 64.24 | 100 |
| FA1 | DW | increase | 18.32 | 31.63 | 13.31 | 72.62 | 100 |
| FA1 | LCP | increase | 17.93 | 20.94 | 3.01 | 16.80 | 100 |
| FA1 | LCF | increase | 4.70 | 5.04 | 0.34 | 7.13 | 100 |
| FA1 | LADF | decrease | 22.04 | 24.62 | 2.58 | 11.71 | 0 |
| FA1 | LNDF | decrease | 29.26 | 31.84 | 2.58 | 8.83 | 0 |
| FA1 | LMois | increase | 11.21 | 12.89 | 1.68 | 14.96 | 100 |
| FA1 | SCP | increase | 13.03 | 13.32 | 0.29 | 2.19 | 100 |
| FA1 | SCF | increase | 4.41 | 4.77 | 0.36 | 8.11 | 100 |
| FA1 | SADF | decrease | 24.02 | 25.64 | 1.62 | 6.73 | 0 |
| FA1 | SNDF | decrease | 34.31 | 36.72 | 2.41 | 7.02 | 0 |
| FA1 | SMois | increase | 12.46 | 14.63 | 2.17 | 17.42 | 100 |
| total increase | 28.8 | ||||||
| total decrease | 8.57 | ||||||
| Factors | Traits | Sense | Xo | Xs | SD | SD% | Goal |
|---|---|---|---|---|---|---|---|
| FA1 | LA | Increase | 34.96 | 35.13 | 0.17 | 0.49 | 100 |
| FA1 | LADF | Decrease | 22.04 | 20.09 | −1.95 | −8.83 | 100 |
| FA1 | LNDF | Decrease | 29.26 | 27.91 | −1.35 | −4.60 | 100 |
| FA1 | SADF | Decrease | 24.02 | 23.55 | −0.48 | −1.98 | 100 |
| FA1 | SMois | Increase | 12.46 | 12.66 | 0.21 | 1.66 | 100 |
| FA2 | PH | Increase | 59.35 | 68.06 | 8.71 | 14.68 | 100 |
| FA2 | LCP | Increase | 17.93 | 18.34 | 0.41 | 2.28 | 100 |
| FA2 | LCF | Increase | 4.70 | 4.70 | 0.00 | 0.04 | 100 |
| FA2 | SNDF | Decrease | 34.31 | 34.89 | 0.58 | 1.70 | 0 |
| FA3 | NoL | Increase | 148.94 | 169.31 | 20.37 | 13.68 | 100 |
| FA3 | Br | Increase | 25.85 | 26.43 | 0.58 | 2.23 | 100 |
| FA3 | FW | Increase | 91.32 | 105.83 | 14.50 | 15.88 | 100 |
| FA3 | DW | Increase | 18.32 | 21.08 | 2.75 | 15.02 | 100 |
| FA4 | LMois | Increase | 11.21 | 11.29 | 0.08 | 0.68 | 100 |
| FA4 | SCP | Increase | 13.03 | 13.27 | 0.24 | 1.81 | 100 |
| FA5 | ID | Increase | 4.62 | 4.54 | −0.07 | −1.61 | 0 |
| FA5 | SCF | Increase | 4.41 | 4.51 | 0.10 | 2.16 | 100 |
| Total Increase | 5.31 | ||||||
| Total Decrease | −3.43 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.R.; Ali, Z.; Ijaz, I.; Khan, Z.; Gul, N.; Pervaiz, S.; Alharby, H.F.; Tan, D.K.Y.; Tariq, M.S.; Ghaffar, M.; et al. Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing. Sustainability 2023, 15, 11446. https://doi.org/10.3390/su151411446
Ahmed SR, Ali Z, Ijaz I, Khan Z, Gul N, Pervaiz S, Alharby HF, Tan DKY, Tariq MS, Ghaffar M, et al. Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing. Sustainability. 2023; 15(14):11446. https://doi.org/10.3390/su151411446
Chicago/Turabian StyleAhmed, Syed Riaz, Zeba Ali, Iram Ijaz, Zafran Khan, Nimra Gul, Soha Pervaiz, Hesham F. Alharby, Daniel K. Y. Tan, Muhammad Sayyam Tariq, Maria Ghaffar, and et al. 2023. "Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing" Sustainability 15, no. 14: 11446. https://doi.org/10.3390/su151411446
APA StyleAhmed, S. R., Ali, Z., Ijaz, I., Khan, Z., Gul, N., Pervaiz, S., Alharby, H. F., Tan, D. K. Y., Tariq, M. S., Ghaffar, M., Bibi, A., & Hakeem, K. R. (2023). Multi-Trait Selection of Quinoa Ideotypes at Different Levels of Cutting and Spacing. Sustainability, 15(14), 11446. https://doi.org/10.3390/su151411446












