Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion
Abstract
:1. Introduction
2. Related Works
2.1. The Definition of Influenza-Like Illnesses
2.2. The Selection of Training Features
2.3. Machine Learning Models for Predicting Outbreaks of Influenza-Like Illnesses
3. Methodology
3.1. Prediction Framework
3.2. Data Pre-Processing
3.3. Keyword Volumes from Google Trends
3.4. Lag Features
3.5. Periodicity
3.6. Machine Learning Models
3.6.1. eXtreme Gradient Boosting (XGBoost)
3.6.2. Random Forest (RF)
3.6.3. Support Vector Regression (SVR)
3.7. Model Evaluation
3.8. Model Testing and Adjustments
3.9. Experimental Design
4. Experiments and Evaluation Results
4.1. Datasets
4.2. Comparisons of Different Combinations of Features
4.3. Evaluation Results and Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Abdelrahman, Z.; Li, M.; Wang, X. Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.T.; Liu, Y.X.; Zhu, Y.C.; Qian, J.; Ye, R.Z.; Li, C.Y.; Ji, X.K.; Li, H.K.; Qi, C.; Wang, Y.; et al. Impacts of transportation and meteorological factors on the transmission of COVID-19. Int. J. Hyg. Environ. Health 2020, 230, 113610. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Wang, L.; Cauchemez, S.; Xu, X.; Wang, X.; Cowling, B.J.; Meyers, L.A. Risk for transportation of coronavirus disease from Wuhan to other cities in China. Emerg. Infect. Dis. 2020, 26, 1049. [Google Scholar] [CrossRef] [Green Version]
- Taiwan Centers for Disease Control. Practical Guidelines for Prevention and Control of Seasonal Influenza; Report; Taiwan Centers for Disease Control: Taipei, Taiwan, 2020. [Google Scholar]
- du Prel, J.B.; Puppe, W.; Gröndahl, B.; Knuf, M.; Weigl, F.; Schaaff, F.; Schaaff, F.; Schmitt, H.J. Are meteorological parameters associated with acute respiratory tract infections? Clin. Infect. Dis. 2009, 49, 861–868. [Google Scholar] [CrossRef] [Green Version]
- Chan, P.K.; Mok, H.; Lee, T.; Chu, I.M.; Lam, W.; Sung, J.J. Seasonal influenza activity in Hong Kong and its association with meteorological variations. J. Med Virol. 2009, 81, 1797–1806. [Google Scholar] [CrossRef]
- Taiwan Centers for Disease Control. Severe Complicated Influenza; Taiwan Centers for Disease Control: Taipei, Taiwan, 2020. [Google Scholar]
- Wang, Y.; Xu, K.; Kang, Y.; Wang, H.; Wang, F.; Avram, A. Regional influenza prediction with sampling Twitter data and PDE model. Int. J. Environ. Res. Public Health 2020, 17, 678. [Google Scholar] [CrossRef] [Green Version]
- Seo, D.W.; Shin, S.Y. Methods using social media and search queries to predict infectious disease outbreaks. Healthc. Inform. Res. 2017, 23, 343. [Google Scholar] [CrossRef] [Green Version]
- Daughton, A.R.; Chunara, R.; Paul, M.J. Comparison of social media, syndromic surveillance, and microbiologic acute respiratory infection data: Observational study. Jmir Public Health Surveill. 2020, 6, e14986. [Google Scholar] [CrossRef]
- Lampos, V.; Zou, B.; Cox, I.J. Enhancing feature selection using word embeddings: The case of flu surveillance. In Proceedings of the 26th International Conference on World Wide Web, Perth, Australia, 3–7 April 2017; pp. 695–704. [Google Scholar]
- Volkova, S.; Ayton, E.; Porterfield, K.; Corley, C.D. Forecasting influenza-like illness dynamics for military populations using neural networks and social media. PLoS ONE 2017, 12, e0188941. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Agrawal, A.; Choudhary, A. Forecasting influenza levels using real-time social media streams. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; pp. 409–414. [Google Scholar]
- Huang, L.H. A Deep Learning Based Approach to Forecasting Influenza-Like Illness Rate. Master’s Thesis, Tzu Chi University, Hualien, Taiwan, 2020. [Google Scholar]
- Ginsberg, J.; Mohebbi, M.H.; Patel, R.S.; Brammer, L.; Smolinski, M.S.; Brilliant, L. Detecting influenza epidemics using search engine query data. Nature 2009, 457, 1012–1014. [Google Scholar] [CrossRef]
- Kang, M.; Zhong, H.; He, J.; Rutherford, S.; Yang, F. Using google trends for influenza surveillance in South China. PLoS ONE 2013, 8, e55205. [Google Scholar] [CrossRef] [Green Version]
- Zeroual, A.; Harrou, F.; Dairi, A.; Sun, Y. Deep learning methods for forecasting COVID-19 time-Series data: A Comparative study. Chaos Solitons Fractals 2020, 140, 110121. [Google Scholar] [CrossRef] [PubMed]
- Paules, C.I.; Subbarao, K. Influenza vaccination and prevention of cardiovascular disease mortality–Authors’ reply. Lancet 2018, 391, 427–428. [Google Scholar] [CrossRef] [Green Version]
- Masters, B.R. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; Elsevier Saunders: Philadelphia, PA, USA, 2016; ISBN 13-978-1-4557-4801-3. [Google Scholar]
- Centers for Disease Control and Prevention. Epidemiology and Prevention of Vaccine-Preventable Diseases; Department of Health & Human Services, Public Health Service, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2005. [Google Scholar]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Sheng, Z.; Huang, C.; Wang, D.; Li, F. Influenza D virus. Curr. Opin. Virol. 2020, 44, 154–161. [Google Scholar] [CrossRef]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of influenza D virus in cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [Green Version]
- Shaman, J.; Kohn, M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl. Acad. Sci. USA 2009, 106, 3243–3248. [Google Scholar] [CrossRef] [Green Version]
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007, 3, e151. [Google Scholar] [CrossRef]
- Cox, N.J.; Subbarao, K. Global epidemiology of influenza: Past and present. Annu. Rev. Med. 2000, 51, 407–421. [Google Scholar] [CrossRef]
- Yap, F.H.; Ho, P.; Lam, K.; Chan, P.K.; Cheng, Y.; Peiris, J.S. Excess hospital admissions for pneumonia, chronic obstructive pulmonary disease, and heart failure during influenza seasons in Hong Kong. J. Med Virol. 2004, 73, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Tian, H.; Lin, X.; Gao, L.; Dai, X.; Zhang, X.; Chen, B.; Zhao, J.; Xu, J. Influence of extreme weather and meteorological anomalies on outbreaks of influenza A (H1N1). Chin. Sci. Bull. 2013, 58, 741–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundell, N.; Andersson, L.M.; Brittain-Long, R.; Lindh, M.; Westin, J. A four year seasonal survey of the relationship between outdoor climate and epidemiology of viral respiratory tract infections in a temperate climate. J. Clin. Virol. 2016, 84, 59–63. [Google Scholar] [CrossRef]
- Peci, A.; Winter, A.L.; Li, Y.; Gnaneshan, S.; Liu, J.; Mubareka, S.; Gubbay, J.B. Effects of absolute humidity, relative humidity, temperature, and wind speed on influenza activity in Toronto, Ontario, Canada. Appl. Environ. Microbiol. 2019, 85, e02426-18. [Google Scholar] [CrossRef] [Green Version]
- Brunekreef, B.; Holgate, S.T. Air pollution and health. Lancet 2002, 360, 1233–1242. [Google Scholar] [CrossRef]
- Lelieveld, J.; Klingmüller, K.; Pozzer, A.; Pöschl, U.; Fnais, M.; Daiber, A.; Münzel, T. Cardiovascular disease burden from ambient air pollution in Europe reassessed using novel hazard ratio functions. Eur. Heart J. 2019, 40, 1590–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannucci, P.M.; Franchini, M. Health effects of ambient air pollution in developing countries. Int. J. Environ. Res. Public Health 2017, 14, 1048. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, L.; Chen, J.; Chen, K.; Liu, Y.; Chen, X.; Tang, F. Acute effects of air pollution on influenza-like illness in Nanjing, China: A population-based study. Chemosphere 2016, 147, 180–187. [Google Scholar] [CrossRef]
- Feng, C.; Li, J.; Sun, W.; Zhang, Y.; Wang, Q. Impact of ambient fine particulate matter (PM 2.5) exposure on the risk of influenza-like-illness: A time-series analysis in Beijing, China. Environ. Health 2016, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Wu, X.; Geng, X.; Zhao, X.; Liu, Q.; Liu, T. The short-term effects of air pollutants on influenza-like illness in Jinan, China. BMC Public Health 2019, 19, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Hu, W.; Williams, G.; Clements, A.C.; Kan, H.; Tong, S. Air pollution, temperature and pediatric influenza in Brisbane, Australia. Environ. Int. 2013, 59, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.Y.; Wu, Y.C.; Lin, M.H.; Liu, Y.L.; Tsai, Y.Y.; Wu, J.H.; Pan, K.H.; Ke, C.J.; Chen, C.M.; Liu, D.P.J. Applying machine learning models with an ensemble approach for accurate real-time influenza forecasting in Taiwan: Development and validation study. J. Med Internet Res. 2020, 22, e15394. [Google Scholar] [CrossRef] [PubMed]
- Darwish, A.; Rahhal, Y.; Jafar, A. A comparative study on predicting influenza outbreaks using different feature spaces: Application of influenza-like illness data from Early Warning Alert and Response System in Syria. BMC Res. Notes 2020, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Leng, K.; Lu, Y.; Wen, L.; Qi, Y.; Gao, W.; Chen, H.; Bai, L.; An, X.; Sun, B.J.E.; et al. Epidemiological features and time-series analysis of influenza incidence in urban and rural areas of Shenyang, China, 2010–2018. Epidemiol. Infect. 2020, 148, e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Wang, H.; Wang, F.; Langley, D.; Avram, A.; Liu, M. Prediction of influenza-like illness based on the improved artificial tree algorithm and artificial neural network. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tapak, L.; Hamidi, O.; Fathian, M.; Karami, M. Comparative evaluation of time series models for predicting influenza outbreaks: Application of influenza-like illness data from sentinel sites of healthcare centers in Iran. BMC Res. Notes 2019, 12, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Central Weather Bureau. Central Meteorological Administration Station Data Description; Report; Central Weather Bureau: Taipei, Taiwan, 2018. [Google Scholar]
- Google. FAQ about Google Trends Data. Available online: https://support.google.com/trends/answer/4365533?hl=en (accessed on 7 February 2022).
- Choi, H.; Varian, H. Predicting the present with Google Trends. Econ. Rec. 2012, 88, 2–9. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H. Xgboost: Extreme gradient boosting. R Package Version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote. Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support vector regression machines. Adv. Neural Inf. Process. Syst. 1997, 9, 155–161. [Google Scholar]
- Awad, M.; Khanna, R. Support vector regression. In Efficient Learning Machines; Springer: Berkeley, CA, USA, 2015; pp. 67–80. [Google Scholar]
- Chakraborty, P.; Lewis, B.; Eubank, S.; Brownstein, J.S.; Marathe, M.; Ramakrishnan, N. What to know before forecasting the flu. PLoS Comput. Biol. 2018, 14, e1005964. [Google Scholar] [CrossRef] [PubMed]
- Suntronwong, N.; Vichaiwattana, P.; Klinfueng, S.; Korkong, S.; Thongmee, T.; Vongpunsawad, S.; Poovorawan, Y. Climate factors influence seasonal influenza activity in Bangkok, Thailand. PLoS ONE 2020, 15, e0239729. [Google Scholar] [CrossRef] [PubMed]
- Kamigaki, T.; Chaw, L.; Tan, A.G.; Tamaki, R.; Alday, P.P.; Javier, J.B.; Olveda, R.M.; Oshitani, H.; Tallo, V.L. Seasonality of influenza and respiratory syncytial viruses and the effect of climate factors in subtropical–tropical asia using influenza-like illness surveillance data, 2010–2012. PLoS ONE 2016, 11, e0167712. [Google Scholar]
- The NYC Health Department. Is It the FLU OR COVID-19? Report; The NYC Health Department: New York, NY, USA, 2020. [Google Scholar]


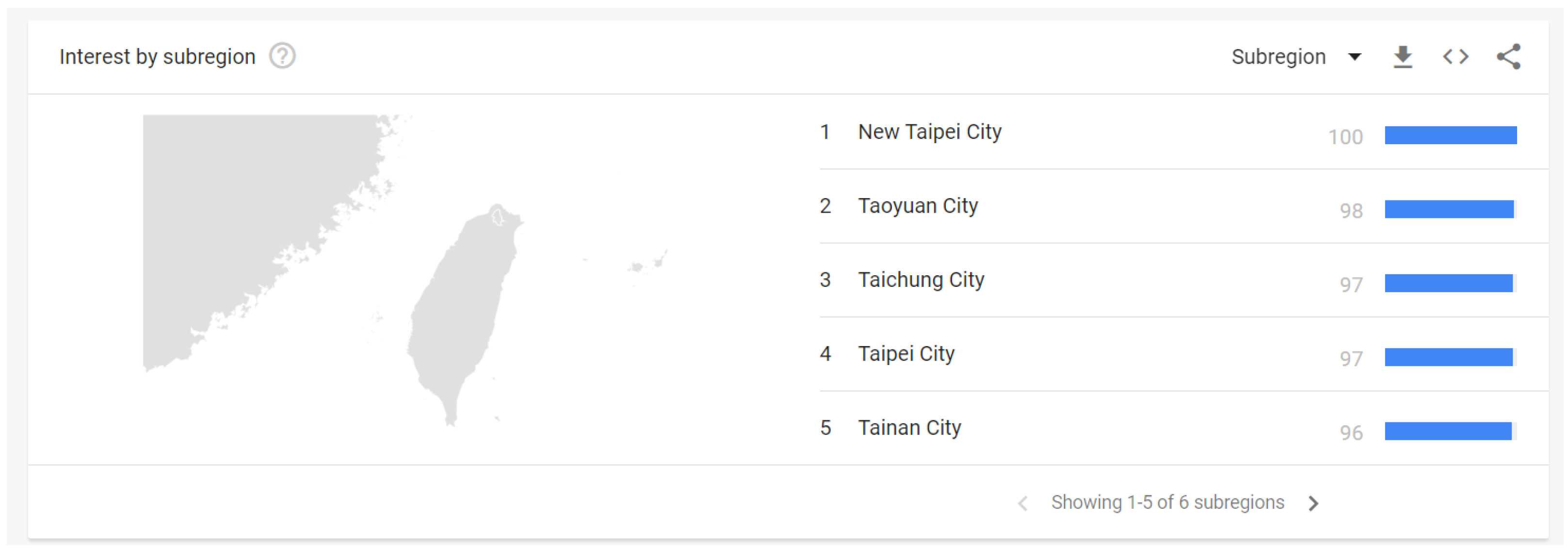
| Influenza | Common Cold | |
|---|---|---|
| Pathogens | Influenza viruses | More than 200 viruses, such as commonly seen respiratory syncytial viruses and adenoviruses, etc. |
| Affected parts of body | Whole body | Respiratory tracts mainly |
| Main clinical symptoms | Fever, cough, muscle aches, fatigue, runny nose, sore throat | Sore throat, sneezing, stuffy nose, runny nose |
| Complications | Pneumonia, myocarditis, encephalitis, neurological symptoms (Reye’s syndrome), and other complications | Less common (otitis media, pneumonia) |
| Modes of transmission | Droplet and contact transmission | Droplet and contact transmission |
| Author | Dataset | Model | Evaluation Metrics |
|---|---|---|---|
| Cheng et al. [39] | ILI dataset, records of patients with severe influenza with complications | ARIMA, Random Forest, SVR, XGBoost | Pearson correlation, MAPE, Hit rate of trend prediction |
| Darwish et al. [40] | EWARS data | GLM, SVR, Gradient boosting, Random Forest, LSTM | MAPE, RMSE |
| Chen et al. [41] | Influenza surveillance data | SARIMA | MAPE, RMSE, R2 (coefficient of determination) |
| Hu et al. [42] | Twitter dataset, ILI dataset | IAT-BPNN | MSE, RMSE, MAPE |
| Tapak et al. [43] | ILI dataset from FluNet | SVM, ANN, random forest | RMSE, MAE, ICC (intra-class correlation coefficient) |
| Item | Specifications and Version Description |
|---|---|
| CPU | Intel i7-8700 3.2 GHz–4.6 GHz |
| GPU | Intel® UHD Graphics 630 |
| RAM | 8 GB DDR4 2400 MHz * 4 |
| SSD | Micron Crucial MX500 500GB SATAIII |
| System OS | Windows Pro 10 1909 |
| Anaconda | 1.9.7 |
| Jupyter Notebook | 6.0.1 |
| Python | 3.7.4 |
| scikit-learn | 0.23.1 |
| XGBoost | 1.1.1 |
| Pandas | 1.1.0 |
| Numpy | 1.19.1 |
| Features | Measurement Units | Descriptions | Notes |
|---|---|---|---|
| PP | Millimeter (mm) | Precipitation | The minimum value of precipitation in this research is 0. All negative values that may be caused by instrumental and human factors are replaced by 0. After excluding outliers of a distance greater than three standard deviations, the average value for a week is calculated (using one week as a unit), and a log value is taken. |
| RH | Percentage (%) | Average relative humidity | After excluding outliers of a distance greater than three standard deviations, the average value for a week is calculated (using one week as a unit), and a log value is taken. |
| TX | Celsius(C) | Average temperature | After excluding any average temperature of a negative value and obvious outliers of a distance greater than three standard deviations, the average value for a week is calculated (using one week as a unit), and a log value is taken. |
| TD | Celsius(C) | Daily temperature differences | A week is taken as a unit. After the data for a week are tallied up, a log value is taken. |
| WD | Meter per second (m/s) | Average wind speeds | After excluding outliers of a distance greater than three standard deviations, the average value for a week is calculated (using one week as a unit), and a log value is taken. |
| ILI | Number of people | The number of influenza-like cases | The number of emergency visits by patients of influenza-like illnesses is obtained and tallied up for all age groups in each county/city. |
| ILI_D | Number of people | Differences in the numbers of influenza-like cases | Weekly changes in the number of emergency visits by patients of influenza-like illnesses are obtained by deducting the number of emergency visits of this week with the number of visits of the week before. |
| GT_I | [0, 100] | Keyword search volumes on Google Trends—influenza (flu) | Values of search volumes of a keyword, “influenza (flu),” on Google Trends by people in Taiwan on a weekly basis within five years. |
| HoC | Number of days | The number of public holidays in Taiwan per week | The weekly number of public holidays in Taiwan in five years |
| Features | Measurement Units | Descriptions | Notes |
|---|---|---|---|
| ILI_LW | Number of people | The number of influenza-like cases from the week before | The number of emergency visits by patients with influenza-like illnesses from the week before is obtained and tallied up in accordance with all age groups in each county/city. |
| GT_IS | [0, 100] | Keyword search volumes on Google Trends—influenza (flu) symptoms | Values of search volumes of keywords, influenza (flu) symptoms, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_C | [0, 100] | Keyword search volumes on Google Trends—common cold | Values of search volumes of keywords, common cold, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_CS | [0, 100] | Keyword search volumes on Google Trends—common cold symptoms | Values of search volumes of keywords, common cold symptoms, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_D | [0, 100] | Keyword search volumes on Google Trends—diarrhea | Values of search volumes of keyword, diarrhea, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_N | [0, 100] | Keyword search volumes on Google Trends—nausea | Values of search volumes of a keyword, nausea, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_FR | [0, 100]] | Keyword search volumes on Google Trends—fever | Values of search volumes of a keyword, fever, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_T | [0, 100] | Keyword search volumes on Google Trends—tiredness | Values of search volumes of a keyword, tiredness, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_FE | A value range of [0, 100] | Keyword search volumes on Google Trends—fatigue | Values of search volumes of a keyword, fatigue, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_SM | A value range of [0, 100] | Keyword search volumes on Google Trends—muscle pain | Values of search volumes of a keywords, muscle pain, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_H | A value range of [0, 100] | Keyword search volumes on Google Trends—headaches | Values of search volumes of a keyword, headaches, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_ST | A value range of [0, 100] | Keyword search volumes on Google Trends—sore throat | Values of search volumes of keywords, sore throat, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_SN | A value range of [0, 100] | Keyword search volumes on Google Trends—stuffy nose | Values of search volumes of keywords, stuffy nose, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_RN | A value range of [0, 100] | Keyword search volumes on Google Trends—runny nose | Values of search volumes of keywords, runny nose, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_CH | A value range of [0, 100] | Keyword search volumes on Google Trends—coughing | Values of search volumes of a keyword, coughing, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_S | A value range of [0, 100] | Keyword search volumes on Google Trends—sneezing | Values of search volumes of a keyword, sneezing, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| GT_DA | A value range of [0, 100] | Keyword search volumes on Google Trends—breathing difficulty | Values of search volumes of keywords, breathing difficulty, on Google Trends by people in Taiwan on a weekly basis within five years are obtained. |
| PIR | Number of people | Total population in each county/city/town/village | The statistics on total population in each county/city/town/village for every four quarters are obtained and converted into the weekly population data. |
| PD | Population density | Population Density | The population indicators in each county/city/town/village for every four quarters are obtained and converted into the weekly population density data. |
| PDoI | Population density | Population density of influenza | The number of patients with influenza this week (ILI) is divided by the total population of a county/city (PIR) and then multiplied by the population density of the county/city (PD) to obtain the weekly population density of influenza cases. |
| PM10 | g/m3 | Air quality index —PM10 | The PM10 data are obtained from the air quality index and computed to obtain weekly averages. |
| PM2.5 | g/m3 | Air quality index —PM2.5 | The PM2.5 data are obtained from the air quality index and computed to obtain weekly averages. |
| SO2 | ppb | Air quality index —SO2 | The PMSO2 data are obtained from the air quality index and computed to obtain weekly averages. |
| O3 | ppb | Air quality index —O3 | The O3 data are obtained from the air quality index and computed to obtain weekly averages. |
| NO2 | ppb | Air quality index —NO2 | The NO2 data are obtained from the air quality index and computed to obtain weekly averages. |
| Cov19 | Number of people | The number of confirmed cases of COVID-19 | The number of confirmed cases of COVID-19 is obtained and tallied up for all age groups in each county/city. |
| Negative Values | Descriptions |
|---|---|
| −9991 | Instrument failures, to be repaired |
| −9996 | Data accumulated later |
| −9997 | No information available due to unknown reasons or malfunctions |
| −9998 | Traces of rain |
| −9999 | No data due to no observation |
| Anomalies | Descriptions |
|---|---|
| # | Indicates an invalid value after instrument checks |
| * | Indicates an invalid value after program checks |
| x | Indicates an invalid value after manual checks |
| NR | Indicates no rain fall |
| blank | Indicates no value |
| 888 | Indicates no wind |
| 999 | Indicates instrument failures |
| Week | City | Flue_Amt | Lag | New | Lag | New | Lag | New | Lag |
|---|---|---|---|---|---|---|---|---|---|
| Flue_Amt | PP | PP | GT_I | GT_I | PM10 | PM10 | |||
| 2021-11 | Nantou County | 158 | 148 | 0 | 0 | 5 | 3 | 50.302 | 54.827 |
| 2021-11 | Taoyuan City | 468 | 459 | 0 | 0.068 | 5 | 3 | 47.811 | 36.383 |
| 2021-12 | Nantou County | 133 | 158 | 1.292 | 0 | 3 | 5 | 45.599 | 50.302 |
| 2021-12 | Taoyuan City | 570 | 468 | 2.047 | 0 | 3 | 5 | 40.372 | 47.811 |
| 2021-13 | Nantou County | Predict | 133 | 0.080 | 1.292 | 3 | 3 | 42.813 | 45.599 |
| 2021-13 | Taoyuan City | Predict | 570 | 0 | 2.047 | 3 | 3 | 43.098 | 40.372 |
| Year | Week | Months | Year | Week | Week | Week | Week | Week | Week | Week |
|---|---|---|---|---|---|---|---|---|---|---|
| 2021 | 01 | 02 | 03 | 04 | 05 | 06 | 07 | |||
| 2021 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2021 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 2021 | 3 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 2021 | 4 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 2021 | 5 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 2021 | 6 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 2021 | 7 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Combinations | Features Used |
|---|---|
| Org_df | Atmospheric hydrological data, the number of influenzalike cases, Google Trends search volumes: influenza, public holidays, as listed in Table 4 |
| GT_df | Similar to Org_df, but adding the features of 17 keywords relating to influenza-like symptoms on Google Trends, such as runny nose, common cold, sore throat, etc., also adding the population data |
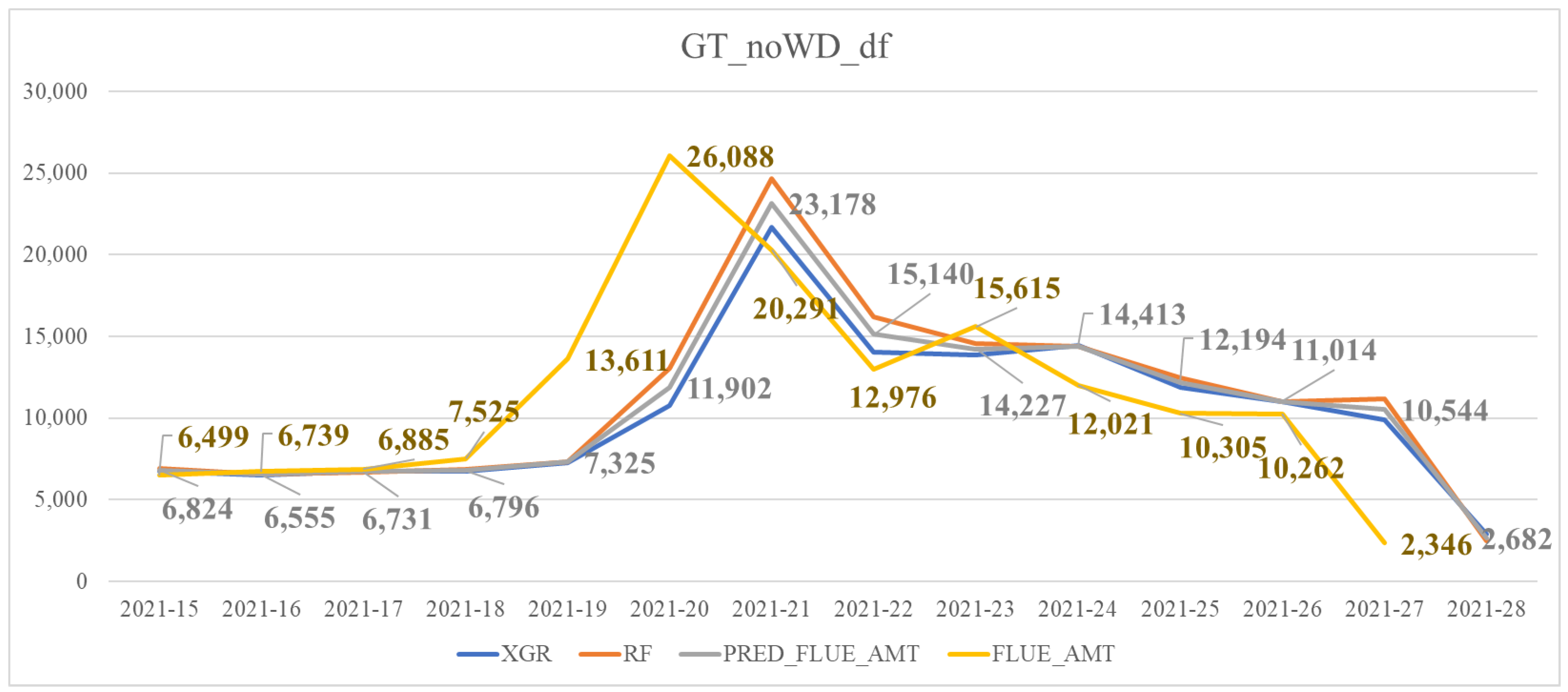
| GT_noWD_df | Similar to GT_df but excluding the feature of wind speed differences (WD) |
| AQI_df | Similar to GT_df but adding air quality indices, e.g., PM10, PM2.5, NO2, SO2 and O3 |
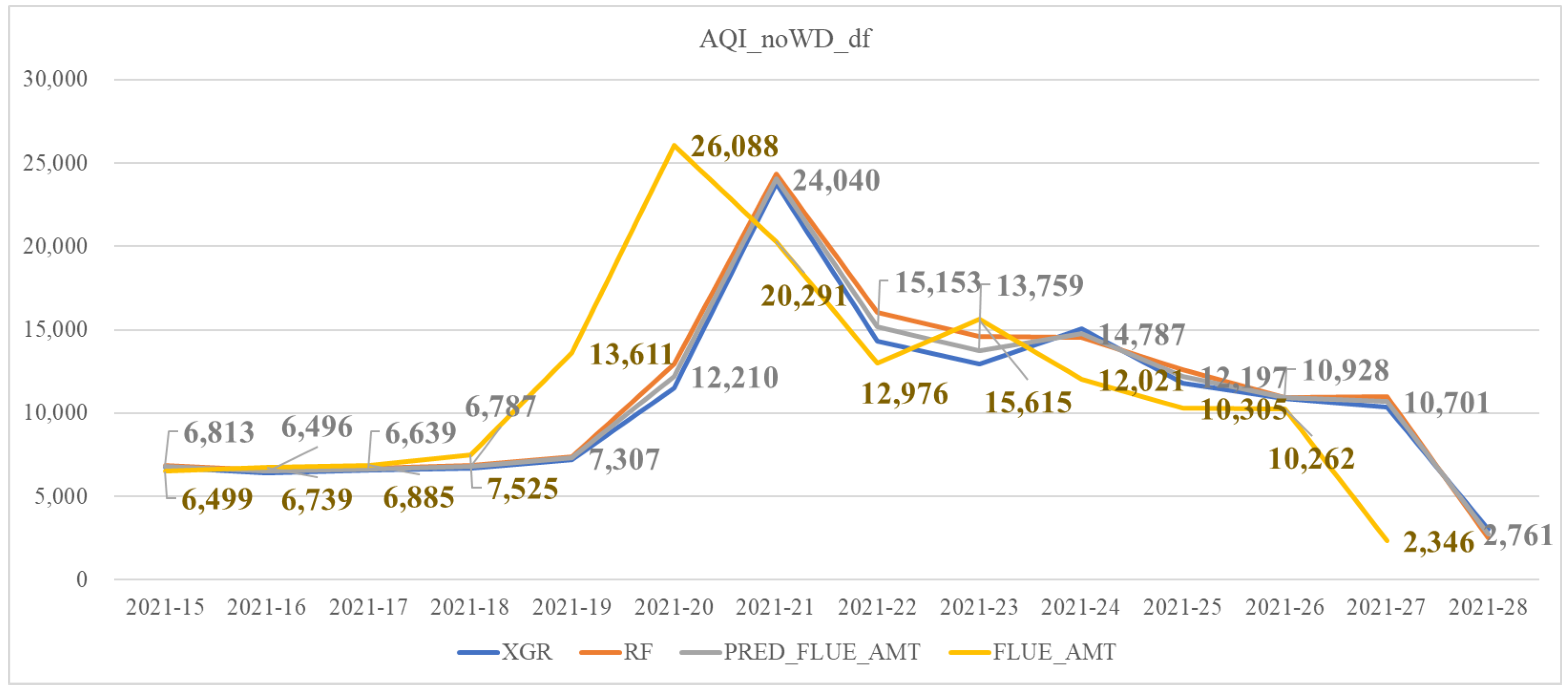
| AQI_noWD_df | Similar to AQI_df but excluding the feature of wind speed differences (WD) |
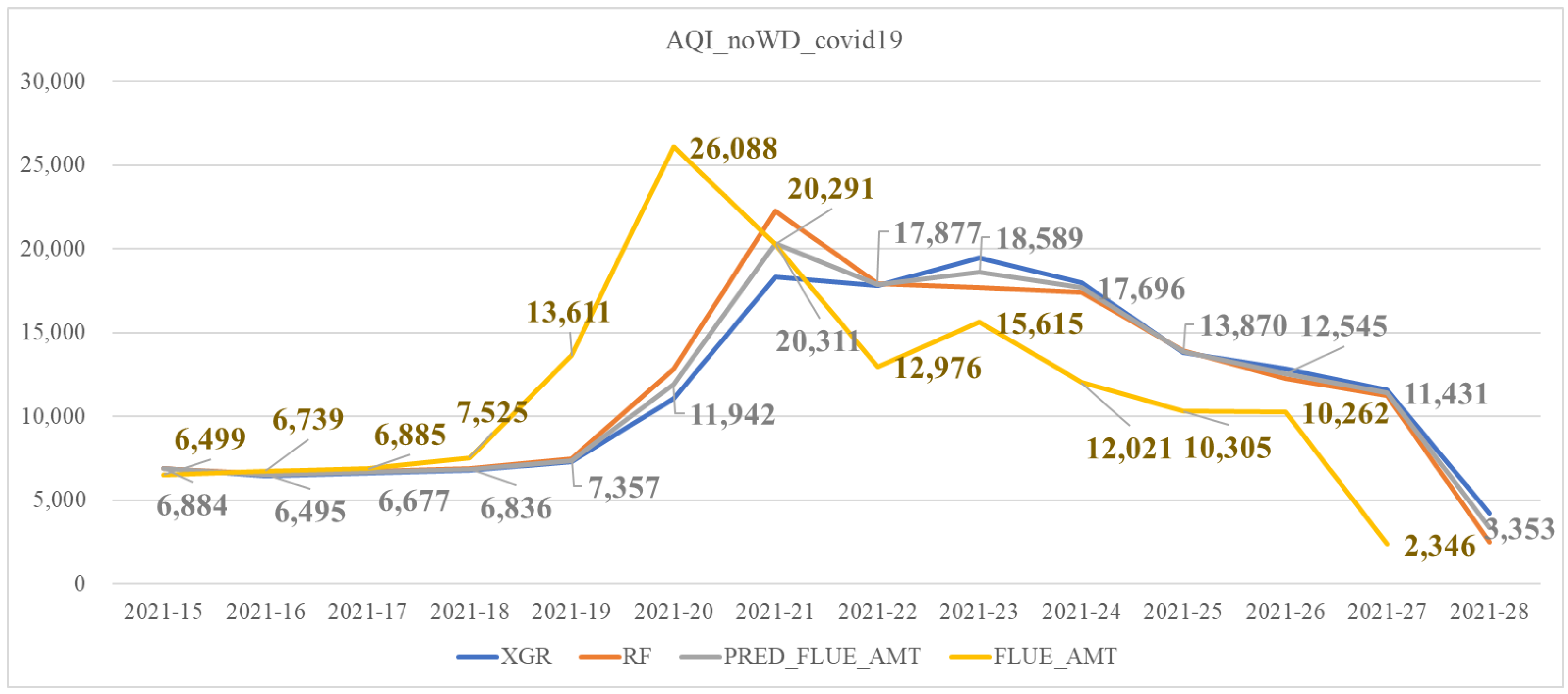
| Covid_noWD_df | Similar to AQI_noWD_df but adding the feature of the number of COVID-19 confirmed cases |
| Week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.234 | 0.226 | 0.247 |
| 2021-16 | 0.244 | 0.200 | 0.232 |
| 2021-17 | 0.164 | 0.182 | 0.198 |
| 2021-18 | 0.215 | 0.194 | 0.220 |
| 2021-19 | 0.222 | 0.198 | 0.235 |
| 2021-20 | 0.203 | 0.187 | 0.205 |
| 2021-21 | 0.247 | 0.223 | 0.230 |
| 2021-22 | 0.187 | 0.198 | 0.224 |
| 2021-23 | 0.192 | 0.212 | 0.229 |
| 2021-24 | 0.159 | 0.193 | 0.218 |
| 2021-25 | 0.198 | 0.208 | 0.237 |
| 2021-26 | 0.203 | 0.187 | 0.218 |
| 2021-27 | 0.248 | 0.224 | 0.242 |
| 2021-28 | 0.259 | 0.234 | 0.244 |
| week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.220 | 0.222 | 0.250 |
| 2021-16 | 0.223 | 0.196 | 0.226 |
| 2021-17 | 0.154 | 0.176 | 0.190 |
| 2021-18 | 0.166 | 0.193 | 0.219 |
| 2021-19 | 0.204 | 0.204 | 0.246 |
| 2021-20 | 0.161 | 0.178 | 0.198 |
| 2021-21 | 0.226 | 0.224 | 0.226 |
| 2021-22 | 0.162 | 0.190 | 0.220 |
| 2021-23 | 0.187 | 0.205 | 0.223 |
| 2021-24 | 0.159 | 0.186 | 0.219 |
| 2021-25 | 0.186 | 0.211 | 0.235 |
| 2021-26 | 0.192 | 0.186 | 0.229 |
| 2021-27 | 0.230 | 0.217 | 0.226 |
| 2021-28 | 0.233 | 0.227 | 0.232 |
| Week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.214 | 0.223 | 0.251 |
| 2021-16 | 0.203 | 0.197 | 0.226 |
| 2021-17 | 0.155 | 0.178 | 0.190 |
| 2021-18 | 0.162 | 0.194 | 0.218 |
| 2021-19 | 0.207 | 0.202 | 0.246 |
| 2021-20 | 0.166 | 0.177 | 0.198 |
| 2021-21 | 0.203 | 0.227 | 0.226 |
| 2021-22 | 0.168 | 0.191 | 0.220 |
| 2021-23 | 0.169 | 0.203 | 0.223 |
| 2021-24 | 0.160 | 0.186 | 0.220 |
| 2021-25 | 0.192 | 0.212 | 0.235 |
| 2021-26 | 0.194 | 0.188 | 0.229 |
| 2021-27 | 0.230 | 0.219 | 0.226 |
| 2021-28 | 0.224 | 0.226 | 0.232 |
| Week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.216 | 0.224 | 0.247 |
| 2021-16 | 0.206 | 0.199 | 0.225 |
| 2021-17 | 0.161 | 0.179 | 0.190 |
| 2021-18 | 0.167 | 0.194 | 0.218 |
| 2021-19 | 0.198 | 0.207 | 0.244 |
| 2021-20 | 0.145 | 0.177 | 0.200 |
| 2021-21 | 0.209 | 0.225 | 0.227 |
| 2021-22 | 0.173 | 0.194 | 0.220 |
| 2021-23 | 0.204 | 0.208 | 0.223 |
| 2021-24 | 0.176 | 0.187 | 0.220 |
| 2021-25 | 0.187 | 0.212 | 0.237 |
| 2021-26 | 0.195 | 0.189 | 0.229 |
| 2021-27 | 0.229 | 0.221 | 0.227 |
| 2021-28 | 0.221 | 0.228 | 0.231 |
| Week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.217 | 0.224 | 0.247 |
| 2021-16 | 0.204 | 0.199 | 0.225 |
| 2021-17 | 0.164 | 0.180 | 0.190 |
| 2021-18 | 0.161 | 0.196 | 0.217 |
| 2021-19 | 0.186 | 0.208 | 0.244 |
| 2021-20 | 0.170 | 0.178 | 0.200 |
| 2021-21 | 0.215 | 0.224 | 0.227 |
| 2021-22 | 0.161 | 0.193 | 0.220 |
| 2021-23 | 0.167 | 0.208 | 0.223 |
| 2021-24 | 0.163 | 0.188 | 0.220 |
| 2021-25 | 0.183 | 0.213 | 0.236 |
| 2021-26 | 0.196 | 0.192 | 0.229 |
| 2021-27 | 0.223 | 0.221 | 0.227 |
| 2021-28 | 0.238 | 0.227 | 0.231 |
| Week | XGR | RF | SVR |
|---|---|---|---|
| 2021-15 | 0.204 | 0.188 | 0.224 |
| 2021-16 | 0.152 | 0.171 | 0.223 |
| 2021-17 | 0.172 | 0.170 | 0.200 |
| 2021-18 | 0.206 | 0.194 | 0.237 |
| 2021-19 | 0.178 | 0.175 | 0.232 |
| 2021-20 | 0.176 | 0.168 | 0.204 |
| 2021-21 | 0.162 | 0.177 | 0.199 |
| 2021-22 | 0.233 | 0.167 | 0.213 |
| 2021-23 | 0.270 | 0.169 | 0.212 |
| 2021-24 | 0.235 | 0.178 | 0.212 |
| 2021-25 | 0.208 | 0.206 | 0.238 |
| 2021-26 | 0.249 | 0.211 | 0.245 |
| 2021-27 | 0.219 | 0.191 | 0.226 |
| 2021-28 | 0.140 | 0.168 | 0.219 |
| Combinations | RMSLE |
|---|---|
| Org_df | 0.2085 |
| GT_df | 0.197 |
| GT_noWD_df | 0.1955 |
| AQI_df | 0.1975 |
| AQI_noWD_df | 0.1965 |
| Covid_noWD_df | 0.1905 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.-C.; Ou, Y.-L.; Li, J.; Wu, W.-C. Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion. Sustainability 2022, 14, 2803. https://doi.org/10.3390/su14052803
Wei Y-C, Ou Y-L, Li J, Wu W-C. Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion. Sustainability. 2022; 14(5):2803. https://doi.org/10.3390/su14052803
Chicago/Turabian StyleWei, Yu-Chih, Yan-Ling Ou, Jianqiang Li, and Wei-Chen Wu. 2022. "Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion" Sustainability 14, no. 5: 2803. https://doi.org/10.3390/su14052803
APA StyleWei, Y.-C., Ou, Y.-L., Li, J., & Wu, W.-C. (2022). Forecasting the Potential Number of Influenza-like Illness Cases by Fusing Internet Public Opinion. Sustainability, 14(5), 2803. https://doi.org/10.3390/su14052803







