Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
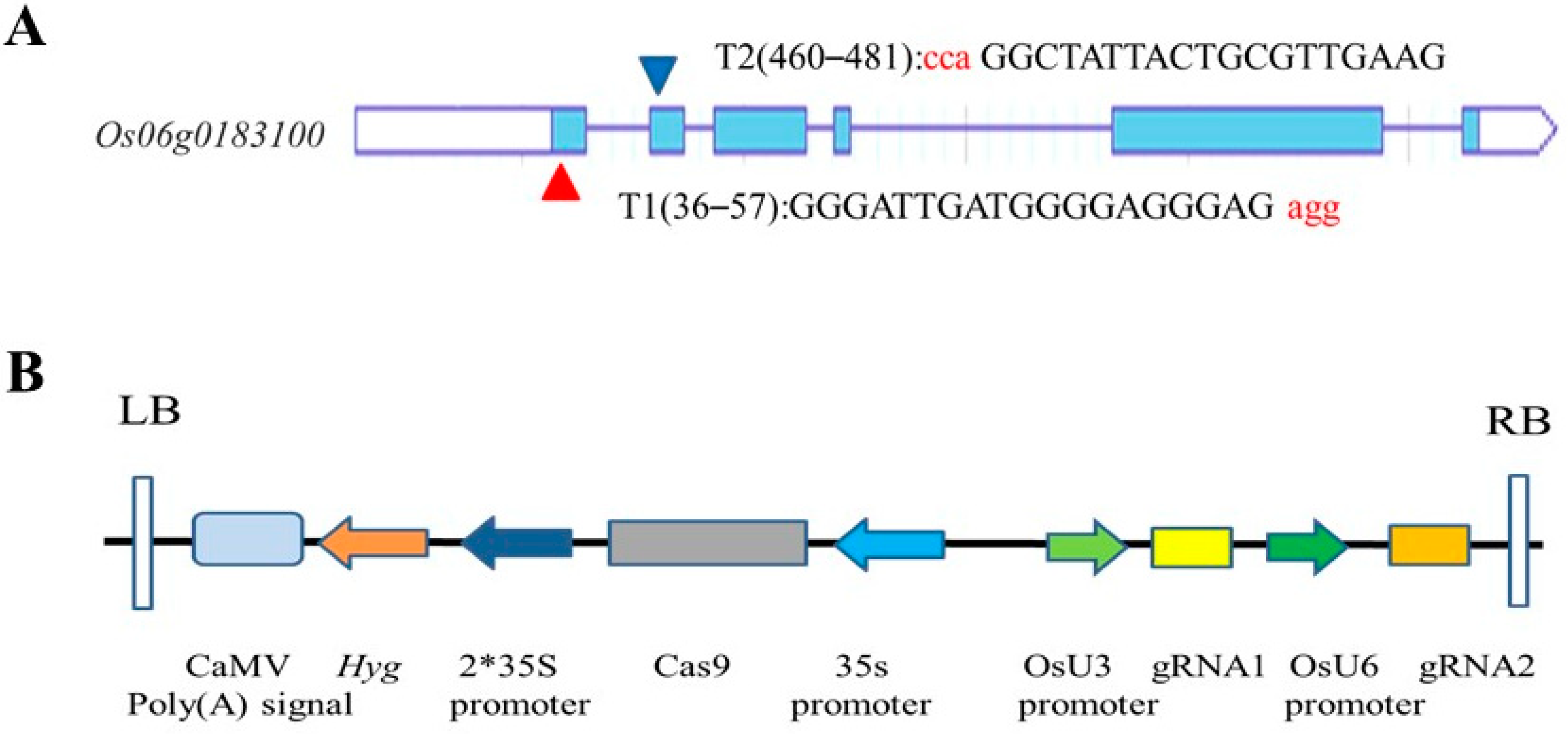
2.2. Vector Construction
2.3. Detection and Identification of Transgenic Plants
2.4. Identification and Evaluation of Salt Tolerance for Mutant Plants
2.5. Data Analysis
3. Results and Analysis
3.1. Construction of the Recombinant Expression Vector of the OsRR22 Gene in Rice
3.2. Acquisition of Transgenic Seedlings and Identification of Homozygotes
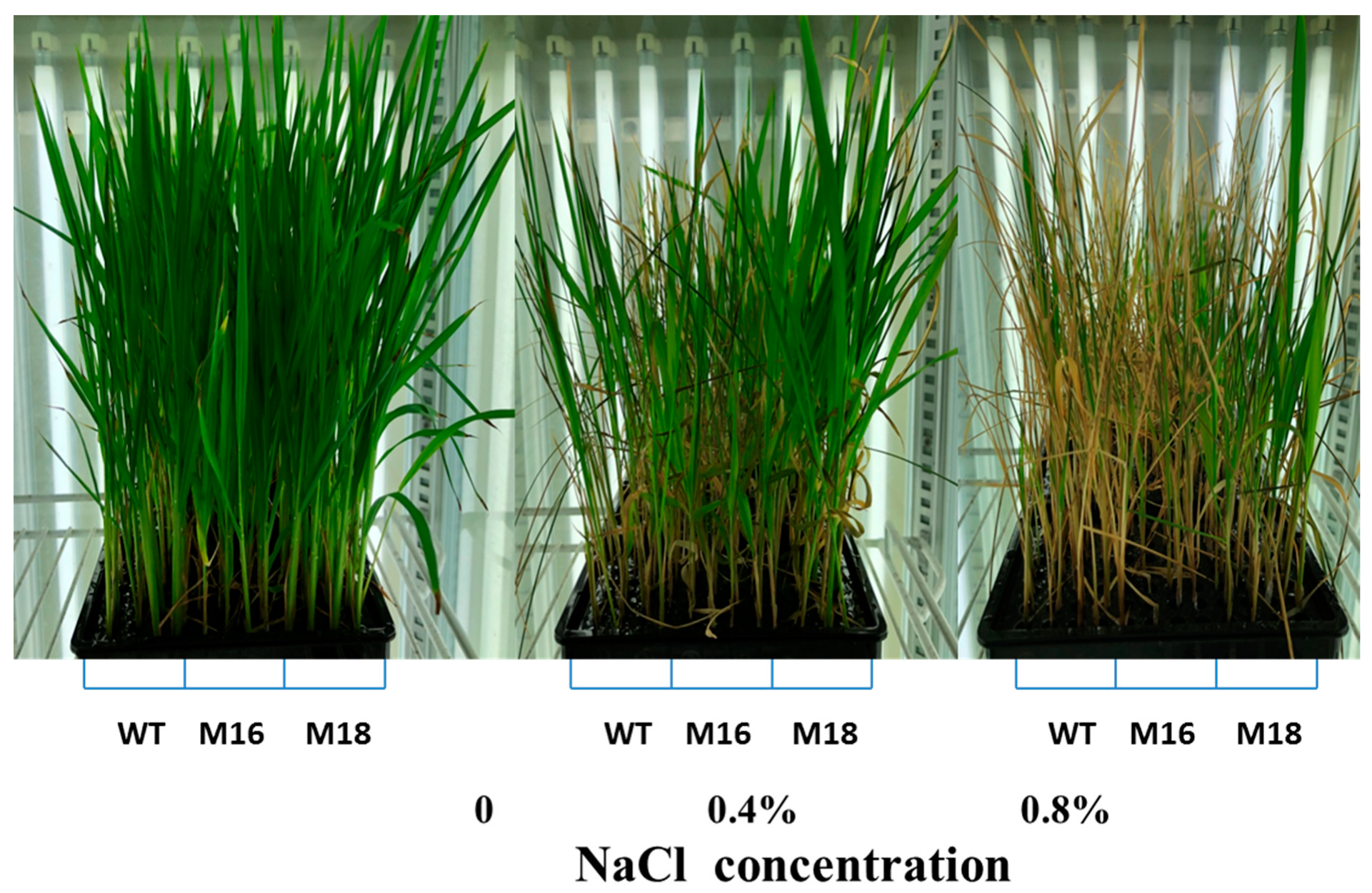
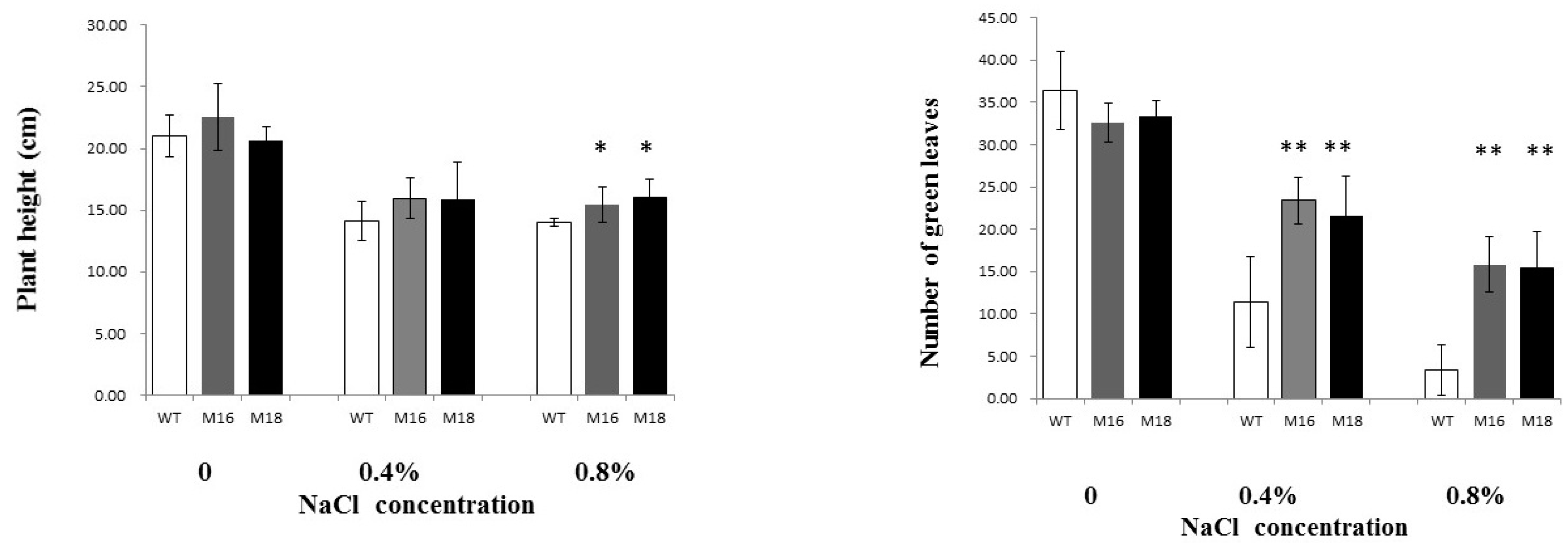
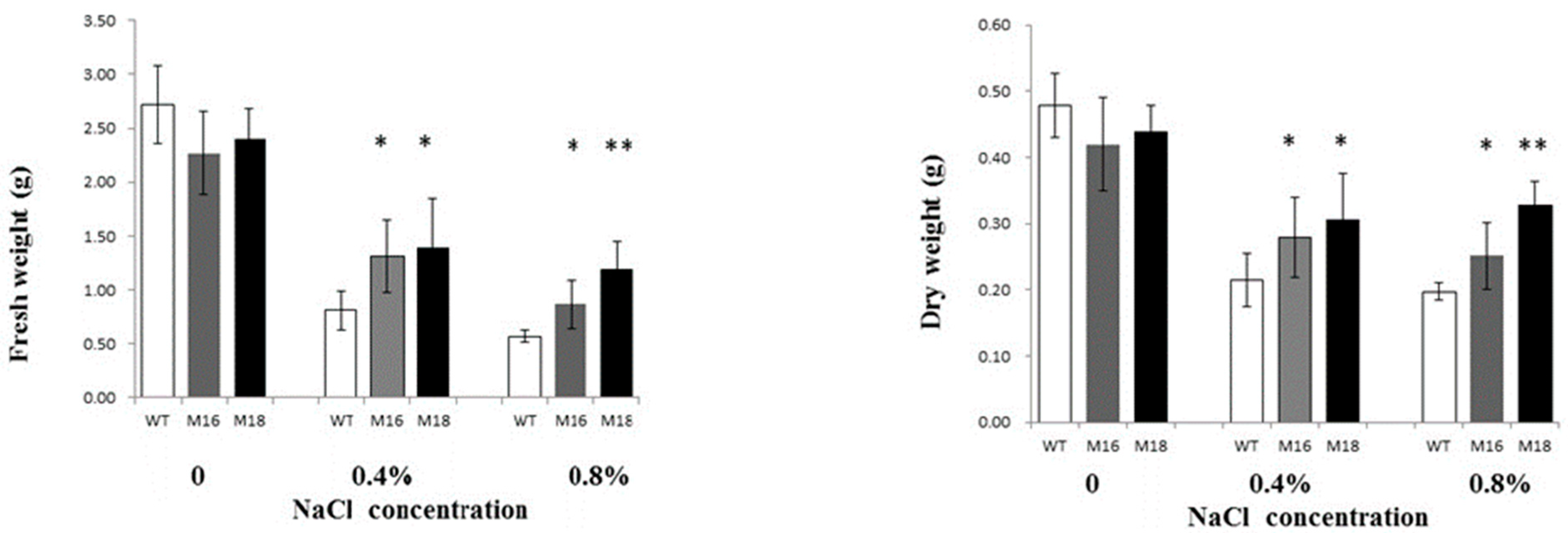
3.3. Salt Tolerance of Rice OsRR22 Gene Mutant Lines
4. Discussion
4.1. Creation of Novel Salt-Tolerant Rice Germplasm with OsRR22 Knockout
4.2. Research on the Physiological Mechanism of Salt Tolerance in Rice
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, B.; Wang, Z.C.; Sun, Z.G.; Chen, Y.; Yang, F. Resources and sustainable resource exploitation of salinized land in China. Agric. Res. Arid. Areas 2005, 23, 154–158, (In Chinese with English abstract). [Google Scholar]
- Mumns, R.; Gilliham, M. Salinity tolerance of crops–what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, A.M.; Heuer, S.; Thomson, M.J.; Wissuwa, M. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol. Biol. 2007, 65, 547–570. [Google Scholar] [CrossRef] [PubMed]
- Grattan, S.R.; Zeng, L.H.; Shannon, M.C.; Roberts, S.R. Rice is more sensitive to salinity than previously thought. Calif. Agric. 2002, 56, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Yeo, A.R.; Flowers, T.J. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Aust. J. Plant Physiol. 1986, 13, 161–173. [Google Scholar] [CrossRef]
- Hu, T.T.; Liu, C.; Wang, J.K.; Ding, C.W.; Guo, R.L.; Wu, Y.L.; Xu, J.A.; Wang, Y.S. Progress of genetic and breeding on salt tolerance in rice. Mol. Plant Breed. 2009, 7, 110–116, (In Chinese with English abstract). [Google Scholar]
- Lang, N.; Buu, B.C.; Ismail, A.M. Enhancing and stabilizing the productivity of salt-affected areas by incorporating genes for tolerance of abiotic stresses in rice. Omonrice 2011, 18, 41–49. [Google Scholar]
- Bimpong, I.K.; Manneh, B.; Sock, M.; Diaw, F.; Amoah, N.K.A.; Ismail, A.M.; Gregorio, G.; Singh, R.K.; Wopereis, M. Improving salt tolerance of lowland rice cultivar ’Rassi’ through marker-aided backcross breeding in West Africa. Plant Sci. 2016, 242, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Jing, W.; Zhang, W. Research progress on gene mapping and cloning for salt tolerance and variety improvement for salt tolerance by molecular marker-assisted selection in rice. Chin. J. Rice Sci. 2017, 31, 111–123, (In Chinese with English abstract). [Google Scholar]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Hu, H.H.; Dai, M.Q.; Yao, J.L.; Xiao, B.Z.; Li, X.H.; Zhang, Q.F.; Xiong, L.Z. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotech. 2015, 33, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Zheng, Y.L.; Su, Z.L.; Yu, S.B.; Song, H.B.; Zheng, X.Y.; Lin, G.G.; Wu, W.R. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 2019, 9, 4107–4114. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.B.; Liu, C.; Tang, D.Y.; Yan, L.; Wang, D.; Yang, Y.Z.; Gui, J.S.; Zhao, X.Y.; Li, L.G.; Tang, X.D.; et al. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CATC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef] [Green Version]
- Mahi, H.E.; Perez-Hormaeche, J.; Luca, A.D.; Villalta, I.; Espartero, J.; Gamez-Arjona, F.; Fernandez, J.L.; Bundo, M.; Mendoza, I.; Mieulet, D.; et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.Z.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Shao, G.N.; Xie, L.H.; Jiao, G.A.; Wei, X.J.; Sheng, Z.H.; Tang, S.Q.; Hu, P.S. CRISPR/CAS9-mediated Editing of the Fragrant Gene Badh2 in Rice. Chin. J. Rice Sci. 2017, 31, 216–222, (In Chinese with English abstract). [Google Scholar]
- Barman, H.N.; Sheng, Z.H.; Fiaz, S.; Zhong, M.; Wu, Y.W.; Cai, Y.C.; Wang, W.; Jiao, G.A.; Tang, S.Q.; Wei, X.J.; et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.H.; Zhu, M.; Shabala, L.; Zhou, M.X.; Shabala, S. K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: A Case study for barley. J. Integr. Plant Biol. 2015, 57, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef]
- Chawla, S.; Jain, S.; Jain, V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). J. Plant Biochem. Biot. 2013, 22, 27–34. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Niwagaba, P. Physiological and Biochemical Mechanisms of Salt Tolerance in Rice Mutants sst and hst1. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2018. [Google Scholar]
- Pan, X.B.; Xie, L.J.; Huang, S.J.; Duan, M.; Chen, J.; Xu, J.L. Salt tolerance and breeding strategy of hybrid rice at different growth stages. Jiangsu Agric. Sci. 2017, 45, 56–60, (In Chinese with English abstract). [Google Scholar]

| Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Purpose |
|---|---|---|---|
| RR22-Y | cagtGGTCTCaggcagggattgatggggagggag | cagtGGTCTCaaaacctccctccccatcaatccc | Vector construct |
| RR22-B | cagtGGTCTCatgtgcttcaacgcagtaatagcc | cagtGGTCTCaaaacggctattactgcgttgaag | Vector construct |
| RR22-1 | TCTTGCTGAGAATTGGCTCGAT | AGTTGAGCATCATGGATAGCTGA | Target site sequencing |
| RR22-2 | TGCTTCTGCTTGCATTGTTCA | TCTGAACTTACTGATGACTGGGAGA | Target site sequencing |
| Cas9 | GAACGGTCGTAAGAGGATGCTG | GGTGATGGACTGGTGGATGAGA | Transgenic analysis |
| HPT | GCTCCATACAAGCCAACCACG | CCTGCCTGAAACCGAACTGC | Transgenic analysis |
| Proportion of Mutant Genotypes | Proportion of Mutation Types | |||
|---|---|---|---|---|
| Bivalent mutation | Heterozygote | Homozygote | Deletion | Insertion |
| 37.5 (3/8) | 50 (4/8) | 12.5 (1/8) | 62.5 (10/16) | 12.5 (2/16) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Chen, Z.; Li, P.; Xu, H.; Liu, K.; Zha, W.; Li, S.; Chen, J.; Yang, G.; Huang, J.; et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability 2022, 14, 2621. https://doi.org/10.3390/su14052621
Han X, Chen Z, Li P, Xu H, Liu K, Zha W, Li S, Chen J, Yang G, Huang J, et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability. 2022; 14(5):2621. https://doi.org/10.3390/su14052621
Chicago/Turabian StyleHan, Xiaoli, Zhijun Chen, Peide Li, Huashan Xu, Kai Liu, Wenjun Zha, Sanhe Li, Junxiao Chen, Guocai Yang, Jianliang Huang, and et al. 2022. "Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9" Sustainability 14, no. 5: 2621. https://doi.org/10.3390/su14052621
APA StyleHan, X., Chen, Z., Li, P., Xu, H., Liu, K., Zha, W., Li, S., Chen, J., Yang, G., Huang, J., You, A., & Zhou, L. (2022). Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability, 14(5), 2621. https://doi.org/10.3390/su14052621








