A Review of Cross-Disciplinary Approaches for the Identification of Novel Industrially Relevant Plastic-Degrading Enzymes
Abstract
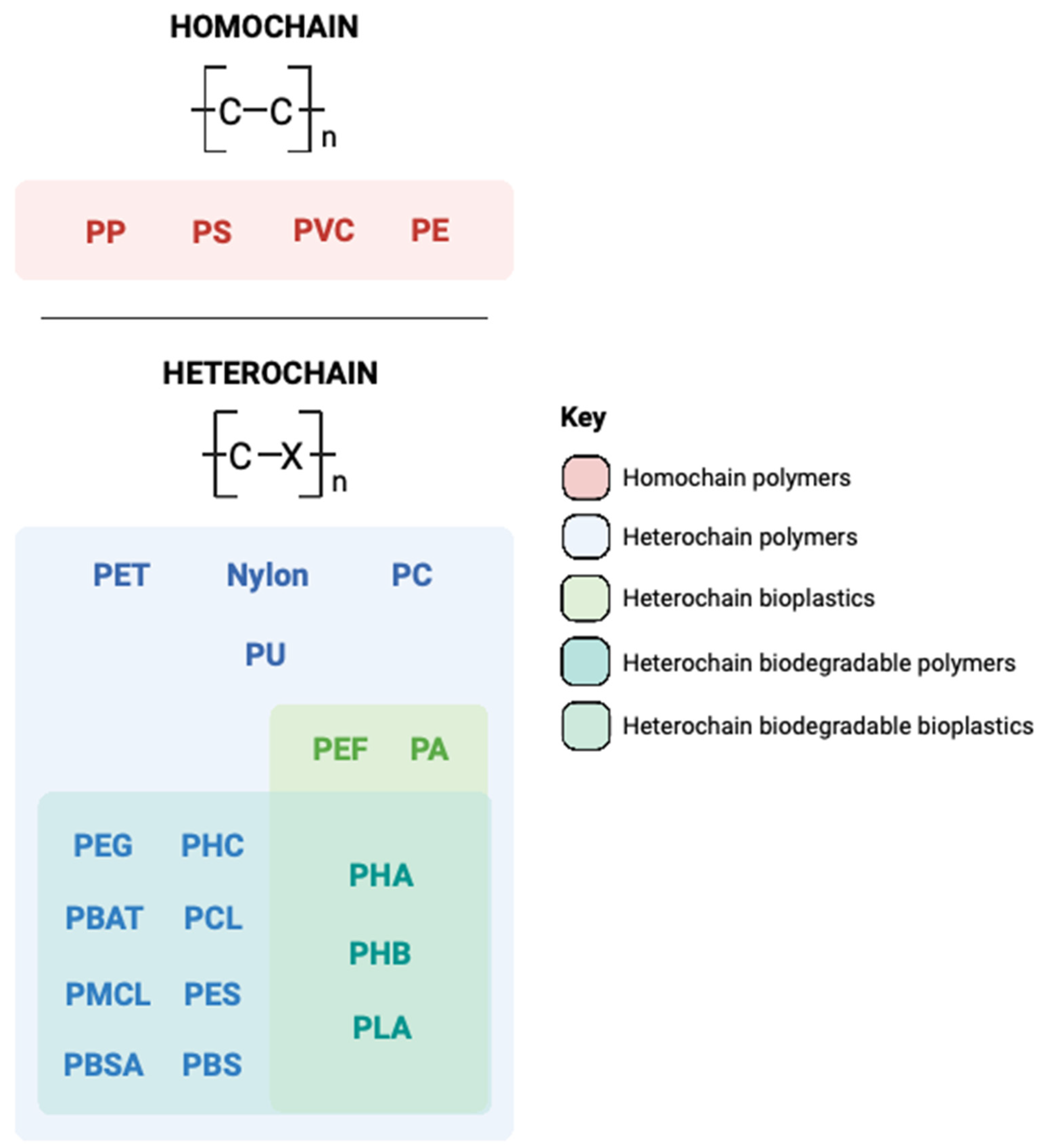
1. Introduction to the Plastic Problem
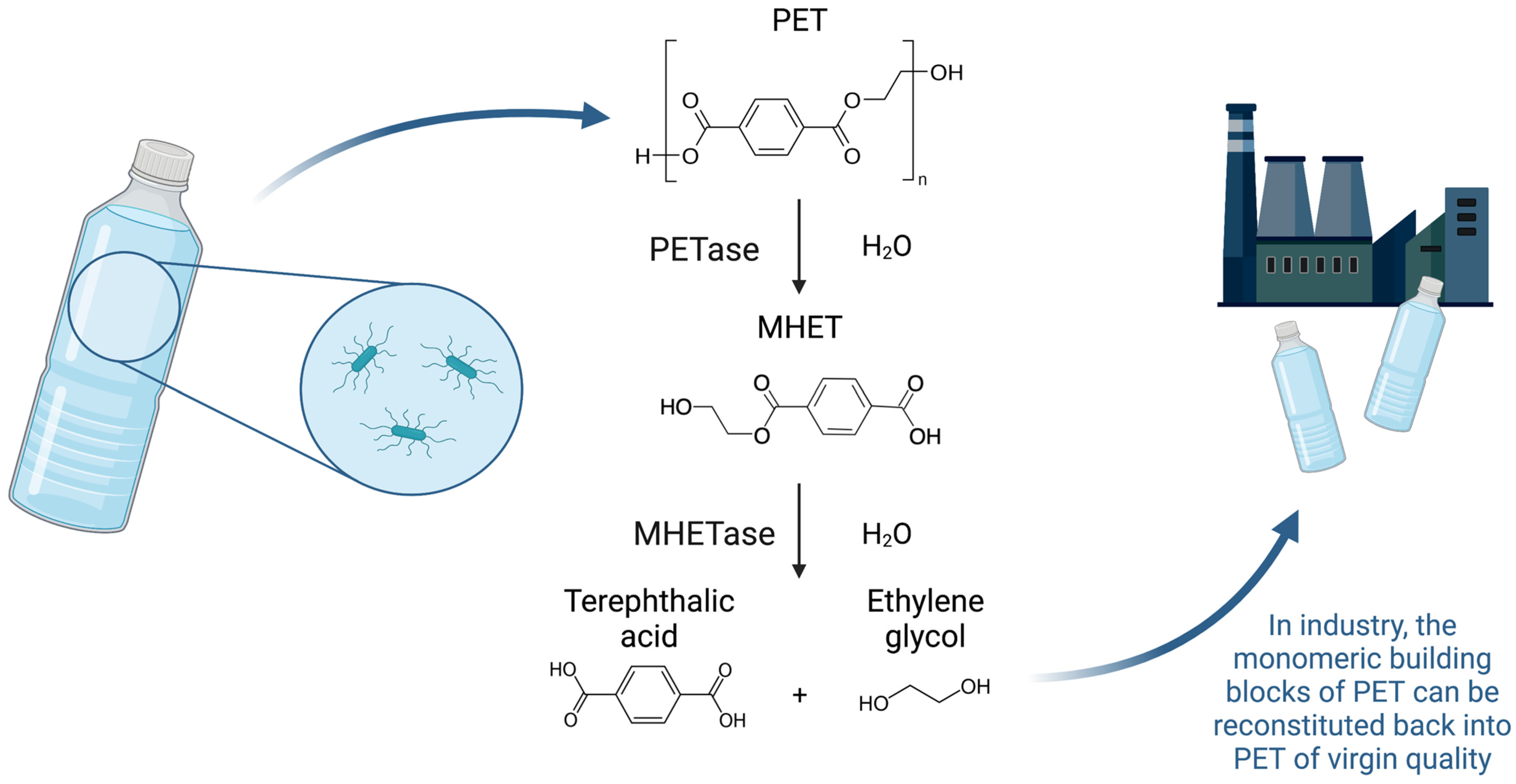
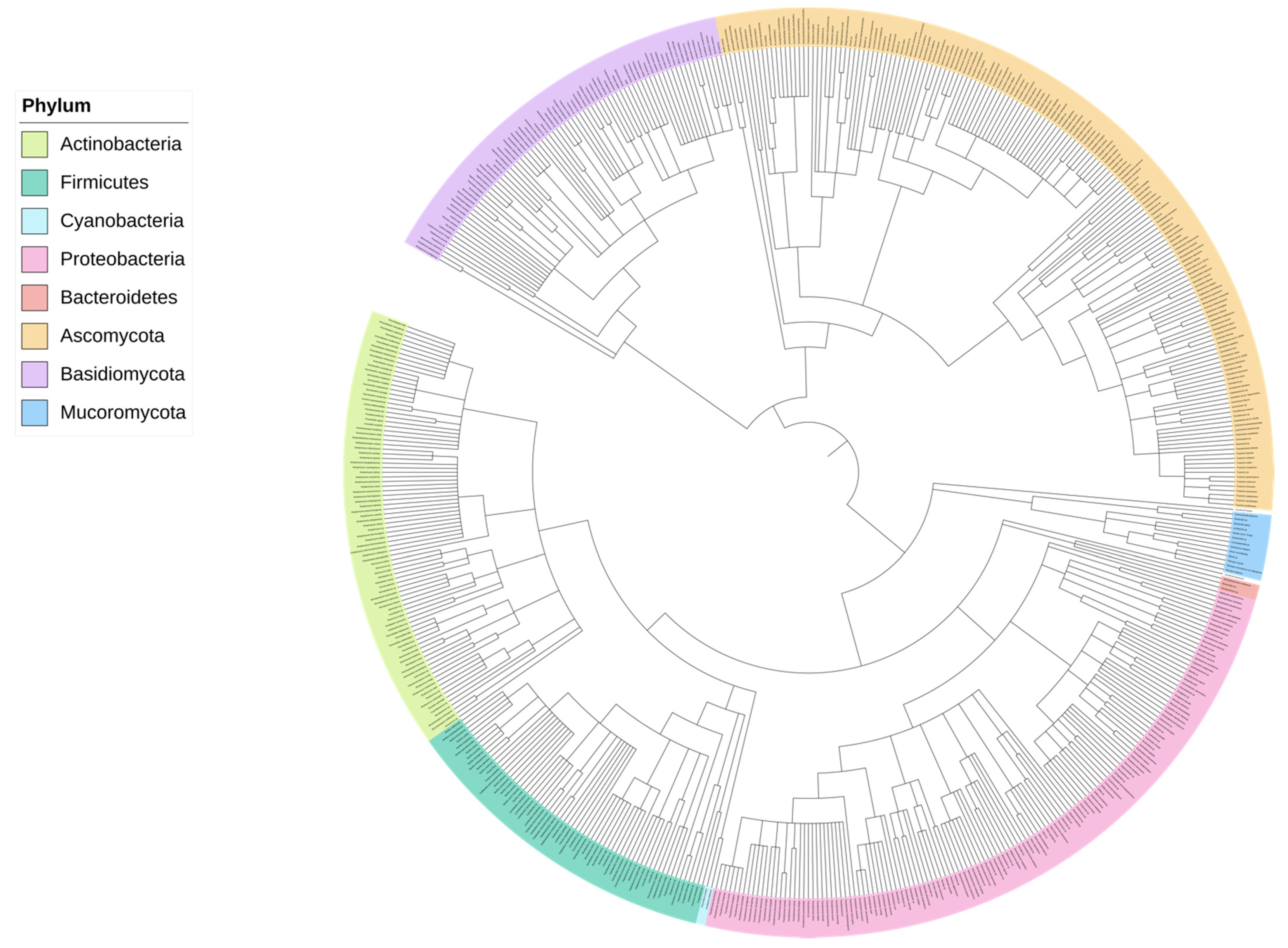
2. A Microbial Solution
3. The Role of Genomics
4. Whole-Genome Sequencing
5. Metagenomics
6. Databases
7. Machine Learning
8. Transcriptomics
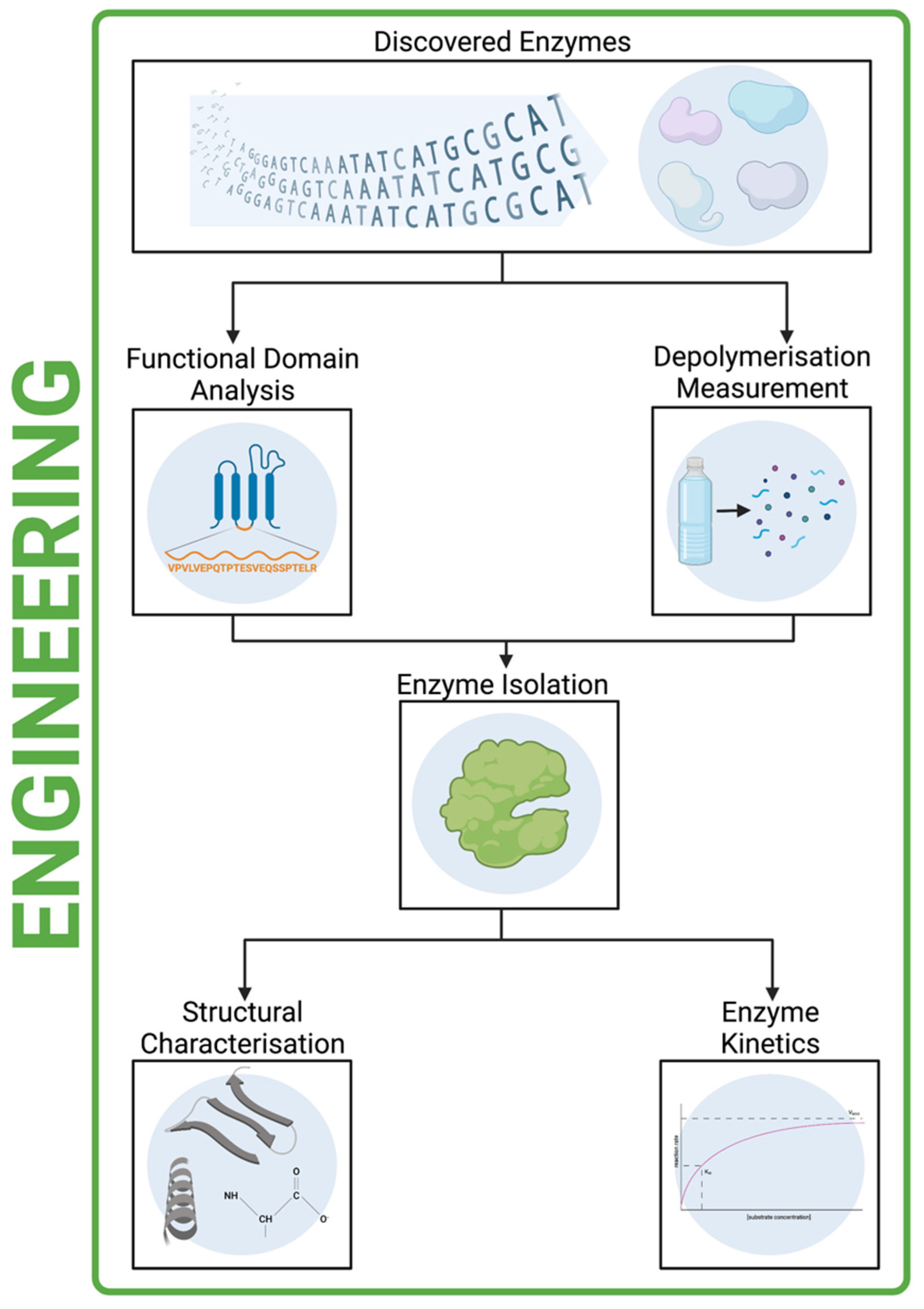
9. Processing of Putative Enzymes
10. Protein Functional Domain Analysis
11. Proving Depolymerisation
12. Enzyme Isolation
13. Structural Characterisation and Enzyme Kinetics
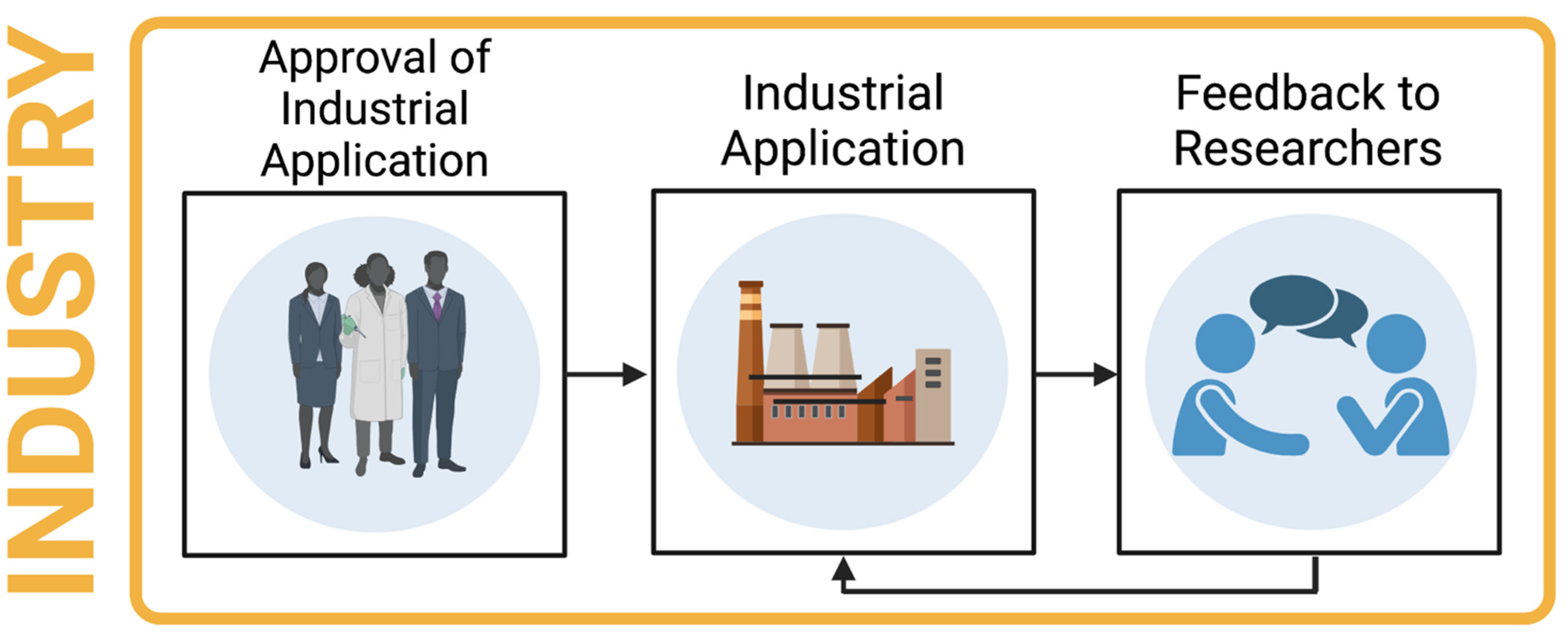
14. Industrial Relevance
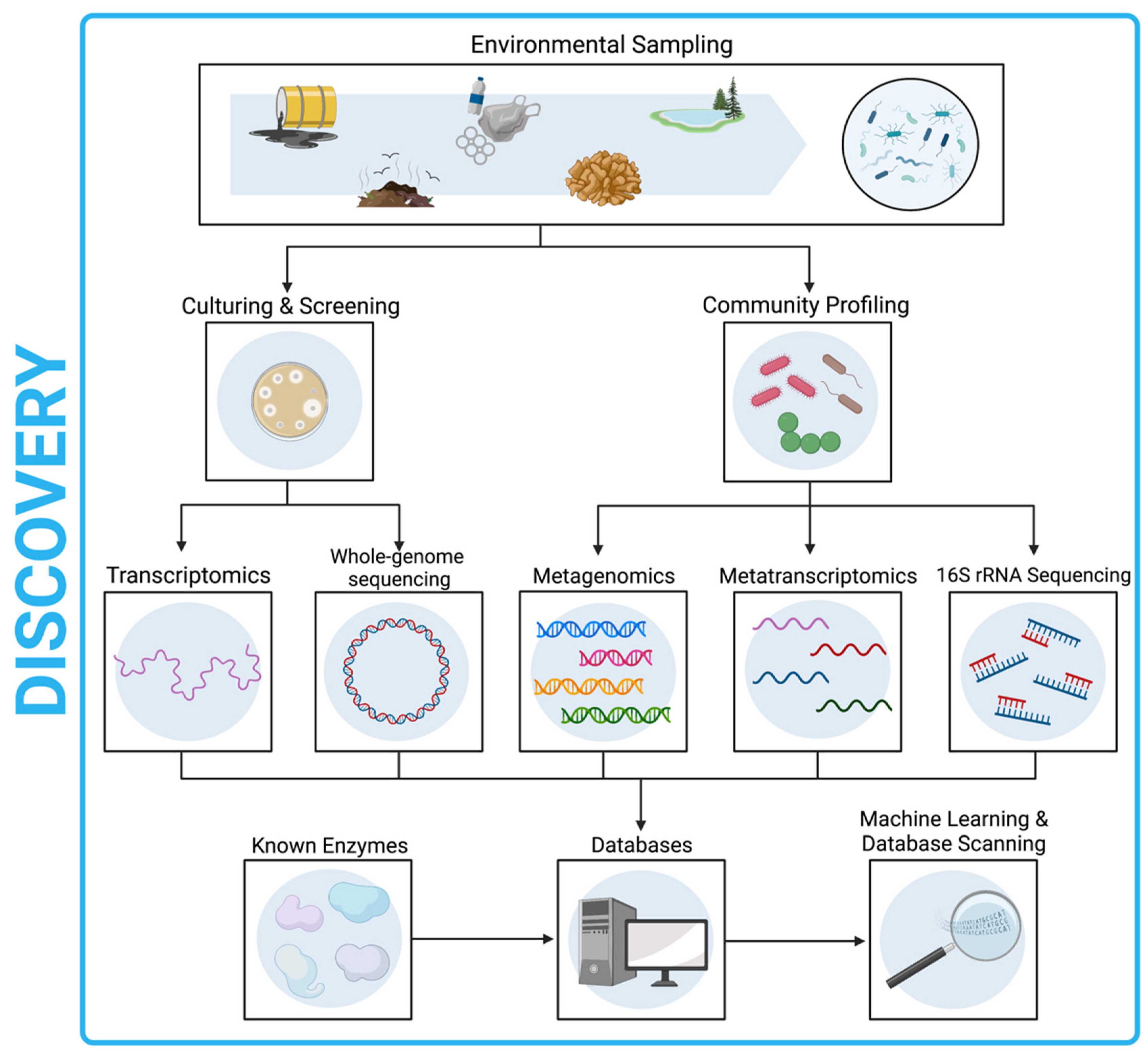
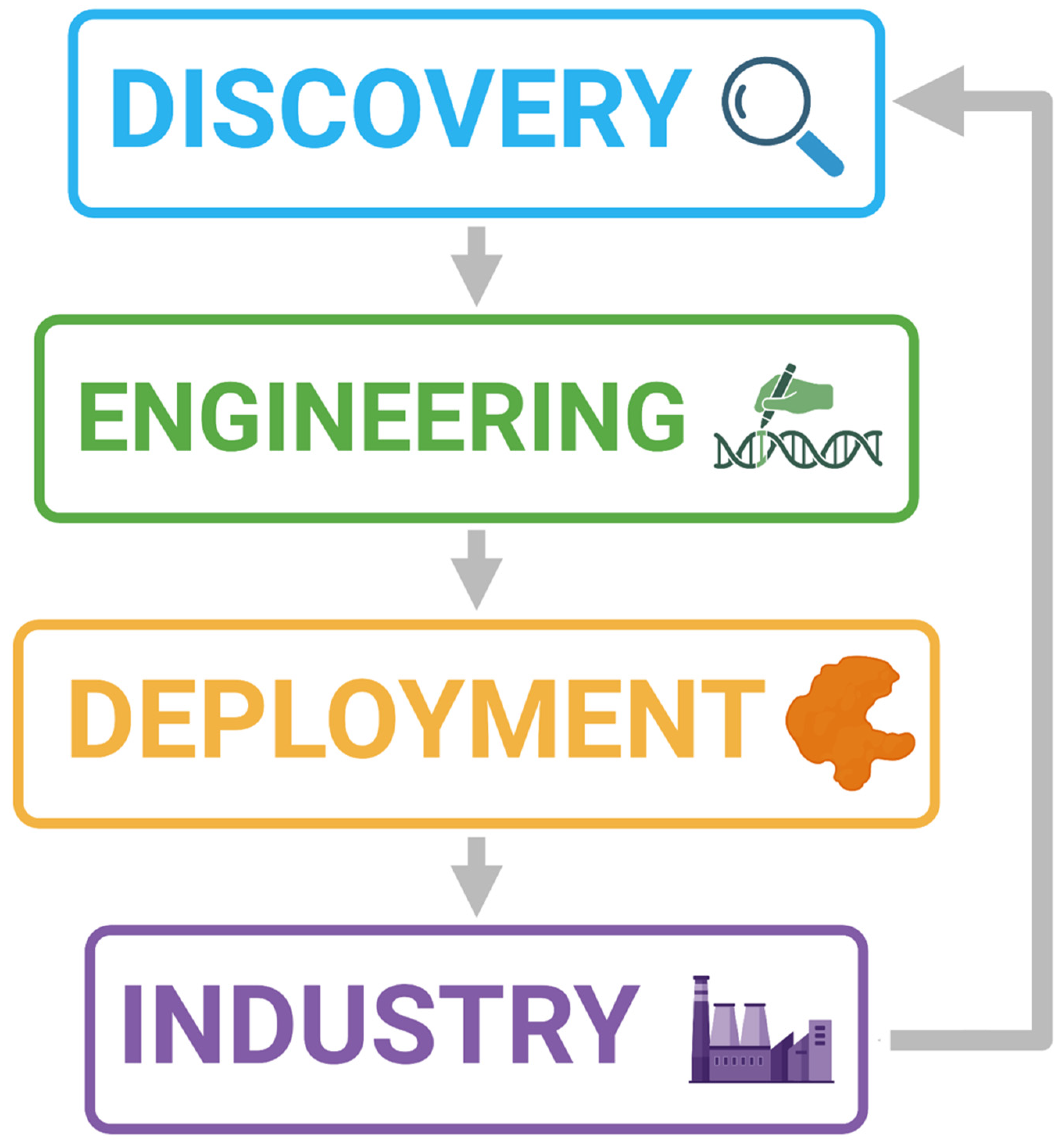
15. Discovery by Sequencing Workflow
- Samples collected from the environment should undergo culturing and screening for plastic-degrading activity followed by WGS, or metagenomic analysis.
- Enzymes of interest may also be discovered through database scanning approaches or ML techniques.
- Species of interest should be identified and submitted to culture databases, provided that the organism has been isolated and is culturable.
- Any identified depolymerase genes should also undergo functional domain analysis to identify likely active site motifs, and transcriptomics used to confirm gene expression.
- Depolymerisation should be measurable and significant. If rapid screening has been performed using methods such as agar clearance zones, especially if screening has been performed using biopolymers, then depolymerisation should be measured using the target plastic. Additionally, the plastic composition and molecular weight should be detailed.
- If depolymerisation has been proven to be significant, then candidates should be taken forward for in-depth enzyme characterisation and kinetic analysis.
- The most promising candidates may then be engineered in vitro with the view to creating an industrially relevant enzyme.
- The most promising putative enzymes should be taken forward for upscaling to industry. This should include exposure to a high substrate load, as would be required in large scale recycling.
16. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Urbanek, A.K.; Rymowicz, W.; Mirończuk, A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 2018, 102, 7669–7678. [Google Scholar] [CrossRef] [PubMed]
- Sheth, M.U.; Kwartler, S.K.; Schmaltz, E.R.; Hoskinson, S.M.; Martz, E.; Dunphy-Daly, M.M.; Schultz, T.F.; Read, A.J.; Eward, W.C.; Somarelli, J.A. Bioengineering a Future Free of Marine Plastic Waste. Front. Mar. Sci. 2019, 6, 624. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current Status and Application Aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Ostle, C.; Thompson, R.; Broughton, D.; Gregory, L.; Wootton, M.; Johns, D.G. The rise in ocean plastics evidenced from a 60-year time series. Nat. Commun. 2019, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lehel, J.; Murphy, S. Microplastics in the Food Chain: Food Safety and Environmental Aspects. Rev. Environ. Contam. Toxicol. 2021, 259, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- UNEP. UN declares war on ocean plastic, United Nation Environment Programme. 2017. Available online: https://www.unenvironment.org/news-and-stories/press-release/un-declares-war-ocean-plastic-0 (accessed on 28 September 2022).
- Liverman, C.T.; Larson, E.L. Preventing Transmission of Pandemic Influenza and Other Viral Respiratory Diseases; The National Academies Press: Washington, DC, USA, 2010. [Google Scholar] [CrossRef]
- Benson, N.U.; Bassey, D.E.; Palanisami, T. COVID pollution: Impact of COVID-19 pandemic on global plastic waste footprint. Heliyon 2021, 7, e06343. [Google Scholar] [CrossRef]
- Vanapalli, K.R.; Sharma, H.B.; Ranjan, V.P.; Samal, B.; Bhattacharya, J.; Dubey, B.K.; Goel, S. Challenges and strategies for effective plastic waste management during and post COVID-19 pandemic. Sci. Total. Environ. 2020, 750, 141514. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current State and Perspectives Related to the Polyethylene Terephthalate Hydrolases Available for Biorecycling. ACS Sustain. Chem. Eng. 2020, 8, 8894–8908. [Google Scholar] [CrossRef]
- Masmoudi, F.; Bessadok, A.; Dammak, M.; Jaziri, M.; Ammar, E. Biodegradable packaging materials conception based on starch and polylactic acid (PLA) reinforced with cellulose. Environ. Sci. Pollut. Res. 2016, 23, 20904–20914. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, B.-H. Poly(butylene succinate) and its copolymers: Research, development and industrialization. Biotechnol. J. 2010, 5, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Aliotta, L.; Seggiani, M.; Lazzeri, A.; Gigante, V.; Cinelli, P. A Brief Review of Poly (Butylene Succinate) (PBS) and Its Main Copolymers: Synthesis, Blends, Composites, Biodegradability, and Applications. Polymers 2022, 14, 844. [Google Scholar] [CrossRef] [PubMed]
- Bhagabati, P. Biopolymers and biocomposites-mediated sustainable high-performance materials for automobile applications. In Sustainable Nanocellulose and Nanohydrogels from Natural Sources; Elsevier: Amsterdam, The Netherlands, 2020; pp. 197–216. [Google Scholar] [CrossRef]
- Li, Z.; Yang, J.; Loh, X.J. Polyhydroxyalkanoates: Opening doors for a sustainable future. NPG Asia Mater. 2016, 8, e265. [Google Scholar] [CrossRef]
- European Bioplastics. Bioplastics Market Data 2018; European Bioplastics: Berlin, Germany, 2018. [Google Scholar]
- Ellis, L.D.; Rorrer, N.A.; Sullivan, K.P.; Otto, M.; McGeehan, J.E.; Román-Leshkov, Y.; Wierckx, N.; Beckham, G.T. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 2021, 4, 539–556. [Google Scholar] [CrossRef]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and Enzymatic Degradation of Synthetic Plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef]
- Letcher, T.M. Environmental impact, societal issues, prevention, and solutions. In Plastic Waste and Recycling; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Zink, T.; Geyer, R. Material Recycling and the Myth of Landfill Diversion. J. Ind. Ecol. 2019, 23, 541–548. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Müller, R.-J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.-D. Enzymatic Degradation of Poly(ethylene terephthalate): Rapid Hydrolyse using a Hydrolase fromT. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Silva, C.M.; Carneiro, F.; O’Neill, A.; Cabral, J.S.M.; Guebitz, G.; Cavaco-Paulo, A. Cutinase—A new tool for biomodification of synthetic fibers. J. Polym. Sci. Part A Polym. Chem. 2005, 43, 2448–2450. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Takehana, T.; Yoshida, S.; Hiraga, K.; Oda, K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). Int. J. Syst. Evol. Microbiol. 2016, 66, 2813–2818. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Aalismail, N.; Martin, C.; Kamau, A.; Guzmán-Vega, F.J.; Jamil, T.; Duarte, C.M. Rapid Evolution of Plastic-degrading Enzymes Prevalent in the Global Ocean. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. Phylogenetic Distribution of Plastic-Degrading Microorganisms. mSystems 2021, 6, e01112-20. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef]
- Pérez-García, P.; Danso, D.; Zhang, H.; Chow, J.; Streit, W.R. Exploring the global metagenome for plastic-degrading enzymes. Methods Enzymol. 2021, 648, 137–157. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Schatz, M.C.; Delcher, A.L.; Salzberg, S.L. Assembly of large genomes using second-generation sequencing. Genome Res. 2010, 20, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore Technology and Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Ma, Y.; Yao, H.; Xu, Z.; Chen, S.; Song, J.; Au, K.F. IDP-denovo: De novo transcriptome assembly and isoform annotation by hybrid sequencing. Bioinformatics 2018, 34, 2168–2176. [Google Scholar] [CrossRef]
- Baeza, J.A. Yes, we can use it: A formal test on the accuracy of low-pass nanopore long-read sequencing for mitophylogenomics and barcoding research using the Caribbean spiny lobster Panulirus argus. BMC Genom. 2020, 21, 882. [Google Scholar] [CrossRef]
- Sharon, B.M.; Hulyalkar, N.V.; Nguyen, V.H.; Zimmern, P.E.; Palmer, K.L.; De Nisco, N.J. Hybrid De Novo Genome Assembly for the Generation of Complete Genomes of Urinary Bacteria using Short- and Long-read Sequencing Technologies. J. Vis. Exp. 2021, 2021, e62872. [Google Scholar] [CrossRef]
- Kchouk, M.; Elloumi, M. An Error Correction and DeNovo Assembly Approach for Nanopore Reads Using Short Reads. Curr. Bioinform. 2017, 13, 241–252. [Google Scholar] [CrossRef]
- Chen, Z.; Erickson, D.L.; Meng, J. Polishing the Oxford Nanopore long-read assemblies of bacterial pathogens with Illumina short reads to improve genomic analyses. Genomics 2021, 113, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.J.; Erni-Cassola, G.; Zadjelovic, V.; Latva, M.; Christie-Oleza, J.A. Marine plastic debris–a new surface for microbial colonization. Environ. Sci. Technol. 2020, 54, 11657–11672. [Google Scholar] [CrossRef] [PubMed]
- Viljakainen, V.; Hug, L. New approaches for the characterization of plastic-associated microbial communities and the discovery of plastic-degrading microorganisms and enzymes. Comput. Struct. Biotechnol. J. 2021, 19, 6191–6200. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, D.; Wei, N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022, 40, 22–37. [Google Scholar] [CrossRef]
- Lear, G.; Kingsbury, J.M.; Franchini, S.; Gambarini, V.; Maday, S.D.M.; Wallbank, J.A.; Weaver, L.; Pantos, O. Plastics and the microbiome: Impacts and solutions. Environ. Microbiome 2021, 16, 2. [Google Scholar] [CrossRef]
- Stamps, B.W.; Zingarelli, S.; Hung, C.-S.; Drake, C.A.; Varaljay, V.A.; Stevenson, B.S.; Crookes-Goodson, W.J. Finished Genome Sequence of a Polyurethane-Degrading Pseudomonas Isolate. Genome Announc. 2018, 6, e00084-18. [Google Scholar] [CrossRef]
- Pinnell, L.J.; Turner, J.W. Shotgun Metagenomics Reveals the Benthic Microbial Community Response to Plastic and Bioplastic in a Coastal Marine Environment. Front. Microbiol. 2019, 10, 1252. [Google Scholar] [CrossRef]
- Molitor, R.; Bollinger, A.; Kubicki, S.; Loeschcke, A.; Jaeger, K.-E.; Thies, S. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb. Biotechnol. 2020, 13, 274–284. [Google Scholar] [CrossRef]
- de Almeida, E.L.; Rincón, A.F.C.; Jackson, S.A.; Dobson, A.D.W. In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-Like Enzyme (SM14est) With Polycaprolactone (PCL)-Degrading Activity, From the Marine Sponge-Derived Strain Streptomyces sp. SM14. Front. Microbiol. 2019, 10, 2187. [Google Scholar] [CrossRef]
- Sameshima-Yamashita, Y.; Ueda, H.; Koitabashi, M.; Kitamoto, H. Pretreatment with an esterase from the yeast Pseudozyma antarctica accelerates biodegradation of plastic mulch film in soil under laboratory conditions. J. Biosci. Bioeng. 2019, 127, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Bardají, D.K.R.; Furlan, J.P.R.; Stehling, E.G. Isolation of a polyethylene degrading Paenibacillus sp. from a landfill in Brazil. Arch. Microbiol. 2019, 201, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Gaytán, I.; Sánchez-Reyes, A.; Burelo, M.; Vargas-Suárez, M.; Liachko, I.; Press, M.; Sullivan, S.; Cruz-Gómez, M.J.; Loza-Tavera, H. Degradation of Recalcitrant Polyurethane and Xenobiotic Additives by a Selected Landfill Microbial Community and Its Biodegradative Potential Revealed by Proximity Ligation-Based Metagenomic Analysis. Front. Microbiol. 2020, 10, 2986. [Google Scholar] [CrossRef]
- Bollinger, A.; Thies, S.; Knieps-Grünhagen, E.; Gertzen, C.; Kobus, S.; Höppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.J.; Jaeger, K.-E. A Novel Polyester Hydrolase From the Marine Bacterium Pseudomonas aestusnigri–Structural and Functional Insights. Front. Microbiol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Sonnendecker, C.; Oeser, J.; Richter, P.K.; Hille, P.; Zhao, Z.; Fischer, C.; Lippold, H.; Blázquez-Sánchez, P.; Engelberger, F.; Ramírez-Sarmiento, C.A.; et al. Low Carbon Footprint Recycling of Post-Consumer PET Plastic with a Metagenomic Polyester Hydrolase. ChemSusChem 2021, 15, e202101062. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-W.; Jo, J.H.; Kim, Y.-B.; Le, T.-K.; Cho, C.-W.; Yun, C.-H.; Chi, W.S.; Yeom, S.-J. Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 2021, 416, 126239. [Google Scholar] [CrossRef]
- Vázquez-Alcántara, L.; Oliart-Ros, R.M.; García-Bórquez, A.; Peña-Montes, C. Expression of a Cutinase of Moniliophthora roreri with Polyester and PET-Plastic Residues Degradation Activity. Microbiol. Spectr. 2021, 9, e00976-21. [Google Scholar] [CrossRef]
- Xi, X.; Ni, K.; Hao, H.; Shang, Y.; Zhao, B.; Qian, Z. Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 2021, 143, 109715. [Google Scholar] [CrossRef]
- Blázquez-Sánchez, P.; Engelberger, F.; Cifuentes-Anticevic, J.; Sonnendecker, C.; Griñén, A.; Reyes, J.; Díez, B.; Guixé, V.; Richter, P.K.; Zimmermann, W.; et al. Antarctic Polyester Hydrolases Degrade Aliphatic and Aromatic Polyesters at Moderate Temperatures. Appl. Environ. Microbiol. 2022, 88, e01842-21. [Google Scholar] [CrossRef]
- Sagong, H.-Y.; Son, H.F.; Seo, H.; Hong, H.; Lee, D.; Kim, K.-J. Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis. J. Hazard. Mater. 2021, 416, 126075. [Google Scholar] [CrossRef]
- Soulenthone, P.; Tachibana, Y.; Suzuki, M.; Mizuno, T.; Ohta, Y.; Kasuya, K.-I. Characterization of a poly(butylene adipate-co-terephthalate) hydrolase from the mesophilic actinobacteria Rhodococcus fascians. Polym. Degrad. Stab. 2021, 184, 109481. [Google Scholar] [CrossRef]
- Zhang, A.; Hou, Y.; Wang, Q.; Wang, Y. Characteristics and polyethylene biodegradation function of a novel cold-adapted bacterial laccase from Antarctic sea ice psychrophile Psychrobacter sp. NJ228. J. Hazard. Mater. 2022, 439, 129656. [Google Scholar] [CrossRef] [PubMed]
- Eiamthong, B.; Meesawat, P.; Wongsatit, T.; Jitdee, J.; Sangsri, R.; Patchsung, M.; Aphicho, K.; Suraritdechachai, S.; Huguenin-Dezot, N.; Tang, S.; et al. Discovery and Genetic Code Expansion of a Polyethylene Terephthalate (PET) Hydrolase from the Human Saliva Metagenome for the Degradation and Bio-Functionalization of PET. Angew. Chem. Int. Ed. 2022, 61, e202203061. [Google Scholar] [CrossRef]
- Edwards, S.; León-Zayas, R.; Ditter, R.; Laster, H.; Sheehan, G.; Anderson, O.; Beattie, T.; Mellies, J.L. Microbial Consortia and Mixed Plastic Waste: Pangenomic Analysis Reveals Potential for Degradation of Multiple Plastic Types via Previously Identified PET Degrading Bacteria. Int. J. Mol. Sci. 2022, 23, 5612. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Wallbank, J.A.; Lear, G.; Kingsbury, J.M.; Weaver, L.; Doake, F.; Smith, D.A.; Audrézet, F.; Maday, S.D.M.; Gambarini, V.; Donaldson, L.; et al. Into the Plastisphere, Where Only the Generalists Thrive: Early Insights in Plastisphere Microbial Community Succession. Front. Mar. Sci. 2022, 9, 841142. [Google Scholar] [CrossRef]
- Parkhill, J. What has high-throughput sequencing ever done for us? Nat. Rev. Genet. 2013, 11, 664–665. [Google Scholar] [CrossRef]
- Popovic, A.; Hai, T.; Tchigvintsev, A.; Hajighasemi, M.; Nocek, B.; Khusnutdinova, A.N.; Brown, G.; Glinos, J.; Flick, R.; Skarina, T.; et al. Activity screening of environmental metagenomic libraries reveals novel carboxylesterase families. Sci. Rep. 2017, 7, srep44103. [Google Scholar] [CrossRef]
- Viljakainen, V.R.; Hug, L.A. The phylogenetic and global distribution of bacterial polyhydroxyalkanoate bioplastic-degrading genes. Environ. Microbiol. 2021, 23, 1717–1731. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, J.; Wu, W.-M.; Zhao, J.; Yang, J. Complete genome sequence of Bacillus sp. YP1, a polyethylene-degrading bacterium from waxworm’s gut. J. Biotechnol. 2015, 200, 77–78. [Google Scholar] [CrossRef]
- León-Zayas, R.; Roberts, C.; Vague, M.; Mellies, J.L. Draft Genome Sequences of Five Environmental Bacterial Isolates That Degrade Polyethylene Terephthalate Plastic. Microbiol. Resour. Announc. 2019, 8, e00237-19. [Google Scholar] [CrossRef] [PubMed]
- Culligan, E.; Sleator, R.D.; Marchesi, J.R.; Hill, C. Metagenomics and novel gene discovery. Virulence 2014, 5, 399–412. [Google Scholar] [CrossRef]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 2017, 364, fnx211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Perez-Garcia, P.; Dierkes, R.F.; Applegate, V.; Schumacher, J.; Chibani, C.M.; Sternagel, S.; Preuss, L.; Weigert, S.; Schmeisser, C.; et al. The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity. Front. Microbiol. 2022, 12, 803896. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Ling, W.; Wang, Z.; Li, Y.; Zhou, M.; Ren, M.; Li, X.; Li, J.; Xia, Z.; Liu, X.; et al. Miniaturized DNA Sequencers for Personal Use: Unreachable Dreams or Achievable Goals. Front. Nanotechnol. 2021, 3, 628861. [Google Scholar] [CrossRef]
- Liu, C.-C.; Dong, S.-S.; Chen, J.-B.; Wang, C.; Ning, P.; Guo, Y.; Yang, T.-L. MetaDecoder: A novel method for clustering metagenomic contigs. Microbiome 2022, 10, 46. [Google Scholar] [CrossRef]
- Latorre-Pérez, A.; Villalba-Bermell, P.; Pascual, J.; Vilanova, C. Assembly methods for nanopore-based metagenomic sequencing: A comparative study. Sci. Rep. 2020, 10, 13588. [Google Scholar] [CrossRef]
- Sereika, M.; Kirkegaard, R.H.; Karst, S.M.; Michaelsen, T.Y.; Sørensen, E.A.; Wollenberg, R.D.; Albertsen, M. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat. Methods 2022, 19, 823–826. [Google Scholar] [CrossRef]
- Olson, N.D.; Treangen, T.J.; Hill, C.M.; Cepeda-Espinoza, V.; Ghurye, J.; Koren, S.; Pop, M. Metagenomic assembly through the lens of validation: Recent advances in assessing and improving the quality of genomes assembled from metagenomes. Briefings Bioinform. 2018, 20, 1140–1150. [Google Scholar] [CrossRef]
- Ko, K.K.K.; Chng, K.R.; Nagarajan, N. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 2022, 7, 486–496. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Buchholz, P.C.F.; Feuerriegel, G.; Zhang, H.; Perez-Garcia, P.; Nover, L.; Chow, J.; Streit, W.R.; Pleiss, J. Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database— PAZy. Proteins: Struct. Funct. Bioinform. 2022, 90, 1443–1456. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Zhang, H. PMBD: A Comprehensive Plastics Microbial Biodegradation Database. Database 2019, 2019, baz119. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Hopgood, A.A. Intelligent Systems for Engineers and Scientists: A Practical Guide to Artificial Intelligence, 4th ed.; CRC Press: Boca Raton, FL, USA, 2022; Volume 1. [Google Scholar]
- Zrimec, J.; Kokina, M.; Jonasson, S.; Zorrilla, F.; Zelezniak, A. Plastic-Degrading Potential across the Global Microbiome Correlates with Recent Pollution Trends. mBio 2021, 12, e02155-21. [Google Scholar] [CrossRef]
- Kumari, A.; Bano, N.; Bag, S.K.; Chaudhary, D.R.; Jha, B. Transcriptome-Guided Insights Into Plastic Degradation by the Marine Bacterium. Front. Microbiol. 2021, 12, 2761. [Google Scholar] [CrossRef]
- Zampolli, J.; Orro, A.; Manconi, A.; Ami, D.; Natalello, A.; Di Gennaro, P. Transcriptomic analysis of Rhodococcus opacus R7 grown on polyethylene by RNA-seq. Sci. Rep. 2021, 11, 21311. [Google Scholar] [CrossRef]
- Kellner, H.; Luis, P.; Portetelle, D.; Vandenbol, M. Screening of a soil metatranscriptomic library by functional complementation of Saccharomyces cerevisiae mutants. Microbiol. Res. 2011, 166, 360–368. [Google Scholar] [CrossRef]
- Takasaki, K.; Miura, T.; Kanno, M.; Tamaki, H.; Hanada, S.; Kamagata, Y.; Kimura, N. Discovery of Glycoside Hydrolase Enzymes in an Avicel-Adapted Forest Soil Fungal Community by a Metatranscriptomic Approach. PLoS ONE 2013, 8, e55485. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.E.; Werner, J.; Jehmlich, N.; Will, S.E.; Neumann-Schaal, M.; Öztürk, B. Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 2020, 11, 5790. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Makarova, K.S.; Natale, D.A.; Spiridonov, A.N.; Tatusov, R.L.; Wolf, Y.I.; Yin, J.; Koonin, E.V. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 2002, 30, 4264–4271. [Google Scholar] [CrossRef] [PubMed]
- Burger, G. Non-functional genes repaired at the RNA level. Comptes Rendus. Biol. 2016, 339, 289–295. [Google Scholar] [CrossRef]
- Penkhrue, W.; Khanongnuch, C.; Masaki, K.; Pathom-Aree, W.; Punyodom, W.; Lumyong, S. Isolation and screening of biopolymer-degrading microorganisms from northern Thailand. World J. Microbiol. Biotechnol. 2015, 31, 1431–1442. [Google Scholar] [CrossRef]
- Lear, G.; Maday, S.D.M.; Gambarini, V.; Northcott, G.; Abbel, R.; Kingsbury, J.M.; Weaver, L.; A Wallbank, J.; Pantos, O. Microbial abilities to degrade global environmental plastic polymer waste are overstated. Environ. Res. Lett. 2022, 17, 043002. [Google Scholar] [CrossRef]
- Rana, K.; Rana, N. Isolation and Screening of Plastic Degrading Bacteria from Dumping Sites of Solid Waste. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 2611–2618. [Google Scholar] [CrossRef]
- Haedar, N.; Clara, T.; Rapak, M.T. Selection of Plastic Degradation Indigenous bacteria Isolated from Tamangapa Landfill Macassar City. J. Physics Conf. Ser. 2019, 1341, 022023. [Google Scholar] [CrossRef]
- Carniel, A.; Waldow, V.D.A.; de Castro, A.M. A comprehensive and critical review on key elements to implement enzymatic PET depolymerization for recycling purposes. Biotechnol. Adv. 2021, 52, 107811. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Wani, S.J.; Alarfaj, A.A.; Syed, A.; El-Enshasy, H.A. Production, purification and evaluation of biodegradation potential of PHB depolymerase of Stenotrophomonas sp. RZS7. PLoS ONE 2020, 15, e0220095. [Google Scholar] [CrossRef] [PubMed]
- Charnock, C. Norwegian Soils and Waters Contain Mesophilic, Plastic-Degrading Bacteria. Microorganisms 2021, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.; Belchior, A.; da Silva, V.D.; Rotter, A.; Petrovski, Ž.; Almeida, P.L.; Lourenço, N.D.; Gaudêncio, S.P. Marine Environmental Plastic Pollution: Mitigation by Microorganism Degradation and Recycling Valorization. Front. Mar. Sci. 2020, 7, 567126. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef]
- Mayumi, D.; Akutsu-Shigeno, Y.; Uchiyama, H.; Nomura, N.; Nakajima-Kambe, T. Identification and characterization of novel poly (DL-lactic acid) depolymerases from metagenome. Appl. Microbiol. Biotechnol. 2008, 79, 743–750. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Karunatillaka, I.; Jaroszewski, L.; Godzik, A. Novel putative polyethylene terephthalate (PET) plastic degrading enzymes from the environmental metagenome. Proteins: Struct. Funct. Bioinform. 2022, 90, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Kellici, T.F.; Mavromoustakos, T.; Jendrossek, D.; Papageorgiou, A.C. Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase. Proteins: Struct. Funct. Bioinform. 2017, 85, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramírez-Sarmiento, C.A. Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.; Blanch, H.W. Technoeconomic analysis of biofuels: A wiki-based platform for lignocellulosic biorefineries. Biomass- Bioenergy 2010, 34, 1914–1921. [Google Scholar] [CrossRef]
- Chen, C.; Han, X.; Ko, T.; Liu, W.; Guo, R. Structural studies reveal the molecular mechanism of PET ase. FEBS J. 2018, 285, 3717–3723. [Google Scholar] [CrossRef]
- Knott, B.C.; Erickson, E.; Allen, M.D.; Gado, J.E.; Graham, R.; Kearns, F.L.; Pardo, I.; Topuzlu, E.; Anderson, J.J.; Austin, H.P.; et al. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. USA 2020, 117, 25476–25485. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, Y.; Wu, P.; Wang, H.; Li, Q.; Gao, J.; Qin, H.-M.; Wei, H.; Bornscheuer, U.T.; Han, X.; et al. Structural insight and engineering of a plastic degrading hydrolase Ple629. Biochem. Biophys. Res. Commun. 2022, 626, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Schmidt, B.; Hildebrandt, A. Deep learning in next-generation sequencing. Drug Discov. Today 2021, 26, 173–180. [Google Scholar] [CrossRef]
- Erickson, E.; Shakespeare, T.J.; Bratti, F.; Buss, B.L.; Graham, R.; Hawkins, M.A.; König, G.; Michener, W.E.; Miscall, J.; Ramirez, K.J.; et al. Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation. ChemSusChem 2022, 15, e202101932. [Google Scholar] [CrossRef]
- Thomsen, T.B.; Hunt, C.J.; Meyer, A.S. Standardized method for controlled modification of poly (ethylene terephthalate) (PET) crystallinity for assaying PET degrading enzymes. MethodsX 2022, 9, 101815. [Google Scholar] [CrossRef]
- Klomklang, W.; Tani, A.; Kimbara, K.; Mamoto, R.; Ueda, T.; Shimao, M.; Kawai, F. Biochemical and molecular characterization of a periplasmic hydrolase for oxidized polyvinyl alcohol from Sphingomonas sp. strain 113P3. Microbiology 2005, 151, 1255–1262. [Google Scholar] [CrossRef]
- Kari, J.; Andersen, M.; Borch, K.; Westh, P. An Inverse Michaelis–Menten Approach for Interfacial Enzyme Kinetics. ACS Catal. 2017, 7, 4904–4914. [Google Scholar] [CrossRef]
- Singh, A.; Rorrer, N.A.; Nicholson, S.R.; Erickson, E.; DesVeaux, J.S.; Avelino, A.F.; Lamers, P.; Bhatt, A.; Zhang, Y.; Avery, G.; et al. Techno-economic, life-cycle, and socioeconomic impact analysis of enzymatic recycling of poly(ethylene terephthalate). Joule 2021, 5, 2479–2503. [Google Scholar] [CrossRef]
- Graham, R.; Erickson, E.; Brizendine, R.K.; Salvachúa, D.; Michener, W.E.; Li, Y.; Tan, Z.; Beckham, G.T.; McGeehan, J.E.; Pickford, A.R. The role of binding modules in enzymatic poly(ethylene terephthalate) hydrolysis at high-solids loadings. Chem Catal. 2022, 2, 2644–2657. [Google Scholar] [CrossRef]
- Uekert, T.; DesVeaux, J.S.; Singh, A.; Nicholson, S.R.; Lamers, P.; Ghosh, T.; McGeehan, J.E.; Carpenter, A.C.; Beckham, G.T. Life cycle assessment of enzymatic poly(ethylene terephthalate) recycling. Green Chem. 2022, 24, 6531–6543. [Google Scholar] [CrossRef]
- Brizendine, R.K.; Erickson, E.; Haugen, S.J.; Ramirez, K.J.; Miscall, J.; Salvachúa, D.; Pickford, A.R.; Sobkowicz, M.J.; McGeehan, J.E.; Beckham, G.T. Particle Size Reduction of Poly(ethylene terephthalate) Increases the Rate of Enzymatic Depolymerization But Does Not Increase the Overall Conversion Extent. ACS Sustain. Chem. Eng. 2022, 10, 9131–9140. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef]
- Kikkawa, Y.; Fujita, M.; Abe, H.; Doi, Y. Effect of Water on the Surface Molecular Mobility of Poly(lactide) Thin Films: An Atomic Force Microscopy Study. Biomacromolecules 2004, 5, 1187–1193. [Google Scholar] [CrossRef]
- Kawai, F.; Oda, M.; Tamashiro, T.; Waku, T.; Tanaka, N.; Yamamoto, M.; Mizushima, H.; Miyakawa, T.; Tanokura, M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2014, 98, 10053–10064. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.R.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.-Y.; Lee, S.H.; Hong, H.; Kim, K.-J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef] [PubMed]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.; Cole, H.; Zhang, Y.J.; et al. Deep learning redesign of PETase for practical PET degrading applications. bioRxiv 2021. [Google Scholar] [CrossRef]
- Baker, T. Waste and Resources Action Programme (WRAP) Non-Technical Challenges to Non-Mechanical Recycling Final Report, UKRI Centre for the Circular Chemical Economy. 2022. Available online: https://wrap.org.uk/sites/default/files/2022-08/Non-Technical%20Challenges%20to%20Non-Mechanical%20Recycling.pdf (accessed on 28 September 2022).

| Enzyme | Origin | Discovery Methodology | No. Characterised Enzymes | Target Plastic | Year | Ref |
|---|---|---|---|---|---|---|
| PET hydrolase, Cutinase, Triaylglycerol lipase | Marine and terrestrial environments | Metagenomics and HMM search | 9 | PET | 2018 | [31] |
| Polyurethanase | Laboratory | Culturing and WGS | 1 | PU | 2018 | [52] |
| PHB deploymerase | Biofilms on marine plastics | Metagenomics | n/a | PHB | 2019 | [53] |
| PET hydrolase | Crude oil-contaminated intertidal sand samples | Metagenomics | 1 | PET | 2019 | [54] |
| Hydrolase | Marine sponge | Database search, screening, and cloning | 1 | PCL | 2019 | [55] |
| Esterase | Rice seeds | Culturing and screening | 1 | PCBS, PBSA, PCL | 2019 | [56] |
| Alkane monooxygenase | Landfill soil | Culturing and screening | 1 | LDPE | 2019 | [57] |
| Polyurethane esterase | Landfill | Screening and metagenomics | n/a | PU | 2020 | [58] |
| Polyester hydrolase | Marine and terrestrial environments | Database search and screening | 1 | PET, PU, PCL | 2020 | [59] |
| PET hydrolase | Compost | Metagenomics | 7 | PET | 2021 | [60] |
| Alkane-1 monooxygenase | Landfill soil | Screening and cloning | 1 | PS | 2021 | [61] |
| Cutinase | Culture collection | Screening and cloning | 1 | PES, PCL, PET | 2021 | [62] |
| PET hydrolase | Geothermal groundwater | Metagenomics and database search | 1 | PET | 2021 | [63] |
| Polyester hydrolase | Antarctic marine samples | Culturing, sequencing, and screening | 1 | PET, PCL, PU, PHB, PBS, PLA, PHA | 2021 | [64] |
| Hydrolase | Soil | Database search | 1 | PET | 2021 | [65] |
| Hydrolase | Soil | Culturing and screening | 1 | PBAT, PBSU, PBSA, PCL, PESU | 2021 | [66] |
| Esterase | Antarctic sources, marine (Japan), seaweed | Metagenomics and HMM search | 4 | PET, PU, PCL | 2022 | [67] |
| PET hydrolase | Human saliva | Metagenomics and HMM search | 1 | PET | 2022 | [68] |
| Esterase | Hydrocarbon-polluted soil | Database search | 1 | PET | 2022 | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herbert, J.; Beckett, A.H.; Robson, S.C. A Review of Cross-Disciplinary Approaches for the Identification of Novel Industrially Relevant Plastic-Degrading Enzymes. Sustainability 2022, 14, 15898. https://doi.org/10.3390/su142315898
Herbert J, Beckett AH, Robson SC. A Review of Cross-Disciplinary Approaches for the Identification of Novel Industrially Relevant Plastic-Degrading Enzymes. Sustainability. 2022; 14(23):15898. https://doi.org/10.3390/su142315898
Chicago/Turabian StyleHerbert, Josephine, Angela H. Beckett, and Samuel C. Robson. 2022. "A Review of Cross-Disciplinary Approaches for the Identification of Novel Industrially Relevant Plastic-Degrading Enzymes" Sustainability 14, no. 23: 15898. https://doi.org/10.3390/su142315898
APA StyleHerbert, J., Beckett, A. H., & Robson, S. C. (2022). A Review of Cross-Disciplinary Approaches for the Identification of Novel Industrially Relevant Plastic-Degrading Enzymes. Sustainability, 14(23), 15898. https://doi.org/10.3390/su142315898







