Monitoring Marine Oil Spills in Hyperspectral and Multispectral Remote Sensing Data by the Spectral Gene Extraction (SGE) Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Oil Spills
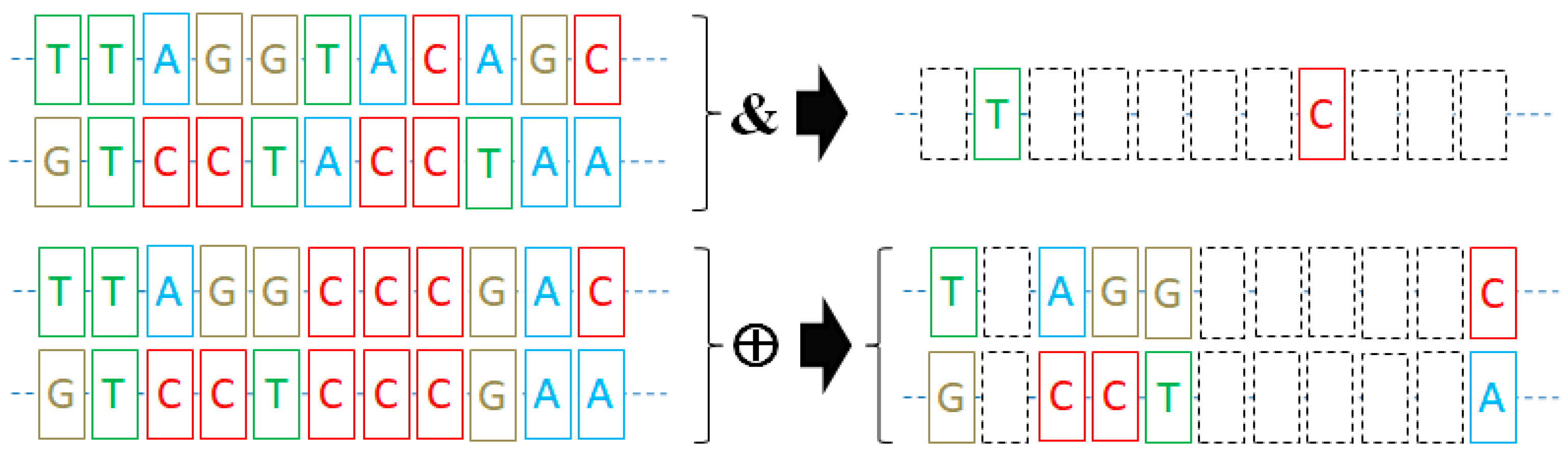
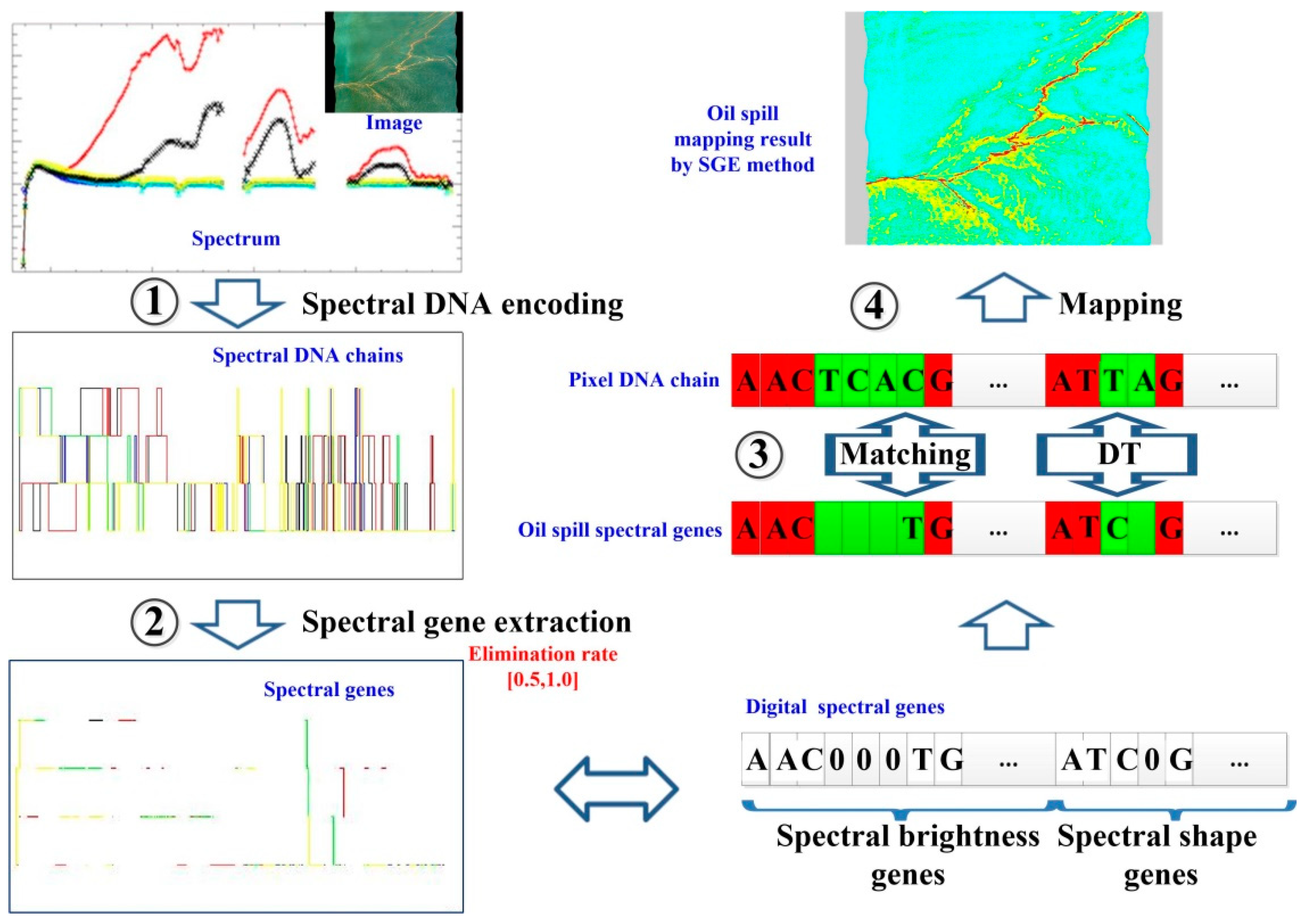
2.2. Spectral DNA Encoding Method
2.3. SGE Method
3. Results
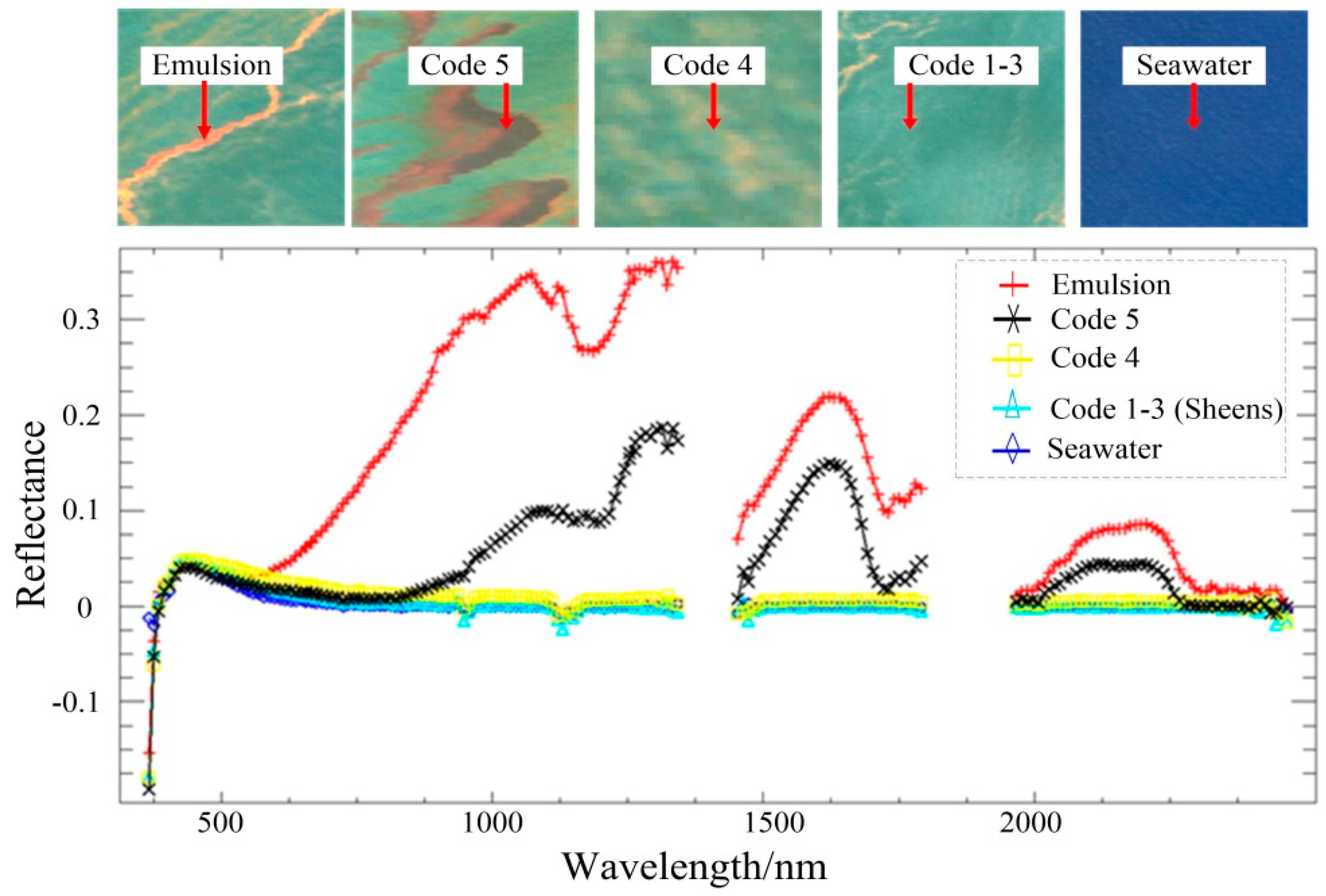
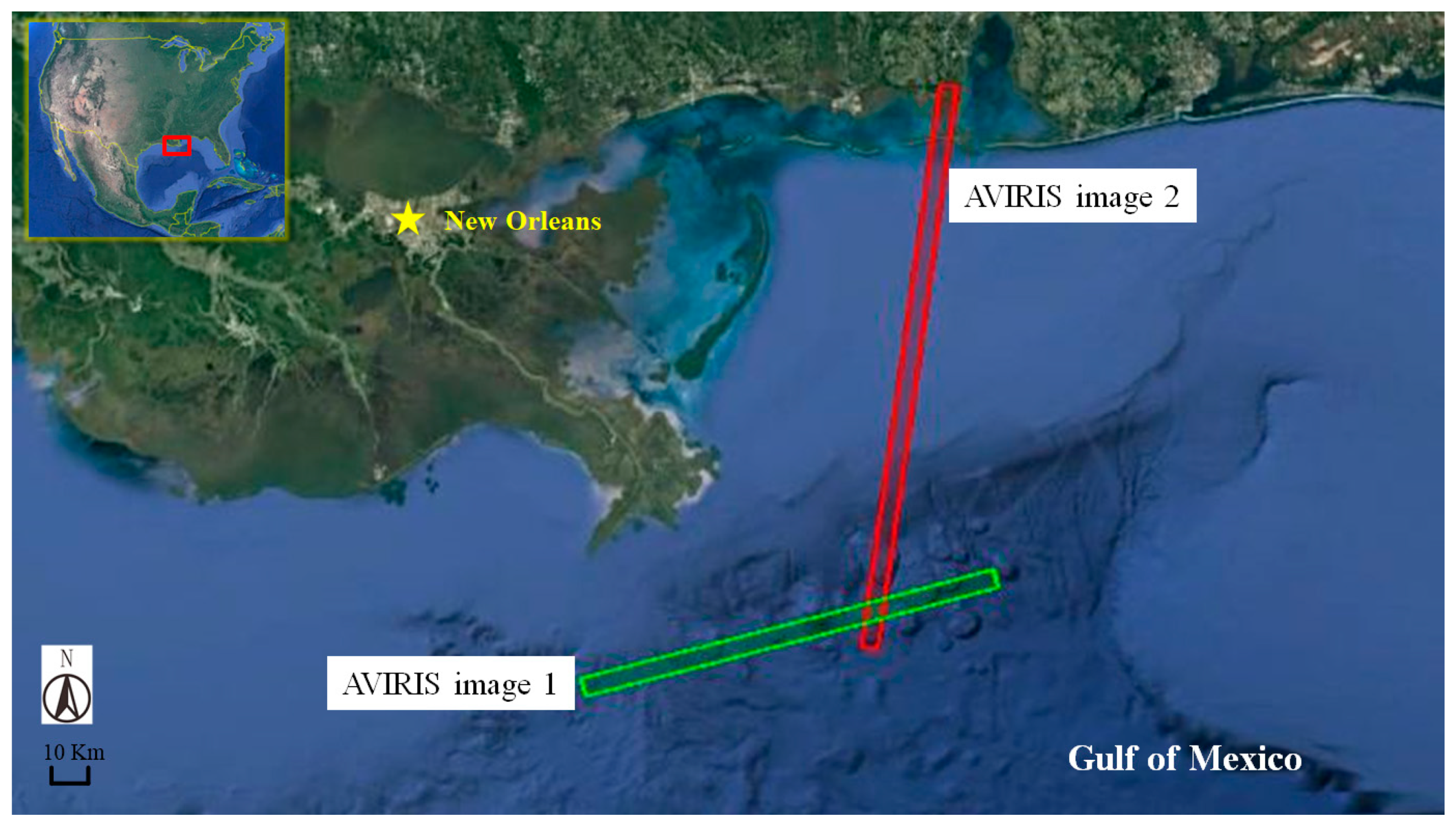
3.1. Experimental Data
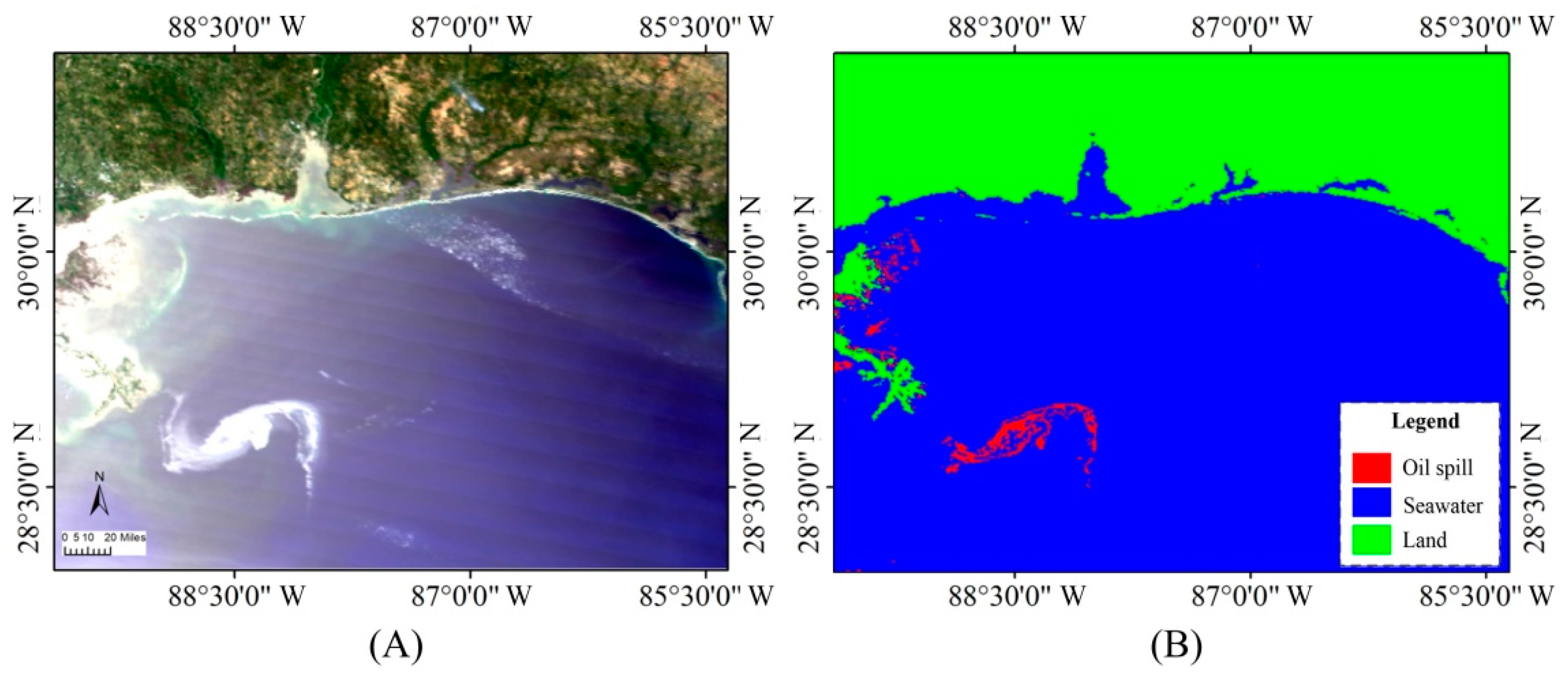
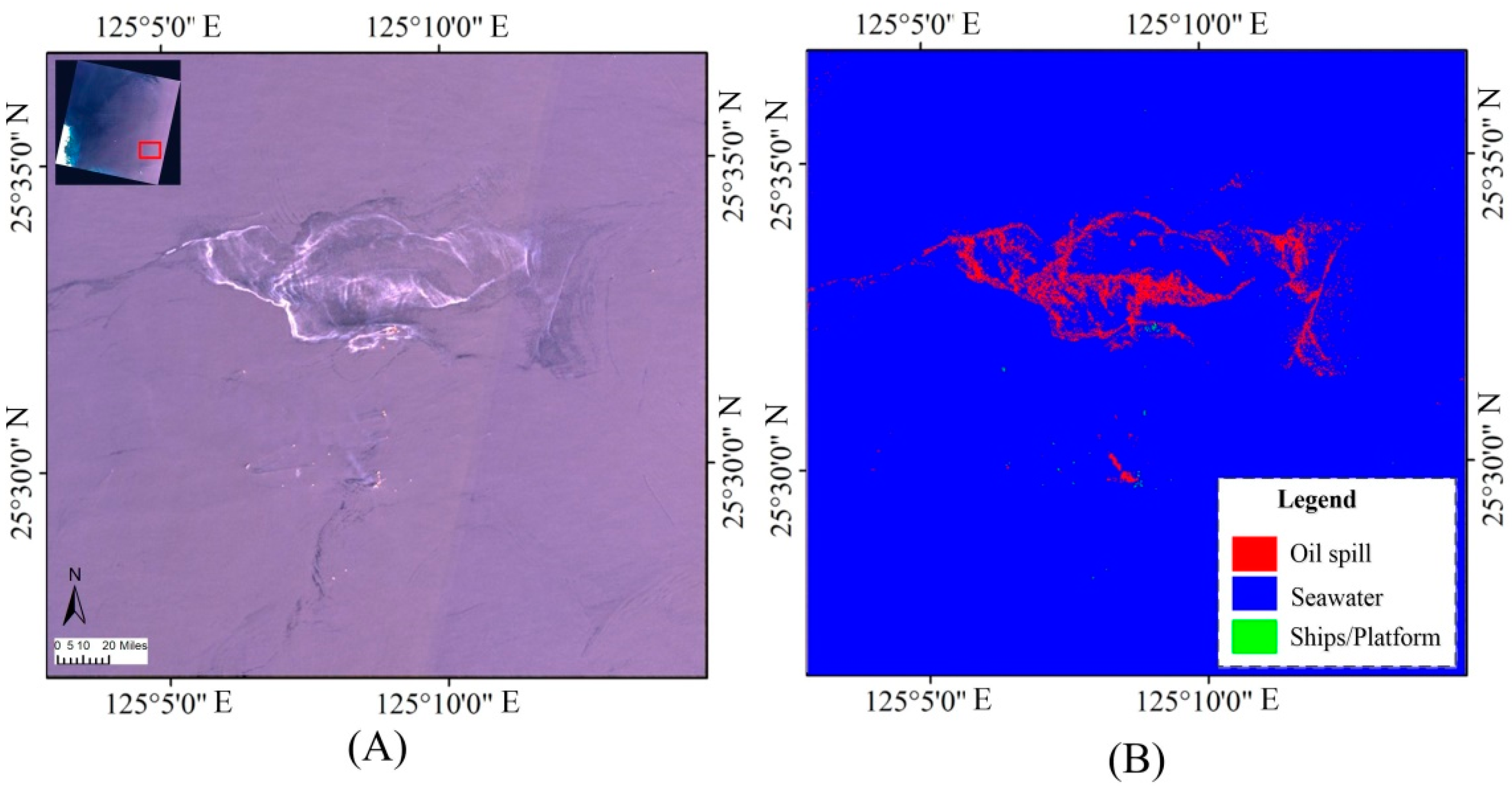
3.2. Experimental Results
4. Discussion
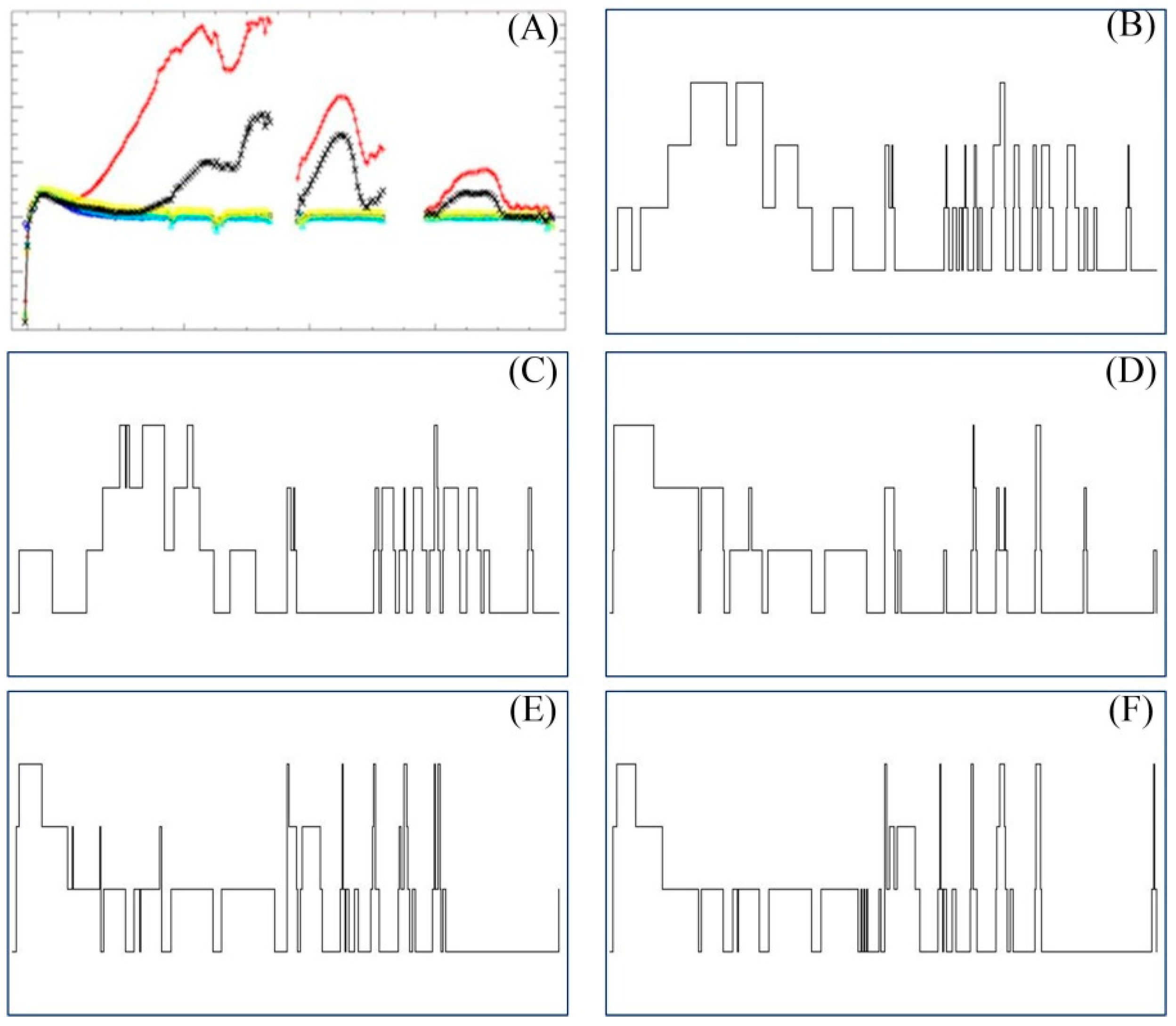
4.1. Spectral Gene Extraction Strategies
4.2. Superiority of the SGE Method
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alloy, M.; Baxter, D.; Stieglitz, J.; Mager, E.; Hoening, R.; Beenetti, D.; Grosell, M.; Oris, J.; Rogerts, A. Ultraviolet Radiation Enhances the Toxicity of Deepwater Horizon Oil to Mahi-mahi (Coryphaena hippurus) embryo. Environ. Sci. Technol. 2016, 50, 2011–2017. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Mager, E.; Grosell, M.; Christina, P.; Schlenker, L.; Stieglitz, J.; Benetti, D.; Hazard, E.; Courtney, S.; Diamante, G.; et al. Time- and oil-dependent transcriptomic and physiological responses to Deepwater Horizon oil in mahi-mahi (coryphaena hippurus) embryos and larvae. Environ. Sci. Technol. 2016, 50, 7842–7851. [Google Scholar] [CrossRef] [PubMed]
- Esbaughet, A.; Mager, E.; Stieglitz, J.; Hoening, R.; Brown, T.; French, B.; Linbo, T.; Lay, C.; Forth, H.; Scholz, N.; et al. The effects of weathering and chemical dispersion on Deepwater Horizon crude oil toxicity to mahi-mahi (coryphaena hippurus) early life stages. Sci. Total Environ. 2016, 543, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Mariano, A.; Kourafalou, V.; Srinivasan, A.; Kang, H.; Halliwell, G.; Ryan, E.; Roffer, M. On the modeling of the 2010 Gulf of Mexico Oil Spill. Dynam. Atmos. Oceans 2011, 52, 322–340. [Google Scholar] [CrossRef]
- Kokaly, R.; Couvillion, B.; Holloway, J.; Roberts, D.; Ustin, S.; Peterson, S.; Khanna, S.; Piazza, S. Spectroscopic remote sensing of the distribution and persistence of oil from the Deepwater Horizon spill in barataria bay marshes. Remote Sens. Environ. 2013, 129, 210–230. [Google Scholar] [CrossRef]
- Shen, H.; Zeng, C.; Zhang, L. Recovering reflectance of AQUA MODIS band 6 based on within-class local fitting. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2010, 4, 185–192. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.; Boardman, J.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, S110–S122. [Google Scholar] [CrossRef]
- Clark, R.; Swayze, G.; Leifer, I.; Livo, K.; Kokaly, R.; Hoeffen, T.; Lundeen, S.; Green, R.; Pearson, N.; Sarture, C.; et al. A method for quantitative mapping of thick oil spills using imaging spectroscopy. US Geological Survey Open-File Report 2010, 1167, 1–51. [Google Scholar] [CrossRef]
- Cong, L.; Nutter, B.; Liang, D. Estimation of oil thickness and aging from hyperspectral signature. In Proceedings of the IEEE Southwest Symposium on Image Analysis and Interpretation, Santa Fe, NM, USA, 22–24 April 2012; pp. 213–216. [Google Scholar] [CrossRef]
- Loos, E.; Brown, L.; Borstad, G.; Mudge, T.; Alvarez, M. Characterization of oil slicks at sea using remote sensing techniques. In Proceedings of the 2012 Oceans, Hampton Roads, VA, USA, 14–19 October 2012. [Google Scholar] [CrossRef]
- Zhao, J.; Temimi, M.; Ghedira, H. Exploring the potential of optical remote sensing for oil spill detection in shallow coastal waters-a case study in the Arabian Gulf. Opt. Express 2014, 22, 13755–13772. [Google Scholar] [CrossRef]
- Liu, B. Spatial–spectral jointed stacked auto-encoder-based deep learning for oil slick extraction from hyperspectral images. J. Indian Soc. Remote Sens. 2019, 47, 1989–1997. [Google Scholar] [CrossRef]
- Zhao, D.; Cheng, X.; Zhang, H.; Niu, Y.; Qi, Y.; Zhang, H. Evaluation of the ability of spectral indices of hydrocarbons and seawater for identifying oil slicks utilizing hyperspectral images. Remote Sens. 2018, 10, 421. [Google Scholar] [CrossRef]
- Chang, C.; Wang, J.; Lin, C. Progressive coding for hyperspectral signature characterization. Opt. Eng. 2006, 45, 097002. [Google Scholar] [CrossRef][Green Version]
- Jiao, H.; Zhong, Y.; Zhang, L. An unsupervised spectral matching classifier based on artificial DNA computing for hyperspectral remote sensing imagery. IEEE Trans. Geosci. Remote Sens. 2014, 52, 4524–4538. [Google Scholar] [CrossRef]
- Wu, K.; Zhao, D.; Zhong, Y.; Du, Q. Multi-probe based artificial DNA encoding and matching classifier for hyperspectral remote sensing imagery. Remote Sens. 2016, 8, 645. [Google Scholar] [CrossRef]
- Carpenter, A. The BONN agreement aerial surveillance programme: Trends in north sea oil pollution 1986-2004. Mar. Pollut. Bull. 2007, 54, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Dave, S.; Hughes, E.; Gair, S. Monitoring and Quantifying Oil Pollution using Aerial Surveillance Problems and Solutions. In Proceedings of the International Conference on Health, Safety and Environment in Oil and Gas Exploration and Production, Perth, Australia, 1–7 September 2012. [Google Scholar] [CrossRef]
- Leifer, I.; Lehr, W.; Simecek-Beatty, D.; Bradley, E.; Clark, R.; Dennison, P.; Hu, Y.; Matheson, S.; Jones, C.; Holt, B.; et al. State of the art satellite and airborne marine oil spill remote sensing: Application to the BP Deepwater Horizon oil spill. Remote Sens. Environ. 2012, 124, 185–209. [Google Scholar] [CrossRef]
- Svejkovsky, J.; Hess, M.; Muskat, J.; Nedwed, T.; McCall, J.; Garcia, O. Characterization of surface oil thickness distribution patterns observed during the Deepwater Horizon (mc-252) oil spill with aerial and satellite remote sensing. Mar. Pollut. Bull. 2016, 110, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Liu, B.; Chen, S.; Ma, Y.; Zhang, Y.; Shen, C.; Cui, J. Classification of the different thickness of the oil film based on wavelet transform spectrum information. Aquat. Procedia 2015, 3, 133–143. [Google Scholar] [CrossRef]
- Otremba, Z. The impact on the reflectance in VIS of a type of crude oil film floating on the water surface. Opt. Express 2000, 7, 129–134. [Google Scholar] [CrossRef]
- Xing, Q.; Li, L.; Lou, M.; Bing, L.; Zhao, R.; Li, Z. Observation of oil spills through Landsat thermal infrared imagery: A case of Deepwater Horizon. Aquat. Procedia 2015, 3, 151–156. [Google Scholar] [CrossRef]
- Kumar, A.; Suresh, G. Weathering of Oil Spill: Modeling and Analysis. Aquat. Procedia 2015, 4, 435–442. [Google Scholar] [CrossRef]
- Riehm, A.; Rokke, D.; McCornick, A. Water-in-oil microstructures formed by marine oil dispersants in a model crude oil. Langmuir 2016, 32, 3954–3962. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Hu, C.; Feng, L.; Swayze, G.; Holmes, J.; Graettinger, G.; Macdonald, I.; Garcia, O.; Leifer, I. Oil slick morphology derived from AVIRIS measurements of the Deepwater Horizon oil spill: Implications for spatial resolution requirements of remote sensors. Mar. Pollut. Bul. 2016, 103, 276–285. [Google Scholar] [CrossRef]
- Xiong, P.; Gu, X.; Yu, T.; Meng, Q.; Li, J.; Shi, J.; Cheng, Y.; Liu, W.; Liu, Q.; Zhao, L. Thermal infrared emissivity spectrum and its characteristics of crude oil slick covered seawater. Spectrosc. Spectr. Anal. 2014, 34, 2953–2960. [Google Scholar] [CrossRef]
- Niclòs, R.; Monzo, C.; Valor, E.; Bisquert, M. Thermal-infrared spectral and angular characterization of crude oil and seawater emissivity for oil slick identification. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5387–5395. [Google Scholar] [CrossRef]
- Lu, Y.; Zhan, W.; Hu, C. Detecting and quantifying oil slick thickness by thermal remote sensing: A ground-based experiment. Remote Sens. Environ. 2016, 181, 207–217. [Google Scholar] [CrossRef]
- Hu, C.; Li, X.; Pichel, W.; Muller-Karger, F. Detection of natural oil slicks in the NW Gulf of Mexico using MODIS imagery. Geophys. Res. Lett. 2009, 36, L01604–L01608. [Google Scholar] [CrossRef]
- Pisano, A.; Bignami, F.; Santoleri, R. Oil spill detection in glint-contaminated near-infrared MODIS imagery. Remote Sens. 2015, 7, 1112–1134. [Google Scholar] [CrossRef]
- Sun, S.; Hu, C. Sun glint requirement for the remote detection of surface oil films. Geophys. Res. Lett. 2016, 43, 309–316. [Google Scholar] [CrossRef]
- Alivisatos, A. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996, 382, 609–611. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qiang, X.; Yang, Y.; Wang, B.; Yang, D.; Luo, L.; Pan, L.; Wang, S. An unenumerative DNA computing model for vertex coloring problem. IEEE Trans. Nanobiosci. 2011, 10, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Jonoska, N.; Mahalingam, K. Languages of DNA based code words. In Proceedings of the International Workshop on DNA-Based Computers, Madison, WI, USA, 7–10 June 2004; pp. 61–73. [Google Scholar] [CrossRef]
- Xiao, K.; Xu, H. Detection of oil spill in Mexico Gulf based on MODIS data. In Proceedings of the International Conference on Multimedia Technology, Ningbo, China, 1–4 October 2010. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, W.; Fang, Z. Multiple kernel-based SVM classification of hyperspectral images by combining spectral, spatial, and semantic information. Remote Sens. 2017, 12, 120. [Google Scholar] [CrossRef]
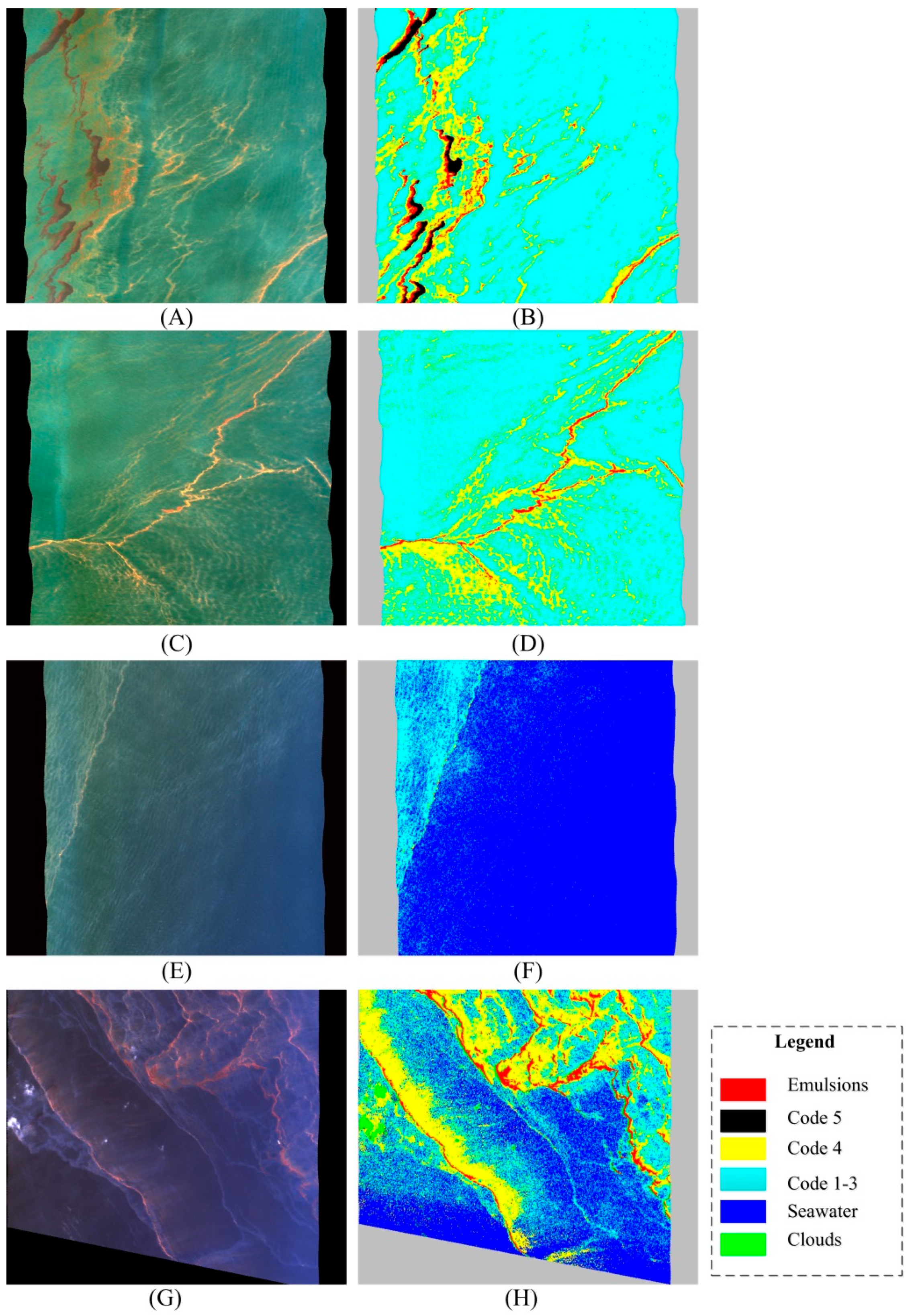

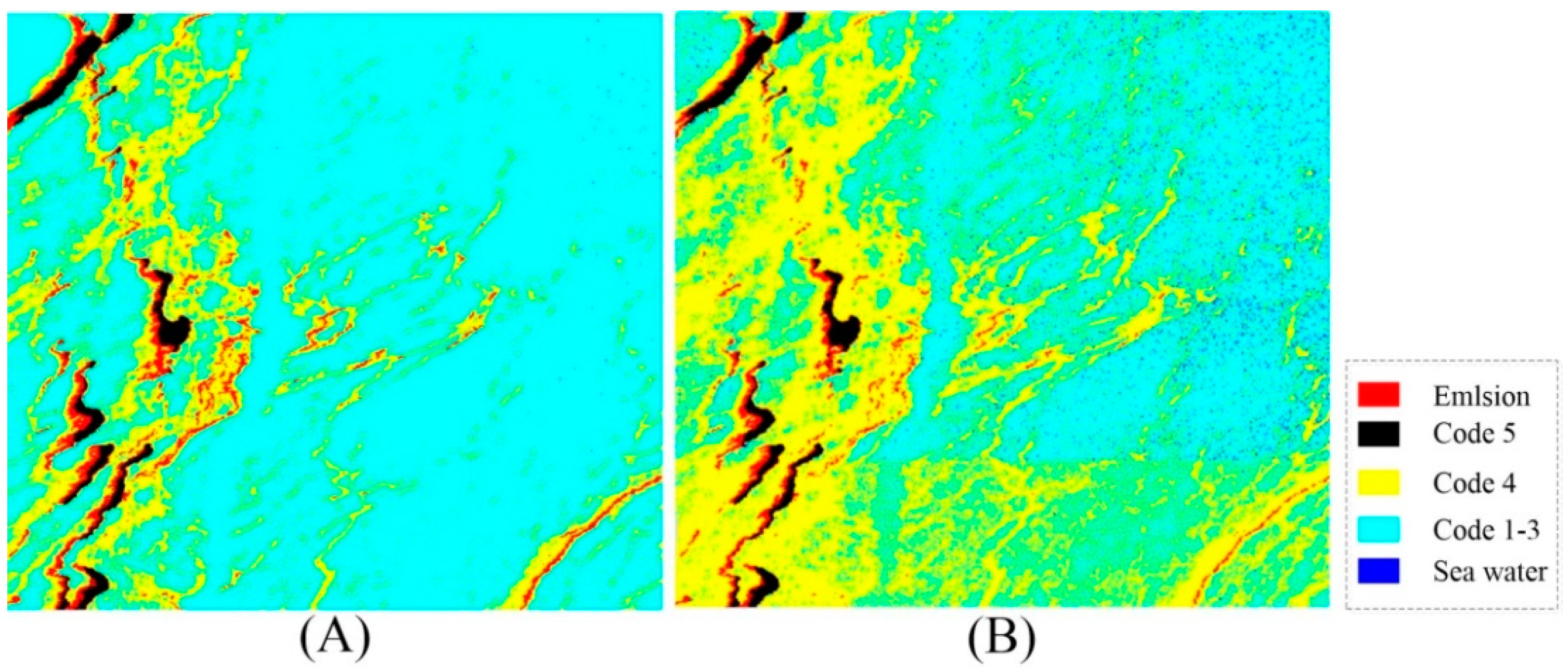
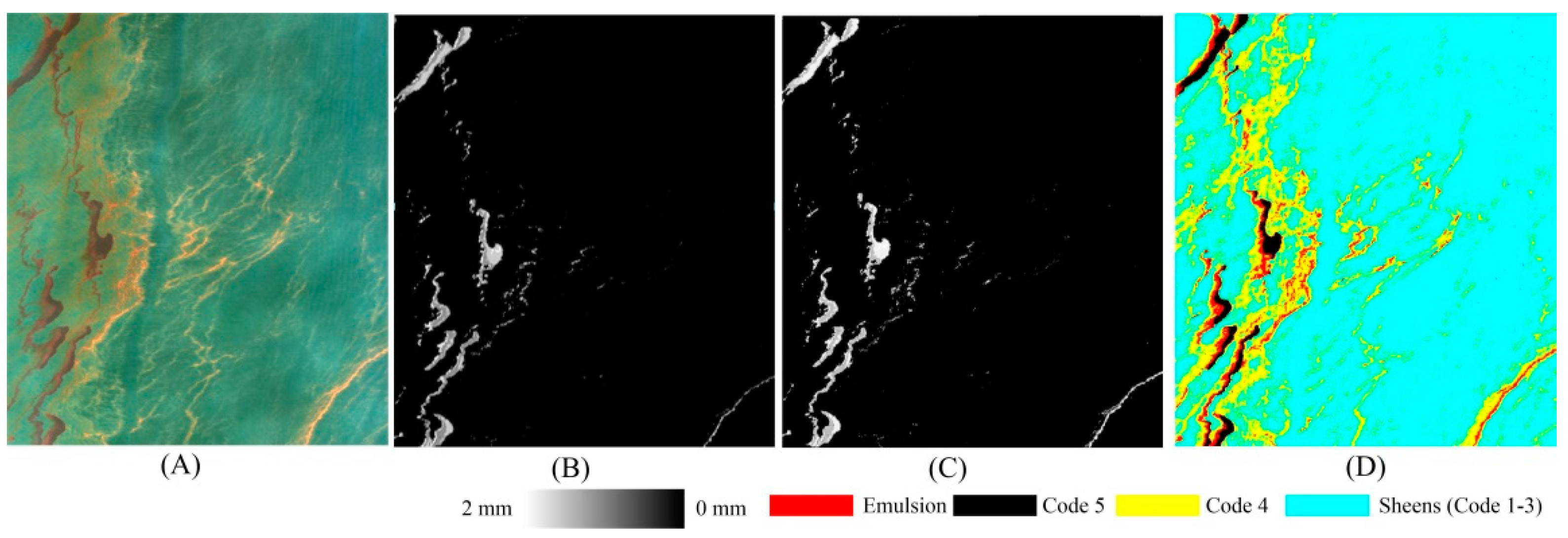
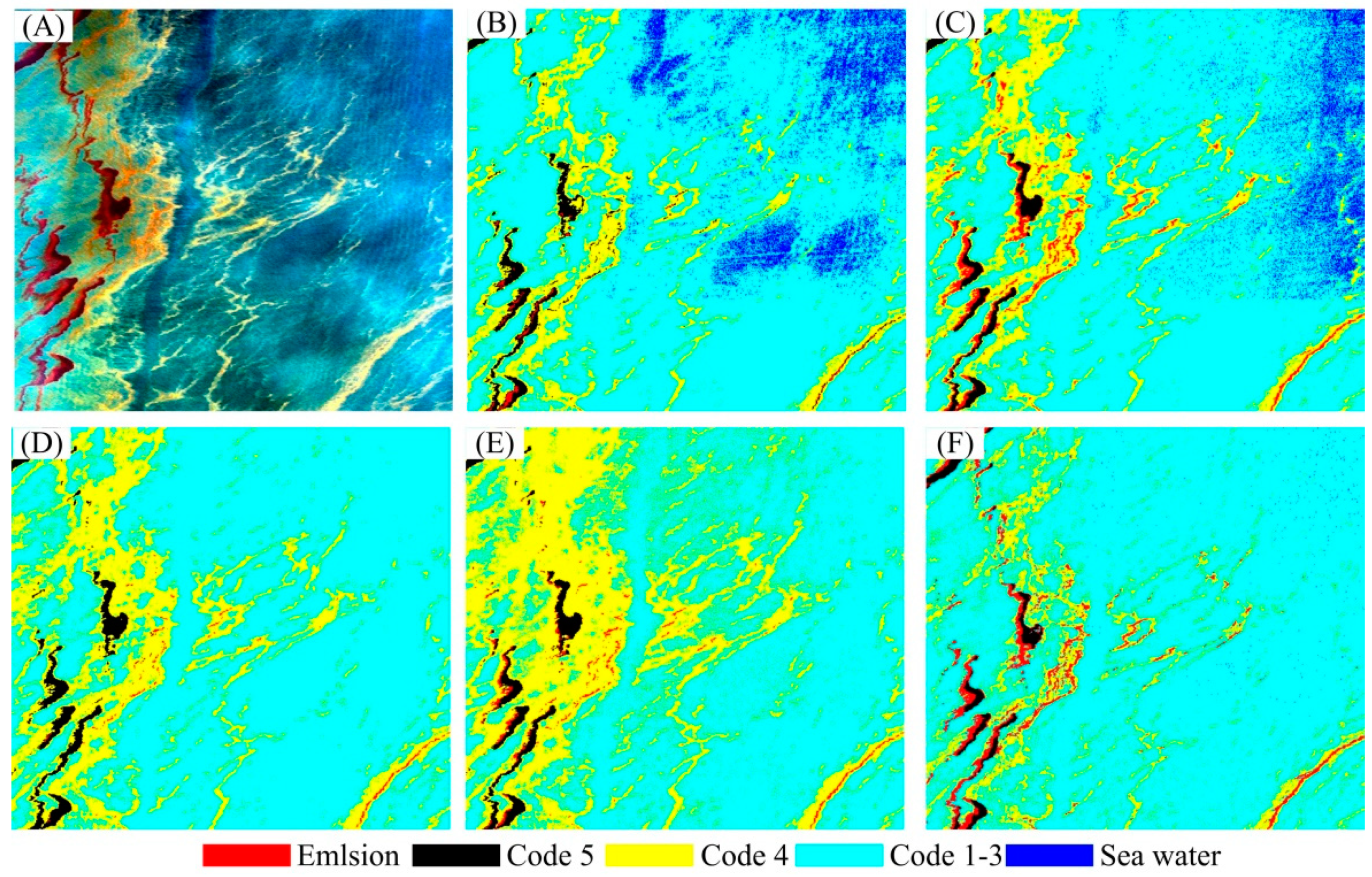
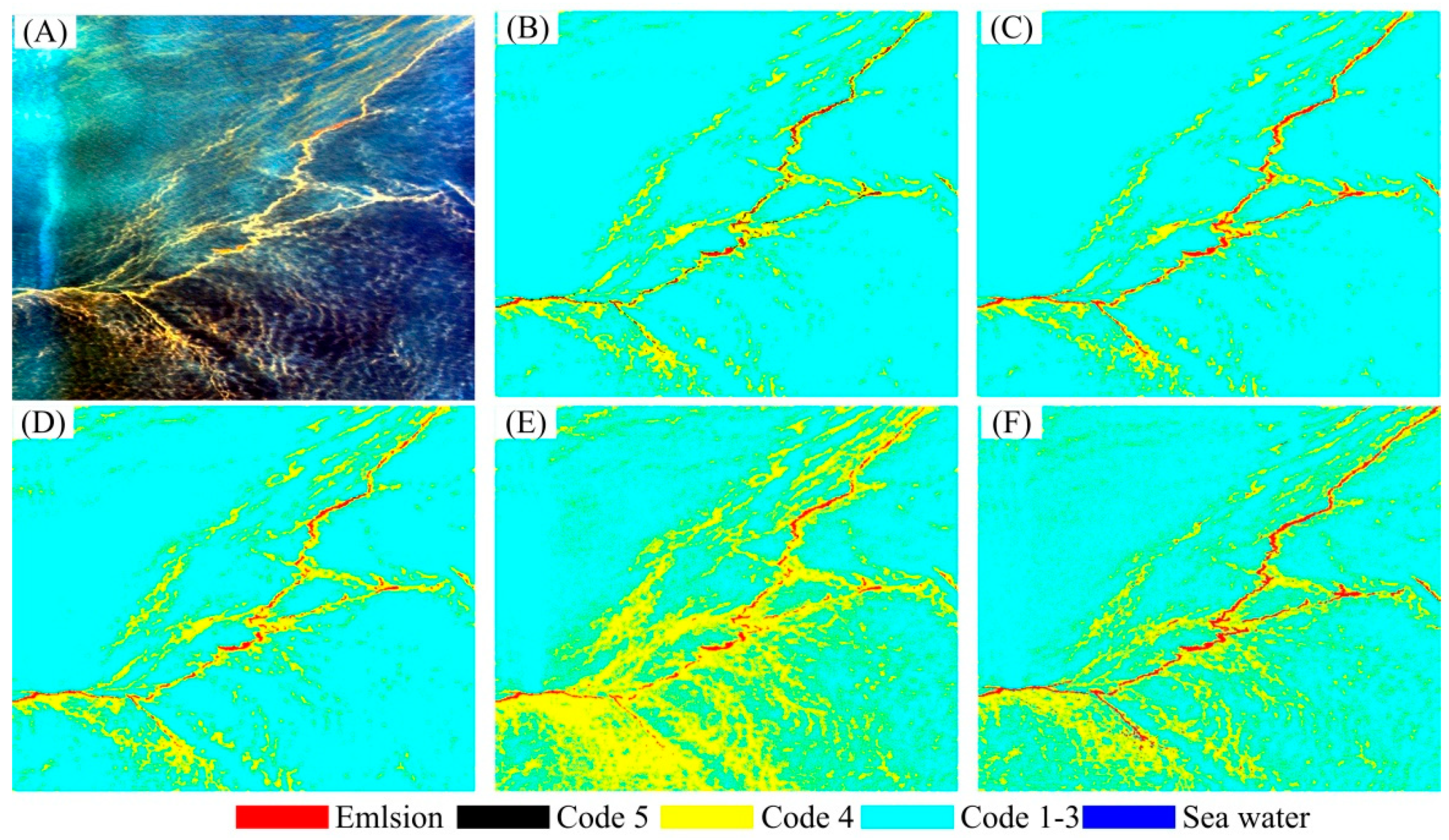
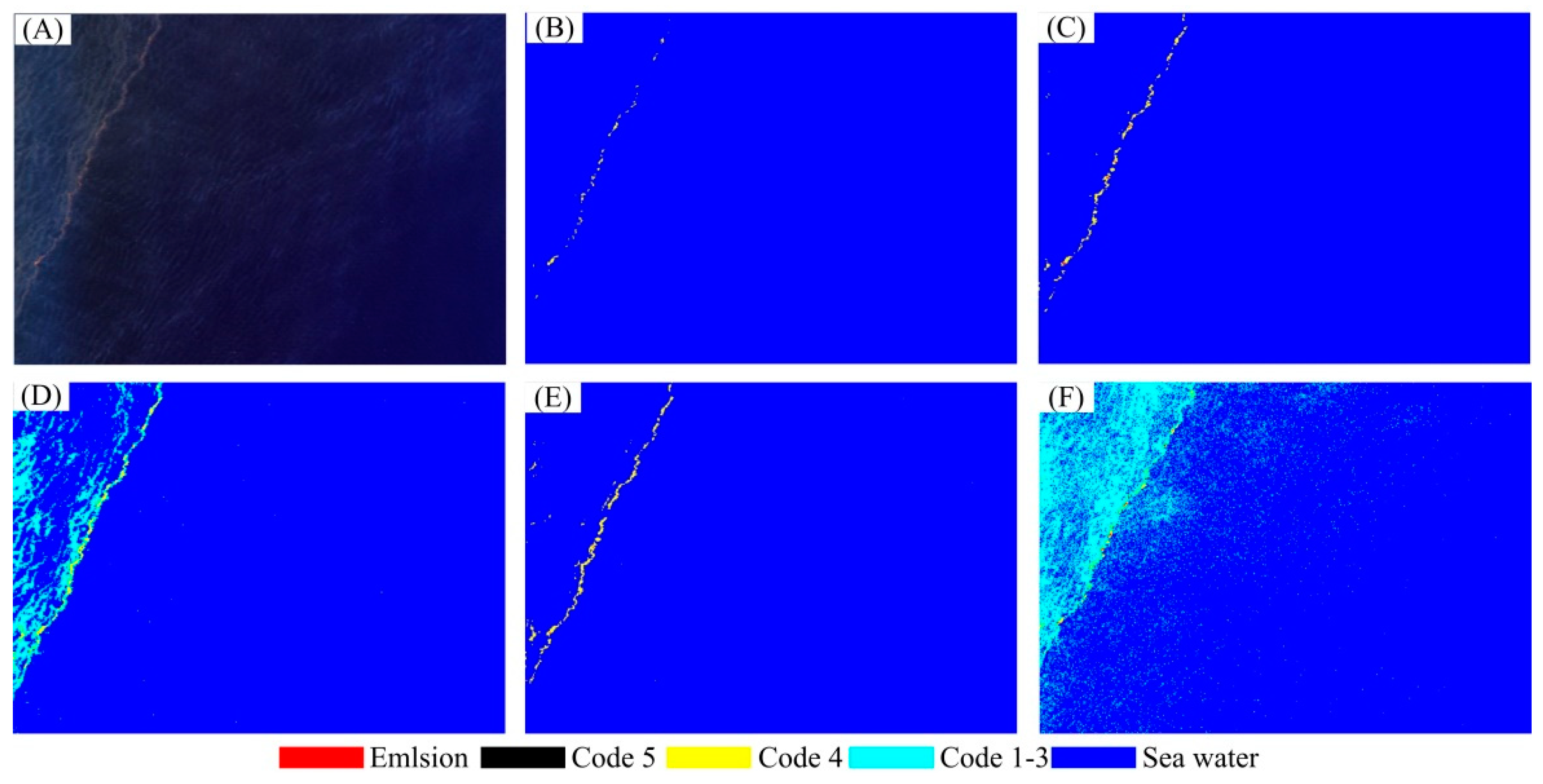
| Emulsion | Code 5 | Code 4 | Code 1–3 | Seawater | |
|---|---|---|---|---|---|
| AVIRIS image 1 | 46 | 63 | 26 | 66 | 27 |
| AVIRIS image 2 | 70 | 14 | 51 | 38 | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.; Tan, B.; Zhang, H.; Deng, R. Monitoring Marine Oil Spills in Hyperspectral and Multispectral Remote Sensing Data by the Spectral Gene Extraction (SGE) Method. Sustainability 2022, 14, 13696. https://doi.org/10.3390/su142013696
Zhao D, Tan B, Zhang H, Deng R. Monitoring Marine Oil Spills in Hyperspectral and Multispectral Remote Sensing Data by the Spectral Gene Extraction (SGE) Method. Sustainability. 2022; 14(20):13696. https://doi.org/10.3390/su142013696
Chicago/Turabian StyleZhao, Dong, Bin Tan, Haitao Zhang, and Rui Deng. 2022. "Monitoring Marine Oil Spills in Hyperspectral and Multispectral Remote Sensing Data by the Spectral Gene Extraction (SGE) Method" Sustainability 14, no. 20: 13696. https://doi.org/10.3390/su142013696
APA StyleZhao, D., Tan, B., Zhang, H., & Deng, R. (2022). Monitoring Marine Oil Spills in Hyperspectral and Multispectral Remote Sensing Data by the Spectral Gene Extraction (SGE) Method. Sustainability, 14(20), 13696. https://doi.org/10.3390/su142013696





