Metal-Tolerant Bacteria of Wastewater Treatment Plant in a Large City
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacteria Cultivation
2.3. PCR Analysis of Target Genes
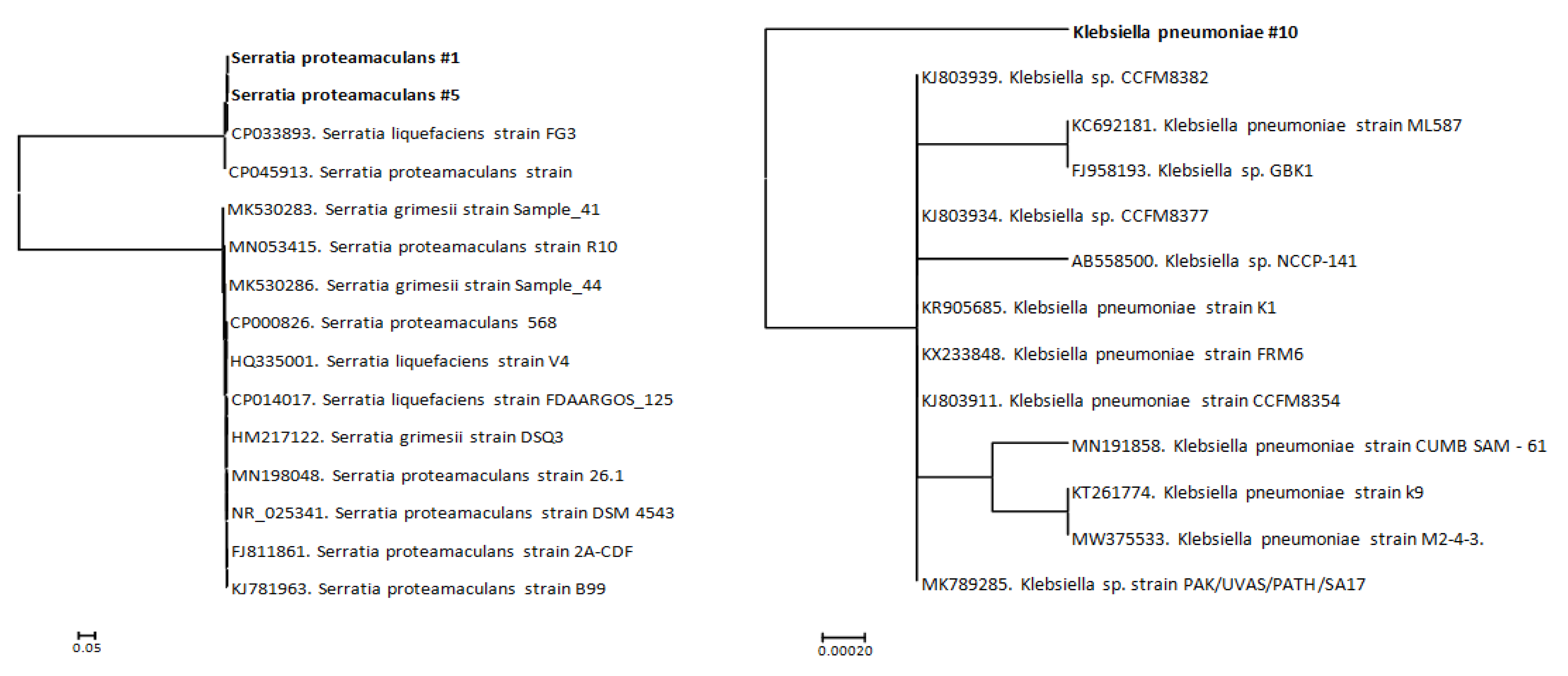
2.4. Anaylsis of the Evolutionary Distances of the Studied Bacteria
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thebo, A.L.; Drechsel, P.; Lambin, E.F.; Nelson, K.L. A global, spatially-explicit assessment of irrigated croplands influenced by urban wastewater flows. Environ. Res. Lett. 2017, 12, 074008. [Google Scholar] [CrossRef]
- Mateo-Sagasta, J.; Raschid-Sally, L.; Thebo, A. Global Wastewater and Sludge Production, Treatment and Use. In Wastewater; Drechsel, P., Qadir, M., Wichelns, D., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 15–38. [Google Scholar] [CrossRef]
- Zamparas, M. The Role of Resource Recovery Technologies in Reducing the Demand of Fossil Fuels and Conventional Fossil-Based Mineral Fertilizers. In Low Carbon Energy Technologies in Sustainable Energy Systems; Kyriakopoulos, G.L., Ed.; Academic Press: New York, NY, USA, 2021; pp. 3–24. [Google Scholar] [CrossRef]
- UN DESA, United Nations, Department of Economic and Social Affairs, Population Division. World Urbanization Prospects: The 2018 Revision, Online Edition UN DESA, New York. 2018. Available online: https://www.un.org/development/desa/publications/2018-revision-of-world-urbanization-prospects.html (accessed on 2 July 2022).
- Crittenden, C.J.; Trussell, R.R.; Hand, D.W.; Howe, K.J.; Tchobanoglous, G. MWH’s Water Treatment: Principles and Design, 3rd ed.; Wiley: Hoboken, NJ, USA, 2012; 1901p. [Google Scholar]
- Sarkar, B.; Mandal, S.; Tsang, Y.F.; Vithanage, M.; Biswas, J.K.; Yi, H.; Dou, X.; Ok, Y.S. Sustainable Sludge Management by Removing Emerging Contaminants from Urban Wastewater Using Carbon Nanotubes. In Industrial and Municipal Sludge. Emerging Concerns and Scope for Resource Recovery; Prasad, M.N.V., de Campos Favas, P.J., Vithanage, M., Mohan, S.V., Eds.; Butterworth-Heinemann: Oxford, UK, 2019; pp. 553–571. [Google Scholar] [CrossRef]
- Fijalkowski, K.; Rorat, A.; Grobelak, A.; Kacprzak, M.J. The presence of contaminations in sewage sludge—the current situation. J. Environ. Manag. 2017, 203, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zheng, X.; Zhang, D.; Iqbal, W.; Liu, C.; Yang, B.; Zhao, X.; Lu, X.; Mao, Y. Microbial characterization of heavy metal resistant bacterial strains isolated from an electroplating wastewater treatment plant. Ecotoxicol. Environ. Saf. 2019, 181, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Ahammad, S.Z.; Graham, D.W.; Dolfing, J. Wastewater Treatment: Biological. In Managing Water Resources and Hydrological Systems; Fath, B.D., Jørgensen, S.E., Megan Cole, M., Eds.; CRC Press: Boca Raton, FL, USA, 2020; pp. 2645–2656. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shin, H.; Han, D.; Hur, H.-G.; Unno, T. Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant. J. Microbiol. 2018, 56, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Vasquez-Medrano, R.; Prato-Garcia, D.; Vedrenne, M. Ferrioxalate-mediated processes. In Advanced Oxidation Processes for Waste Water Treatment; Ameta, S.C., Ameta, R., Eds.; Academic Press: New York, NY, USA, 2018; pp. 89–113. [Google Scholar] [CrossRef]
- Gottschalk, G. Bacterial Metabolism, 2nd ed.; Springer: New York, NY, USA, 1986; 281p. [Google Scholar]
- Cyprowski, M.; Stobnicka-Kupiec, A.; Ławniczek-Wałczyk, A.; Bakal-Kijek, A.; Gołofit-Szymczak, M.; Górny, R.L. Anaerobic bacteria in wastewater treatment plant. Int. Arch. Occup. Environ. Health 2018, 91, 571–579. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Trevors, J.T.; Oddie, K.M.; Belliveau, B.H. Metal resistance in bacteria. FEMS Microbiol. Rev. 1985, 1, 39–54. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- HS 2.1.5.1315-03; Maximum Permissible Concentrations of Chemical Substances in Water Objects of Household and Cultural Water Use. Russian Register of Potentially Hazardous Chemical and Biological Substances of the Ministry of Health of the Russian Federation: Moscow, Russia, 2003; 154p.
- Bahiru, D.B. Determination of heavy metals in wastewater and their toxicological implications around Eastern Industrial Zone, Central Ethiopia. JECE 2020, 12, 72–79. [Google Scholar] [CrossRef]
- Zhou, Y.; Lei, J.; Zhang, Y.; Zhu, J.; Lu, Y.; Wu, X.; Fang, H. Determining discharge characteristics and limits of heavy metals and metalloids for wastewater treatment plants (WWTPs) in China based on statistical methods. Water 2018, 10, 1248. [Google Scholar] [CrossRef]
- Lima de Silva, A.A.; de Carvalho, M.A.; de Souza, S.A.; Dias, P.M.; da Silva Filho, R.G.; de Meirelles Saramago, C.S.; de Melo Bento, C.A.; Hofer, E. Heavy metal tolerance (Cr, Ag and Hg) in bacteria isolated from sewage. Braz. J. Microbiol. 2012, 43, 1620–1631. [Google Scholar] [CrossRef]
- Mandal, M.; Das, S.N.; Mandal, S. Principal component analysis exploring the association between antibiotic resistance and heavy metal tolerance of plasmid-bearing sewage wastewater bacteria of clinical relevance. Access. Microbiol. 2020, 2, acmi000095. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Alspaugh, J.A.; Reller, M.E.; Lagunoff, M.; Reller, L.B.; Drew, W.L.; Ryan, K.J.; Sterling, C.R.; Ahmad, N.; Weissman, S.; Pottinger, P. Ryan & Sherris Medical Microbiology, 8th ed.; McGraw-Hill Education: New York, NY, USA, 2022; 1008p. [Google Scholar]
- Ajmal, A.W.; Saroosh, S.; Mulk, S.; Hassan, M.N.; Yasmin, H.; Jabeen, Z.; Nosheen, A.; Shah, S.M.U.; Naz, R.; Hasnain, Z.; et al. Bacteria Isolated fromWastewater Irrigated Agricultural Soils Adapt to Heavy Metal Toxicity While Maintaining Their Plant Growth Promoting Traits. Sustainability 2021, 13, 7792. [Google Scholar] [CrossRef]
- Sinha, S.; Mukherjee, S.K. Pseudomonas aeruginosa KUCD1, a possible candidate for cadmium bioremediation. Braz. J. Microbiol. 2009, 40, 655–662. [Google Scholar] [CrossRef]
- Malik, A.; Jaiswal, R. Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J. Microbiol. Biotechnol. 2000, 16, 177–182. [Google Scholar] [CrossRef]
- Baïda, N.; Yazourh, A.; Singer, E.; Izard, D. Pseudomonas brenneri sp. nov., a new species isolated from natural mineral waters. Res. Microbiol. 2001, 152, 493–502. [Google Scholar] [CrossRef]
- Banerjee, S.; Kamila, B.; Barman, S.; Joshi, S.R.; Mandal, T.; Halder, G. Interlining Cr(VI) remediation mechanism by a novel bacterium Pseudomonas brenneri isolated from coalmine wastewater. J. Environ. Manag. 2019, 233, 271–282. [Google Scholar] [CrossRef]
- Pereira, J.N.; Morgan, M.E. Nutrition and physiology of P. fragi. J. Bacteriol. 1957, 74, 710–713. [Google Scholar] [CrossRef]
- Champomier-Vergès, M.C.; Stintzi, A.; Meyer, J.M. Acquisition of iron by the non-siderophore-producing Pseudomonas fragi. Microbiology 1996, 142, 1191–1199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hebraud, M.; Dubois, E.; Potier, P.; Labadie, J. Effect of growth temperatures on the protein levels in a psychrotrophic bacterium, Pseudomonas fragi. J. Bacteriol. 1994, 176, 4017–4024. [Google Scholar] [CrossRef]
- Anzai, K.H.; Park, J.Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.A. Eubacteria show their true colors: Genetics of carotenoid pigment biosynthesis from microbes to plants. J. Bacteriol. 1994, 176, 4795–4802. [Google Scholar] [CrossRef] [PubMed]
- Hassen, A.; Saidi, N.; Cherif, M.; Boudabous, A. Effects of heavy metals on Pseudomonas aeruginosa and Bacillus thuringiensis. Bioresourse Technol. 1998, 65, 73–82. [Google Scholar] [CrossRef]
- Fugimore, H.; Kiyono, M.; Nobuhara, K.; Pan-Hou, H. Possible involvement of red pigments in defense against mercury in Pseudomonas K-62. FEMS Microbiol. Lett. 1996, 135, 317–321. [Google Scholar] [CrossRef]
- Verhille, S.; Baïda, N.; Dabboussi, F.; Hamze, M.; Izard, D.; Leclerc, H. Pseudomonas gessardii sp. nov. and Pseudomonas migulae sp. nov., two new species isolated from natural mineral waters. Int. J. Syst. Bacteriol. 1999, 49, 1559–1572. [Google Scholar] [CrossRef]
- Huang, H.; Wu, K.; Khan, A.; Jiang, Y.; Ling, Z.; Liu, P.; Chen, Y.; Tao, X.; Li, X. A novel Pseudomonas gessardii strain LZ-E simultaneously degrades naphthalene and reduces hexavalent chromium. Bioresour. Technol. 2016, 207, 370–378. [Google Scholar] [CrossRef]
- Silverio, M.P.; Kraychete, G.B.; Rosado, A.S.; Bonelli, R.R. Pseudomonas fluorescens Complex and Its Intrinsic, Adaptive, and Acquired Antimicrobial Resistance Mechanisms in Pristine and Human-Impacted Sites. Antibiotics 2022, 11, 985. [Google Scholar] [CrossRef]
- Postgate, J. Nitrogen Fixation, 3rd ed.; Cambridge University Press: Cambridge, UK, 1998; 112p. [Google Scholar]
- Riggs, P.J.; Chelius, M.K.; Iniguez, A.L.; Kaeppler, S.M.; Triplett, E.W. Enhanced maize productivity by inoculation with diazotrophic bacteria. Aust. J. Plant Physiol. 2001, 29, 829–836. [Google Scholar] [CrossRef]
- Mohan, A.K.; Martis, B.S.; Chiplunkar, S.; Kamath, S.; Goveas, L.C.; Rao, C.V. Heavy metal tolerance of Klebsiella pneumoniae Kpn555 isolated from coffee pulp waste. BJRST 2019, 9, 101–106. [Google Scholar]
- Orji, O.U.; Awoke, J.N.; Aja, P.M.; Aloke, C.; Obasi, O.D.; Alum, E.U.; Udu-Ibiam, O.E.; Oka, G.O. Halotolerant and metalotolerant bacteria strains with heavy metals biorestoration possibilities isolated from Uburu Salt Lake, Southeastern, Nigeria. Heliyon 2021, 7, e07512. [Google Scholar] [CrossRef]
- Zagui, G.S.; Moreira, N.G.; Santos, D.V.; Costa Darini, A.L.; Domingo, J.L.; Segura-Muñoz, S.I.; Andrade, L.N. High occurrence of heavy metal tolerance genes in bacteria isolated from wastewater: A new concern? Environ. Res. 2021, 196, 110352. [Google Scholar] [CrossRef]
- Khanna, A.; Khanna, M.; Aggarwal, A. Serratia marcescens—a rare opportunistic nosocomial pathogen and measures to limit its spread in hospitalized patients. J. Clin. Diagn. Res. 2013, 7, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Abriouel, H.; Benomar, N.; Kabisch, J.; Chieffi, D.; Cho, G.-S.; Franz, C. Opportunistic Food-Borne Pathogens. In Food Safety and Preservation; Grumezescu, A.M., Holban, A.M., Eds.; Academic Press: New York, NY, USA, 2018; pp. 269–306. [Google Scholar] [CrossRef]
- Greenberg, L. Serratia marcescens in human affairs. Drug Intell. 1978, 12, 674–679. [Google Scholar] [CrossRef]
- Mahlen, S.D. Serratia infections: From military experiments to current practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Kwak, J.; Lee, K.; Shin, D.H.; Maeng, J.S.; Park, D.S.; Oh, H.W.; Son, K.H.; Bae, K.S.; Park, H.Y. Biochemical and genetic characterization of arazyme, an extracellular metalloprotease produced from Serratia proteamaculans HY-3. J. Microbiol. Biotechnol. 2007, 17, 761–768. [Google Scholar]

| Heavy Metal | Concentration, mg/L | |||
|---|---|---|---|---|
| Maximum Permissible Concentrations for Waters of Household and Cultural Water Use [20] | Secondary Sedimentation Tank (Water) | Secondary Sedimentation Tank (Suspension) | Digester (Suspension) | |
| Cd | 0.001 | 0.0053 | 0.0037 | 0.0186 |
| Cu | 1.0 | 0.0056 | 0.0037 | 0.0248 |
| Ni | 0.02 | 0.0089 | 0.0041 | 0.1204 |
| Pb | 0.01 | <0.0001 | 0.0038 | 0.0050 |
| Zn | 1.0 | 0.0002 | 0.00001 | 0.0002 |
| N | Species (According to Sequencing Results) | Source | Element | Maximal Tolerance Concentration |
|---|---|---|---|---|
| 1 | Serratia proteamaculans | Digester | Ni | 5 mM |
| 2 | Pseudomonas gessardii | Secondary sedimentation tank (sludge) | Ni | 3 mM |
| 3 | Pseudomonas fragi | Secondary sedimentation tank (sludge) | Cd | 3 mM |
| 4 | Pseudomonas fragi | Secondary sedimentation tank (water) | Cd | 3 mM |
| 5 | Serratia proteamaculans | Secondary sedimentation tank (sludge) | Pb | 5 mM |
| 6 | Pseudomonas fragi | Secondary sedimentation tank (water) | Pb | 3 mM |
| 7 | Pseudomonas brenneri | Secondary sedimentation tank (water) | Pb | 3 mM |
| 8 | Pseudomonas gessardii | Secondary sedimentation tank (sludge) | Zn | 5 mM |
| 9 | Pseudomonas gessardii | Secondary sedimentation tank (water) | Zn | 5 mM |
| 10 | Klebsiella pneumonia | Secondary sedimentation tank (sludge) | Cu | 3 mM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perelomov, L.; Sizova, O.; Rahman, M.M.; Perelomova, I.; Minkina, T.; Sokolov, S.; Atroshchenko, Y. Metal-Tolerant Bacteria of Wastewater Treatment Plant in a Large City. Sustainability 2022, 14, 11335. https://doi.org/10.3390/su141811335
Perelomov L, Sizova O, Rahman MM, Perelomova I, Minkina T, Sokolov S, Atroshchenko Y. Metal-Tolerant Bacteria of Wastewater Treatment Plant in a Large City. Sustainability. 2022; 14(18):11335. https://doi.org/10.3390/su141811335
Chicago/Turabian StylePerelomov, Leonid, Olga Sizova, Mohammad Mahmudur Rahman, Irina Perelomova, Tatiana Minkina, Sergei Sokolov, and Yury Atroshchenko. 2022. "Metal-Tolerant Bacteria of Wastewater Treatment Plant in a Large City" Sustainability 14, no. 18: 11335. https://doi.org/10.3390/su141811335
APA StylePerelomov, L., Sizova, O., Rahman, M. M., Perelomova, I., Minkina, T., Sokolov, S., & Atroshchenko, Y. (2022). Metal-Tolerant Bacteria of Wastewater Treatment Plant in a Large City. Sustainability, 14(18), 11335. https://doi.org/10.3390/su141811335










