Changes in Temporal Dynamics and Factors Influencing the Environment of the Bacterial Community in Mangrove Rhizosphere Sediments in Hainan
Abstract
:1. Introduction
2. Materials and Methods
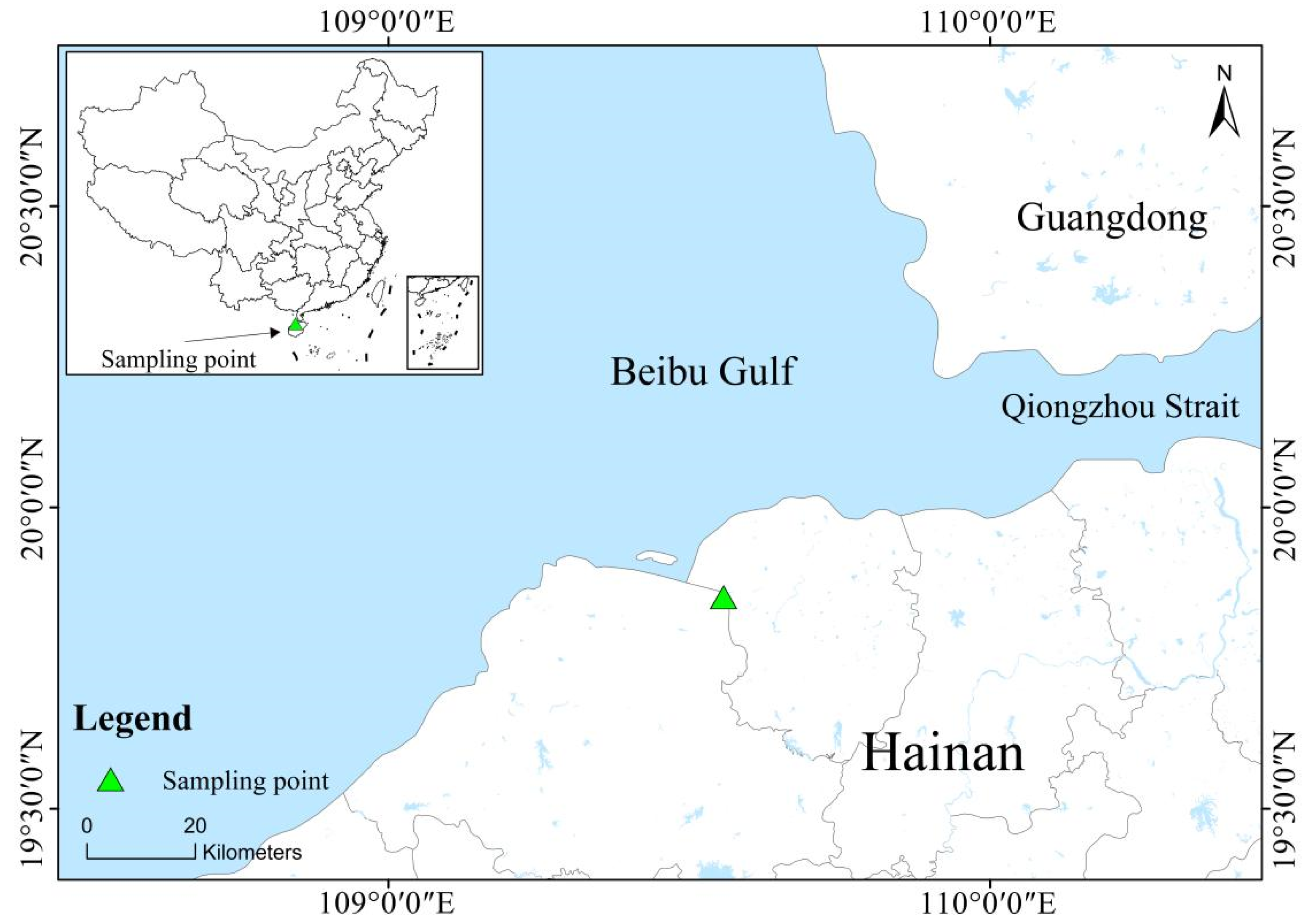
2.1. Overview of the Study Area
2.2. Sample Collection
2.3. Method of Analysis for Sediment Physicochemical Indexes
2.4. High-Throughput Sequencing Method for Sediment Bacteria 16SrRNA
2.5. Statistical Analysis
3. Results
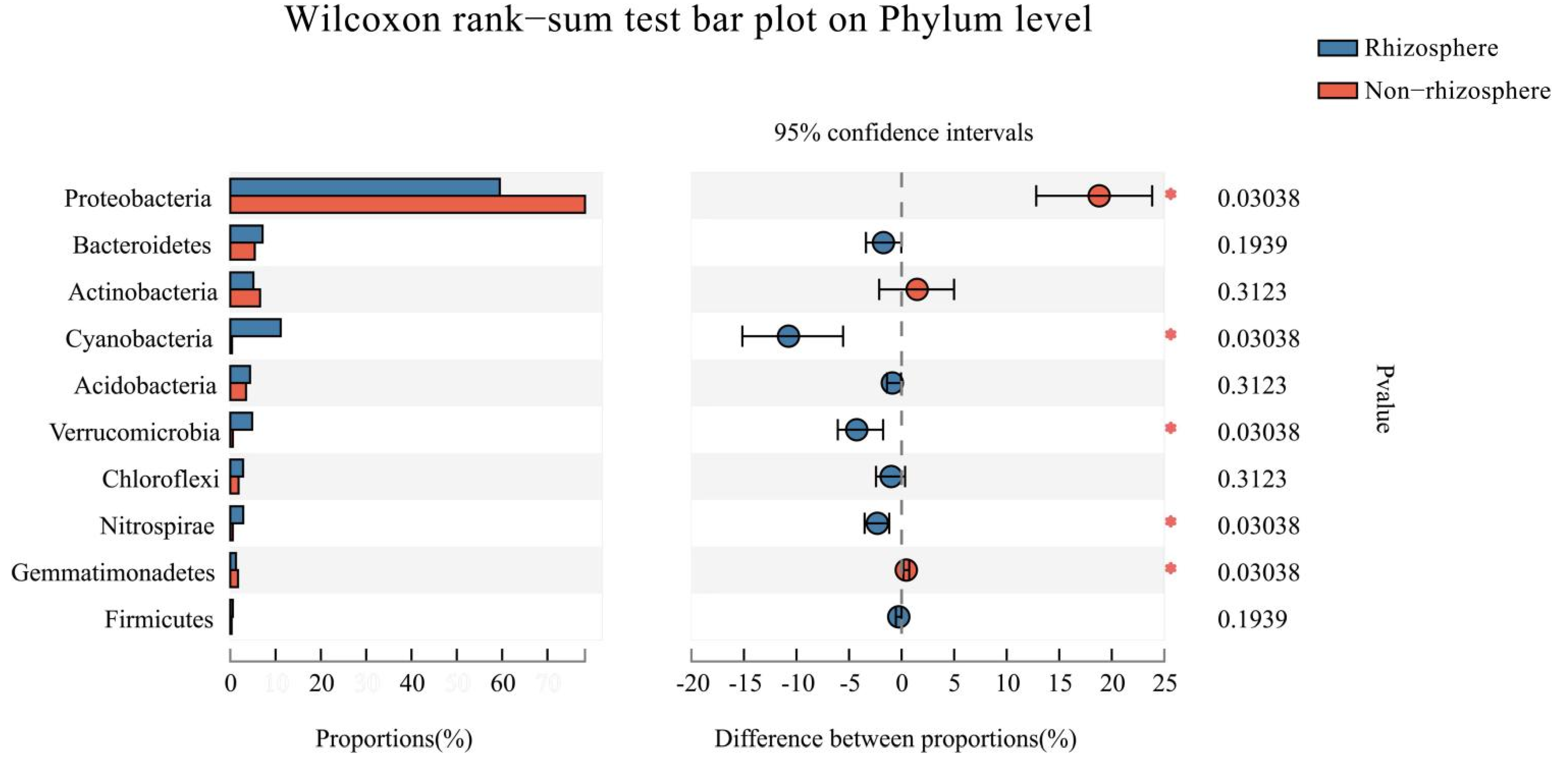
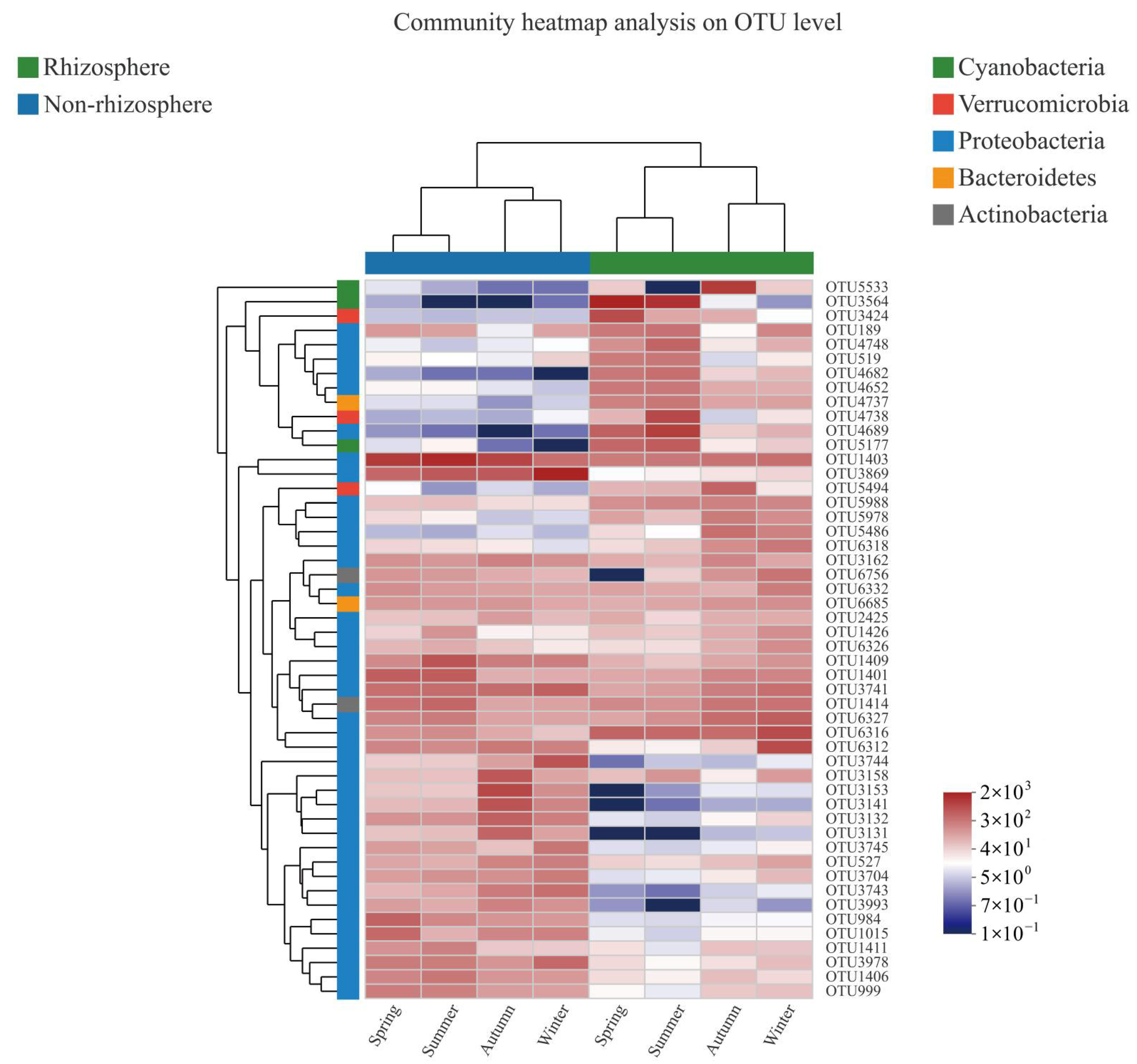
3.1. Bacterial α-Diversity and Its Taxonomic Composition
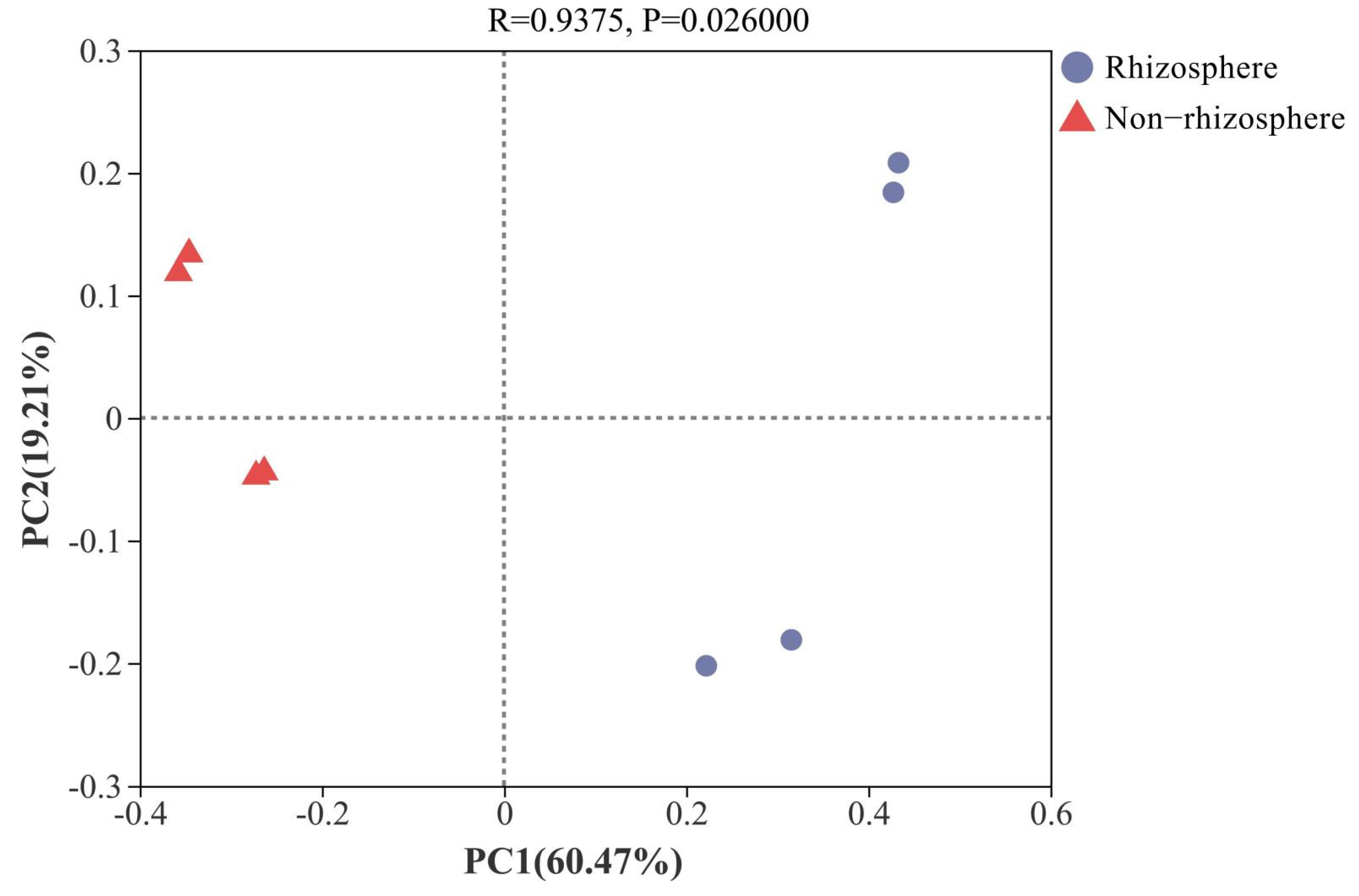
3.2. Diversity Characteristics and the Influential Factors of Bacterial Community β
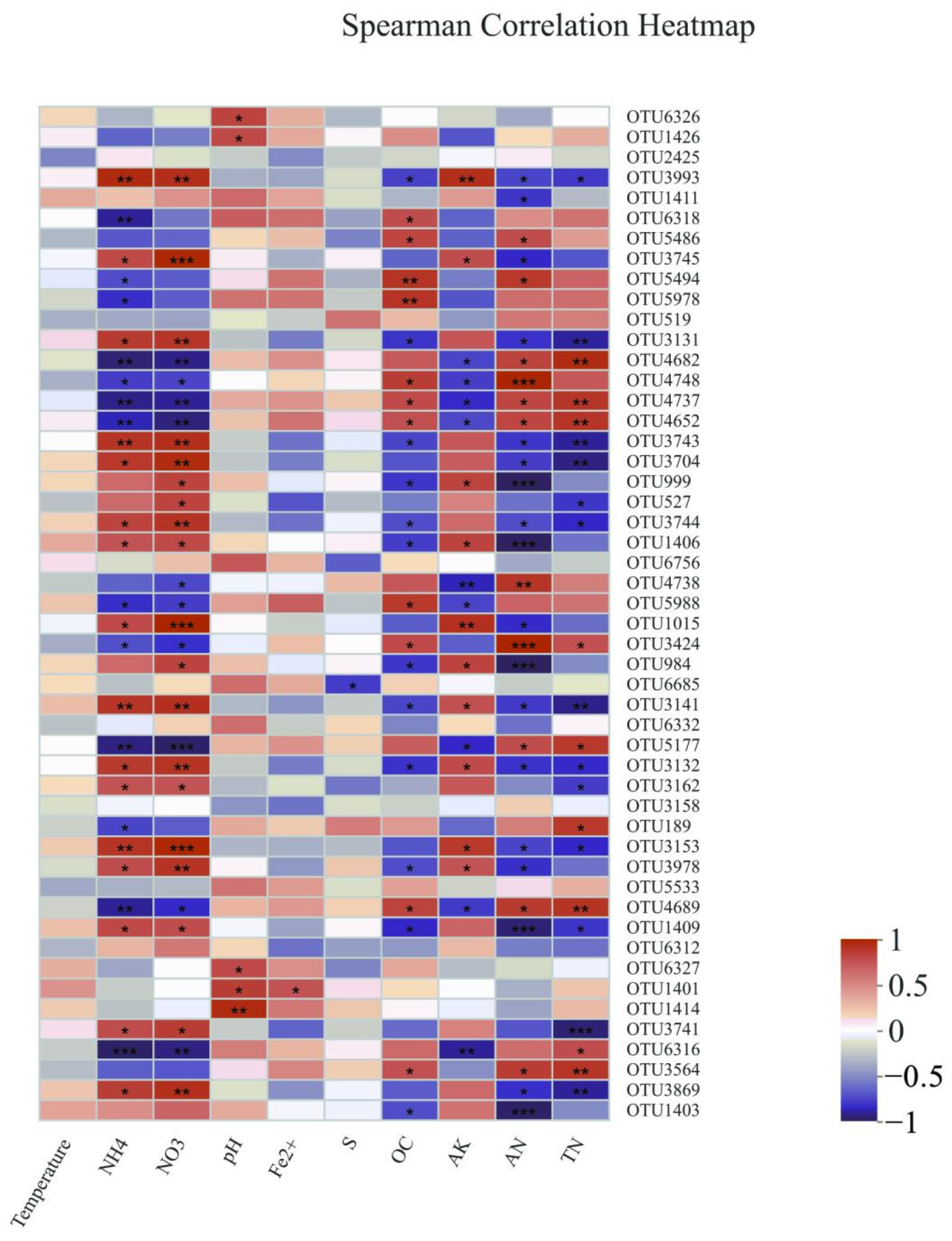
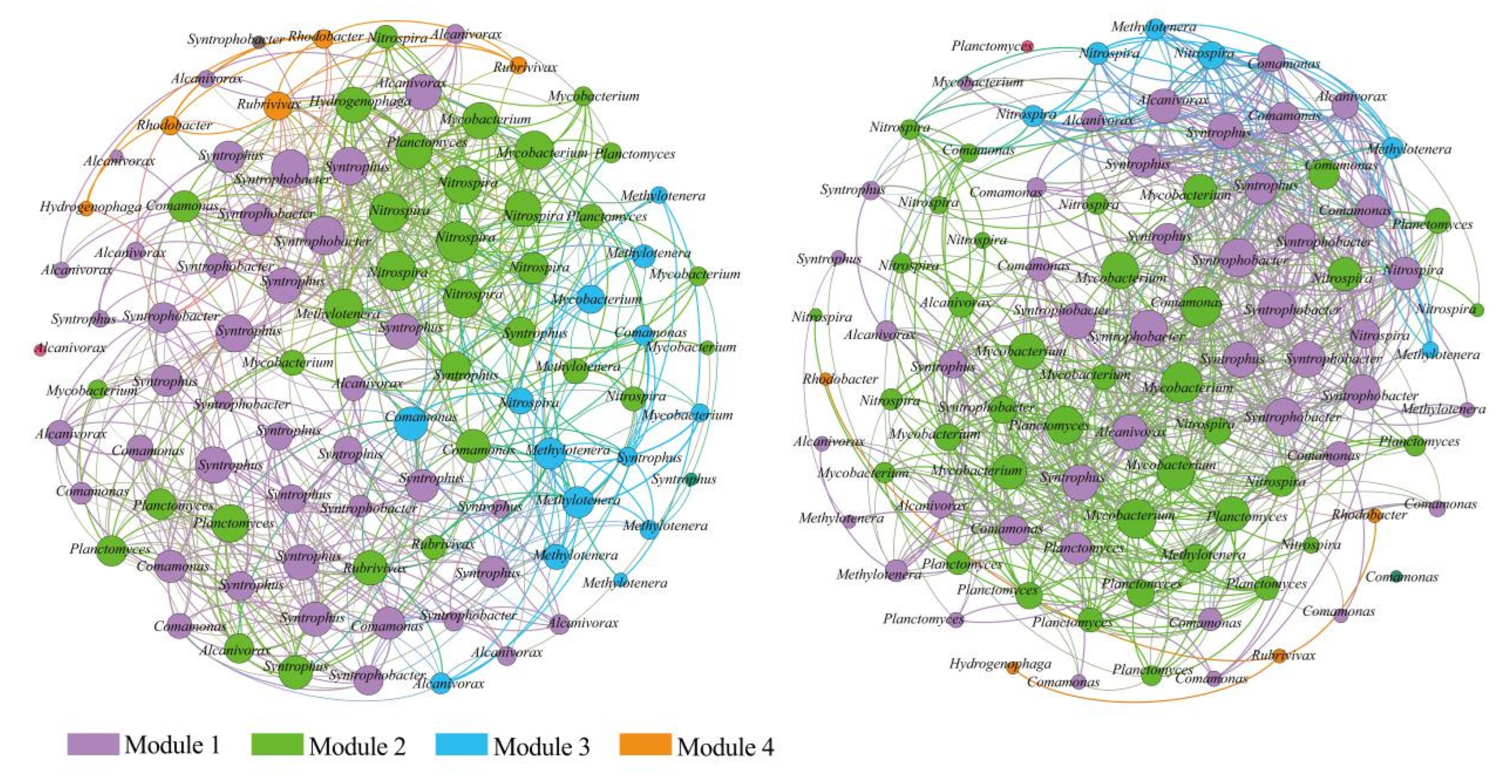
3.3. Interspecific Interactions of Bacterial Communities
4. Discussion
4.1. Characteristics of Bacterial α-Diversity in Rhizosphere and Non-Rhizosphere Soils
4.2. Important Factors Influencing Sediment Bacterial Community Structure
4.3. Co-occurrence Pattern Reveals Interspecific Relationships of Bacterial Communities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbier, E.B.; Koch, E.W.; Silliman, B.R.; Hacker, S.D.; Wolanski, E.; Primavera, J.; Granek, E.F.; Polasky, S.; Aswani, S.; Cramer, L.A.; et al. Coastal Ecosystem-Based Management with Nonlinear Ecological Functions and Values. Science 2008, 319, 321–323. [Google Scholar] [CrossRef]
- Twilley, R.R.; Chen, R.H.; Hargis, T. Carbon sinks in mangroves and their implications to carbon budget of tropical coastal ecosystems. Water Air Soil Pollut. 1992, 64, 265–288. [Google Scholar] [CrossRef]
- Pijush, B.; Arnab, P.; Sohan, S.; Sudip, N.; Anish, B.; Debojyoti, R.; Rudradip, P.; Abhrajyoti, G.; Dhrubajyoti, C.; Maitree, B. Bacterial diversity assessment of pristine mangrove microbial community from Dhulibhashani, Sundarbans using 16S rRNA gene tag sequencing. Genome Data 2016, 7, 76–78. [Google Scholar]
- Gomes, N.C.M.; Flocco, C.G.; Costa, R.; Junca, H.; Vilchez, R.; Pieper, D.H.; Krögerrecklenfort, E.; Paranhos, R.; Mendonça-Hagler, L.C.S.; Smalla, K. Mangrove microniches determine the structural and functional diversity of enriched petroleum hydrocarbon-degrading consortia. FEMS Microbiol. Ecol. 2010, 74, 276–290. [Google Scholar] [CrossRef] [Green Version]
- Alongi, D.M. Bacterial productivity and microbial biomass in tropical mangrove sediments. Microb. Ecol. 1988, 15, 59–79. [Google Scholar] [CrossRef]
- Feller, I.C.; Friess, D.A.; Krauss, K.W.; Lewis, R.R. The state of the world’s mangroves in the 21st century under climate change. Hydrobiologia 2017, 803, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Trevathan-Tackett, S.M.; Sherman, C.D.; Huggett, M.J.; Campbell, A.; Laverock, B.; Hurtado-McCormick, V.; Seymour, J.; Firl, A.; Messer, L.F.; Ainsworth, T.; et al. A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 2019, 3, 1509–1520. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Yao, M. Research progress on assembly of plant rhizosphere microbial community. Acta Microbiol. Sin. 2021, 61, 231–248. [Google Scholar]
- Lin, X.; Hetharua, B.; Lin, L.; Xu, H.; Zheng, T.; He, Z.; Tian, Y. Mangrove Sediment Microbiome: Adaptive Microbial Assemblages and Their Routed Biogeochemical Processes in Yunxiao Mangrove National Nature Reserve, China. Microb. Ecol. 2018, 78, 57–69. [Google Scholar] [CrossRef]
- Cao, H.; Li, M.; Hong, Y.; Gu, J.D. Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst. Appl. Microbiol. 2011, 34, 513–523. [Google Scholar] [CrossRef]
- Zhou, H.W.; Wong, A.H.Y.; Yu, R.M.K.; Park, Y.D.; Wong, Y.S.; Tam, N.F.Y. Polycyclic Aromatic Hydrocarbon-Induced Structural Shift of Bacterial Communities in Mangrove Sediment. Microb. Ecol. 2009, 58, 153–160. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.O.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef]
- Bharathkumar, S.; Rameshkumar, N.; Paul, D.; Prabavathy, V.R.; Nair, S. Characterization of the predominant bacterial population of different mangrove rhizosphere soils using 16S rRNA gene-based single-strand conformation polymorphism (SSCP). World J. Microbiol. Biotechnol. 2008, 24, 387–394. [Google Scholar] [CrossRef]
- Dias, A.C.F.; Andreote, F.D.; Dini-Andreote, F.; Lacava, P.T.; Sá, A.L.B.; Melo, I.S.; Azevedo, J.L.; Araújo, W.L. Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World J. Microbiol. Biotechnol. 2009, 25, 1305–1311. [Google Scholar] [CrossRef]
- Chakraborty, A.; Bera, A.; Mukherjee, A.; Basak, P.; Khan, I.; Mondal, A.; Roy, A.; Bhattacharyya, A.; Sengupta, S.; Roy, D.; et al. Changing bacterial profile of Sundarbans, the world heritage mangrove: Impact of anthropogenic interventions. World J. Microbiol. Biotechnol. 2015, 31, 593–610. [Google Scholar] [CrossRef]
- Basak, P.; Pramanik, A.; Roy, R.; Chattopadhyay, D.; Bhattacharyya, M. Cataloguing the bacterial diversity of the Sundarbans mangrove, India in the light of metagenomics. Genome Data 2015, 4, 90–92. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Genet. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Jiao, S.; Liu, Z.; Lin, Y.; Yang, J.; Chen, W.; Wei, G. Bacterial communities in oil contaminated soils: Biogeography and co-occurrence patterns. Soil Biol. Biochem. 2016, 98, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Beman, J.M.; Steele, J.A.; Fuhrman, J.A. Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. ISME J. 2011, 5, 1077–1085. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [Green Version]
- Chu, J.Y.; Zhang, J.P.; Zhou, X.H.; Liu, B.; Li, Y.M. A comparisonof an ammox bacterial abundance and community structures in three different emerged plants-related sediments. Curr. Microbiol. 2015, 71, 421–427. [Google Scholar] [CrossRef]
- Liu, D.; Fang, S.; Tian, Y.; Sx, C. Nitrogen transformations in the rhizosphere of different tree types in a seasonally flooded soil. Plant Soil Environ. 2014, 60, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; Agriculture Publication: Beijing, China, 2000; pp. 355–356. [Google Scholar]
- Rodriguez, A.G. Simultaneous determination of iron, cobalt, nickel and copper by UV-visible spectrophotometry with multivariate calibration. Talanta 1998, 47, 463–470. [Google Scholar] [CrossRef]
- Niu, Z.-S.; Pan, H.; Guo, X.-P.; Lu, D.-P.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Liu, M.; Yang, Y. Sulphate-reducing bacteria (SRB) in the Yangtze Estuary sediments: Abundance, distribution and implications for the bioavailibility of metals. Sci. Total Environ. 2018, 634, 296–304. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.T.; Su, J.Q.; Zheng, T.L.; Yang, X.R. Impacts of vegetation, tidal process, and depth on the activities, abundances, and community compositions of denitrifiers in mangrove sediment. Appl. Microbiol. Biotechnol. 2014, 98, 9375–9387. [Google Scholar] [CrossRef]
- Ghiglione, J.-F.; Galand, P.E.; Pommier, T.; Pedrós-Alió, C.; Maas, E.W.; Bakker, K.; Bertilson, S.; Kirchman, D.L.; Lovejoy, C.; Yager, P.L.; et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. USA 2012, 109, 17633–17638. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, B.F.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.; Gonzalez, J.; Pedrós-Alió, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lücker, S.; DeLong, E.F. A Nitrospirametagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Affe, H.M.J.; Rigonato, J.; Nunes, J.M.C.; Menezes, M. Metagenomic Analysis of Cyanobacteria in an Oligotrophic Tropical Estuary, South Atlantic. Front. Microbiol. 2018, 9, 1393. [Google Scholar] [CrossRef]
- Chiang, E.; Schmidt, M.L.; Berry, M.A.; Biddanda, B.A.; Burtner, A.; Johengen, T.H.; Palladino, D.; Denef, V.J. Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS ONE 2018, 13, e0195112. [Google Scholar]
- Sardans, J.; Peñuelas, J. The Role of Plants in the Effects of Global Change on Nutrient Availability and Stoichiometry in the Plant-Soil System. Plant Physiol. 2012, 160, 1741–1761. [Google Scholar] [CrossRef] [Green Version]
- Doornbos, R.F.; Loon, L.C.V.; Bakker, P.A.H.M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Boone, D.R.; Bryant, M.P. Propionate-Degrading Bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from Methanogenic Ecosystems. Appl. Environ. Microbiol. 1980, 40, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Felföldi, T.; Székely, A.J.; Gorál, R.; Barkács, K.; Scheirich, G.; András, J.; Rácz, A.; Márialigeti, K. Polyphasic bacterial community analysis of an aerobic activated sludge removing phenols and thiocyanate from coke plant effluent. Bioresour. Technol. 2010, 101, 3406–3414. [Google Scholar] [CrossRef]
- Jackson, B.E.; Bhupathiraju, V.K.; Tanner, R.S.; Woese, C.R.; McInerney, M.J. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch. Microbiol. 1999, 171, 107–114. [Google Scholar] [CrossRef]
- Kasai, Y.; Kishira, H.; Sasaki, T.; Syutsubo, K.; Watanabe, K.; Harayama, S. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 2002, 4, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.C.; Maas, B.; Dapena, A.; van de Pas-Schoonen, K.; van de Vossenberg, J.; Kartal, B.; van Niftrik, L.; Schmidt, I.; Cirpus, I.; Kuenen, J.G.; et al. Biomarkers for In Situ Detection of Anaerobic Ammonium-Oxidizing (Anammox) Bacteria. Appl. Environ. Microbiol. 2005, 71, 1677–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Sar, P.; Sarkar, J.; Dutta, A.; Sarkar, P.; Gupta, A.; Mohapatra, B.; Pal, S.; Kazy, S.K. Petroleum hydrocarbon rich oil refinery sludge of North-East India harbours anaerobic, fermentative, sulfate-reducing, syntrophic and methanogenic microbial populations. BMC Microbiol. 2018, 18, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Fuente Cantó, C.; Simonin, M.; King, E.; Moulin, L.; Bennett, M.J.; Castrillo, G.; Laplaze, L. An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 2020, 103, 951–964. [Google Scholar] [CrossRef] [Green Version]
- Brune, A. Life at the oxic–anoxic interface: Microbial activities and adaptations. FEMS Microbiol. Rev. 2000, 24, 691–710. [Google Scholar] [CrossRef]


| Temperature (°C) | NH4+ (μg/g) | NO3− (μg/g) | pH | Fe2+ (mg/g) | S (mg/kg) | OC (g/kg) | AK (mg/kg) | AN (mg/kg) | TN (mg/kg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Rhizosphere | ||||||||||
| Spring | 17.5000 ± 1.3200 | 2.0000 ± 0.0400 | 2.5100 ± 0.2100 | 7.8100 ± 0.2600 | 0.8300 ± 0.0800 | 0.1500 ± 0.0300 | 0.8200 ± 0.3500 | 85.3300 ± 2.3400 | 158.6500 ± 5.3300 | 1456.2500 ± 36.6500 |
| Summer | 32.0000 ± 2.0600 | 1.8900 ± 0.3600 | 2.7700 ± 0.4500 | 7.6200 ± 0.1800 | 1.0700 ± 0.1100 | 0.0700 ± 0.0100 | 0.9900 ± 0.0400 | 66.2500 ± 4.2600 | 179.62 ± 6.3600 | 1362.2600 ± 40.0500 |
| Autumn | 25.0000 ± 1.0500 | 3.300 ± 0.4300 | 8.4900 ± 0.7600 | 7.9500 ± 0.400 | 1.2400 ± 0.4700 | 0.0400 ± 0.0400 | 1.2000 ± 0.2300 | 85.4600 ± 1.5700 | 132.5200 ± 2.1900 | 599.3600 ± 29.3200 |
| Winter | 18.0000 ± 2.3300 | 1.7000 ± 0.0800 | 7.6900 ± 0.1700 | 8.0200 ± 0.3300 | 0.7100 ± 0.7900 | 0.0500 ± 0.0100 | 0.9600 ± 0.1300 | 33.2500 ± 3.2600 | 122.0500 ± 3.3500 | 722.3600 ± 32.0600 |
| Non-rhizosphere | ||||||||||
| Spring | 19.2500 ± 2.3600 | 8.8200 ± 0.5900 | 15.5100 ± 0.2700 | 8.0100 ± 0.2300 | 1.1800 ± 0.0300 | 0.0700 ± 0.0200 | 0.4000 ± 0.0200 | 123.8900 ± 6.9700 | 65.3200 ± 5.2100 | 933.4600 ± 42.3600 |
| Summer | 36.5200 ± 0.3000 | 11.9500 ± 0.0300 | 11.6800 ± 0.3700 | 7.9800 ± 0.4600 | 1.0400 ± 0.2200 | 0.1000 ± 0.0200 | 0.3500 ± 0.1400 | 97.0500 ± 7.3600 | 59.6300 ± 4.3700 | 567.3100 ± 39.4400 |
| Autumn | 21.0000 ± 2.1100 | 12.9100 ± 0.0600 | 13.6600 ± 0.6300 | 7.0100 ± 0.3900 | 0.6100 ± 0.2700 | 0.0500 ± 0.0100 | 0.2200 ± 0.0500 | 102.6500 ± 4.2400 | 76.9200 ± 2.770 | 316.4500 ± 28.3700 |
| Winter | 19.0000 ± 3.3700 | 14.7500 ± 0.4400 | 16.5500 ± 0.2800 | 7.4000 ± 0.6600 | 0.5300 ± 0.1100 | 0.0700 ± 0.0200 | 0.5200 ± 0.1600 | 99.3600 ± 3.3100 | 100.2900 ± 5.5800 | 231.6400 ± 19.2600 |
| Sobs | Shannon | Simpson | Ace | Chao | ||
|---|---|---|---|---|---|---|
| Rhizosphere | Spring | 736.0000 | 5.2915 | 0.0181 | 795.0025 | 816.8154 |
| Summer | 714.0000 | 5.1772 | 0.0167 | 770.4432 | 777.3733 | |
| Autumn | 861.0000 | 5.6708 | 0.0090 | 922.3090 | 945.0000 | |
| Winter | 853.0000 | 5.6463 | 0.0087 | 910.7371 | 930.8571 | |
| Non-rhizosphere | Spring | 835.0000 | 5.6025 | 0.0098 | 867.7590 | 896.1579 |
| Summer | 825.0000 | 5.4611 | 0.0143 | 858.1264 | 870.3088 | |
| Autumn | 750.0000 | 5.3282 | 0.0123 | 815.6761 | 843.9231 | |
| Winter | 762.0000 | 5.2844 | 0.0187 | 814.8643 | 829.8082 |
| Estimators | Non-Rhizosphere Mean | Non-Rhizosphere SD | Rhizosphere Mean | Rhizosphere SD | p-Value |
|---|---|---|---|---|---|
| Sobs | 793.0000 | 43.1970 | 791.0000 | 76.8070 | 0.9653 |
| Shannon | 5.4191 | 0.1435 | 5.4465 | 0.2495 | 0.8552 |
| Simpson | 0.0138 | 0.0037 | 0.0131 | 0.0049 | 0.8334 |
| ACE | 839.1100 | 27.8050 | 849.6200 | 78.0410 | 0.8081 |
| Chao | 860.0500 | 29.3470 | 867.5100 | 83.0910 | 0.8711 |
| Nodes | Edges | Average Path Length (APL) | Modularity Index (MD) | Graph Density (GD) | Average Degree (AD) | Network Diameter (ND) | Clustering Coefficient (CC) | |
|---|---|---|---|---|---|---|---|---|
| Rhizosphere samples | 100.0000 | 782.0000 | 5.7580 | 0.4790 | 0.1170 | 10.2900 | 11.0000 | 0.9550 |
| Non-rhizosphere samples | 100.0000 | 693.0000 | 2.4560 | 0.5860 | 0.1250 | 9.6400 | 6.0000 | 0.6580 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, K.; Yang, Q.; Jiang, Y.; Liu, W. Changes in Temporal Dynamics and Factors Influencing the Environment of the Bacterial Community in Mangrove Rhizosphere Sediments in Hainan. Sustainability 2022, 14, 7415. https://doi.org/10.3390/su14127415
Lu K, Yang Q, Jiang Y, Liu W. Changes in Temporal Dynamics and Factors Influencing the Environment of the Bacterial Community in Mangrove Rhizosphere Sediments in Hainan. Sustainability. 2022; 14(12):7415. https://doi.org/10.3390/su14127415
Chicago/Turabian StyleLu, Kaiqi, Qiu Yang, Yamin Jiang, and Wenjie Liu. 2022. "Changes in Temporal Dynamics and Factors Influencing the Environment of the Bacterial Community in Mangrove Rhizosphere Sediments in Hainan" Sustainability 14, no. 12: 7415. https://doi.org/10.3390/su14127415
APA StyleLu, K., Yang, Q., Jiang, Y., & Liu, W. (2022). Changes in Temporal Dynamics and Factors Influencing the Environment of the Bacterial Community in Mangrove Rhizosphere Sediments in Hainan. Sustainability, 14(12), 7415. https://doi.org/10.3390/su14127415






