Analysis of Microbial Communities in Aged Refuse Based on 16S Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sample Acquisition
2.2. Physical and Chemical Indicator Measurements
2.3. DNA Extraction and High-Throughput Sequencing
2.4. High-Throughput Sequencing Data Analysis
2.5. Statistical Analysis
3. Results
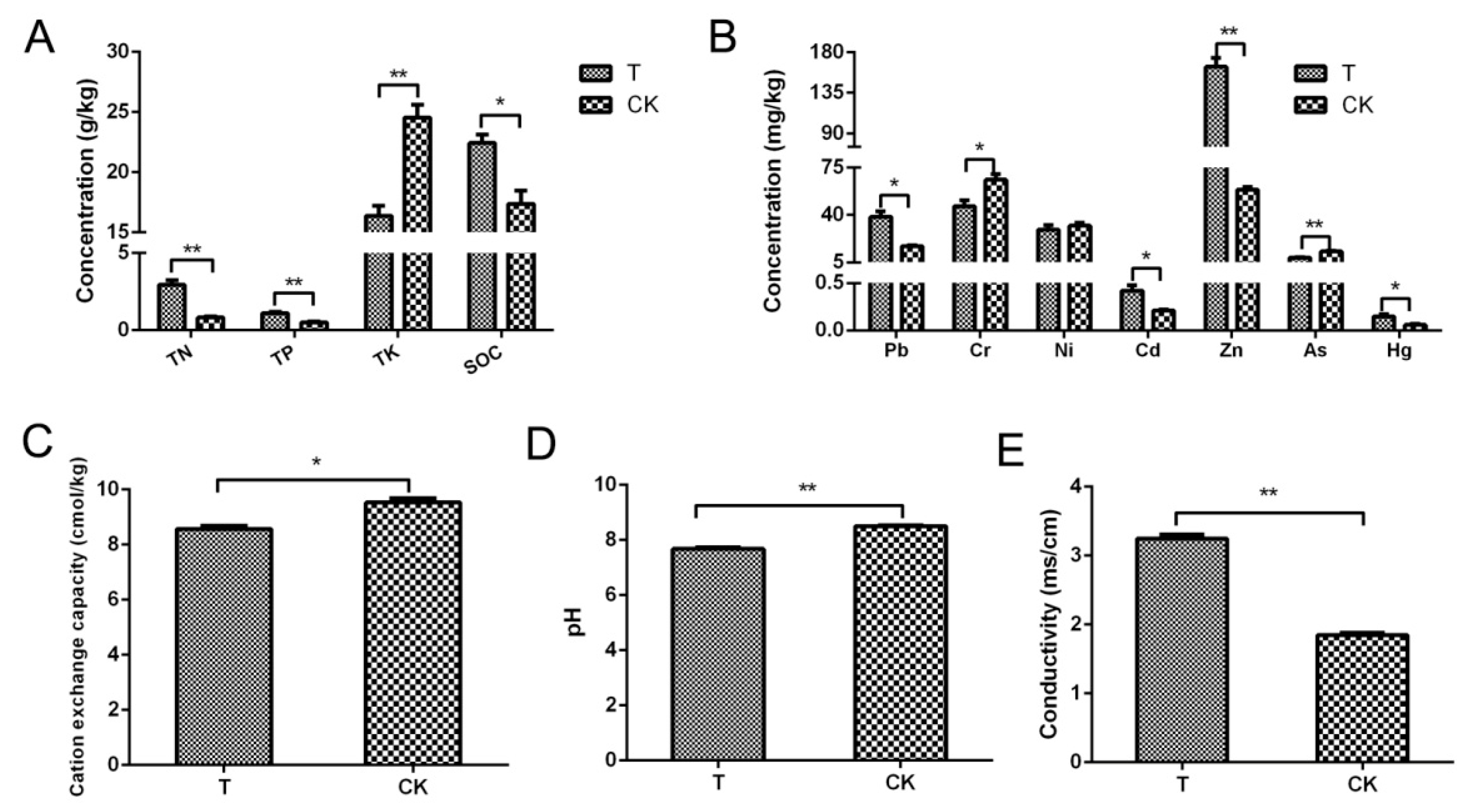
3.1. Comparison of Physical and Chemical Factors
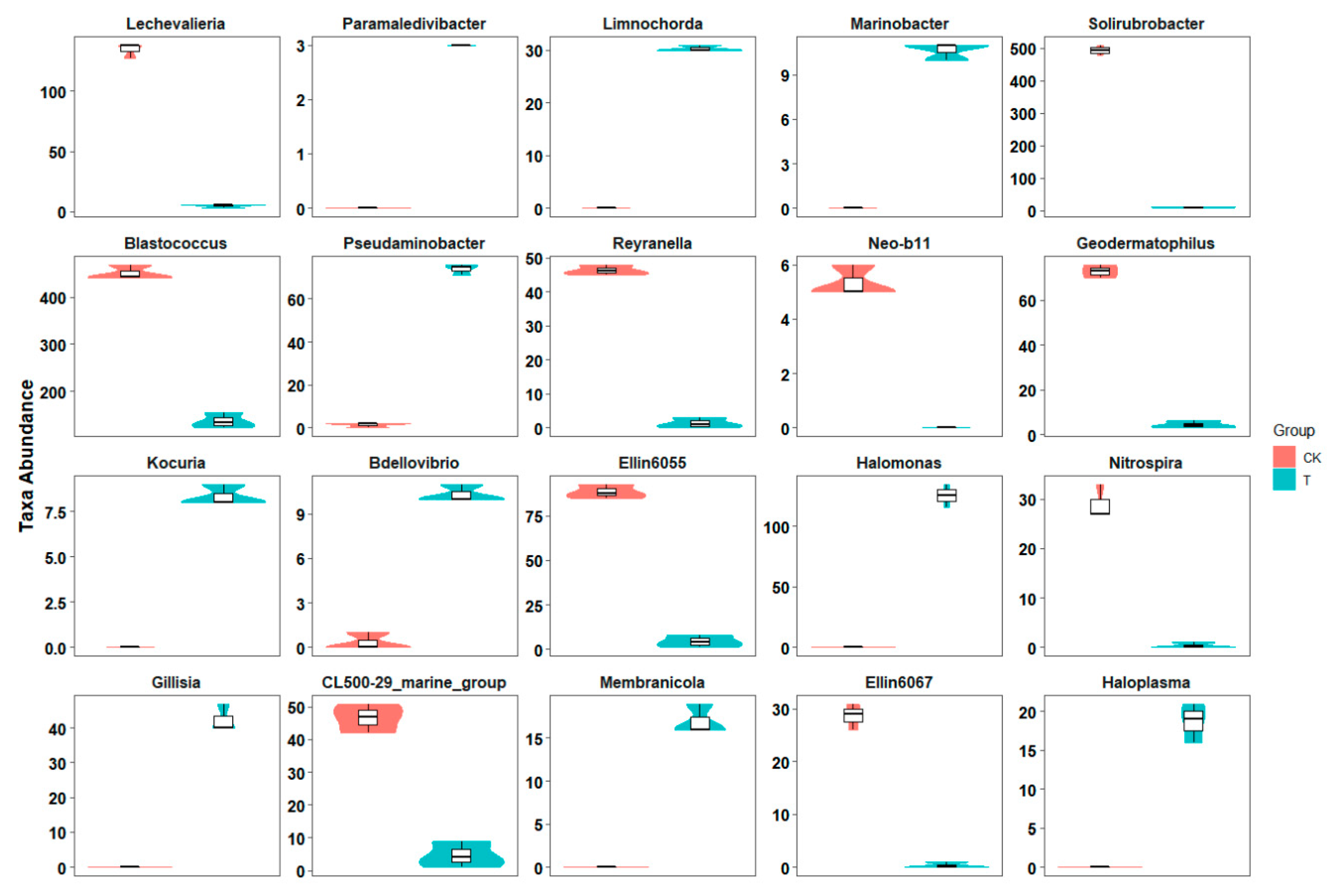
3.2. Microbial Community Structure Composition
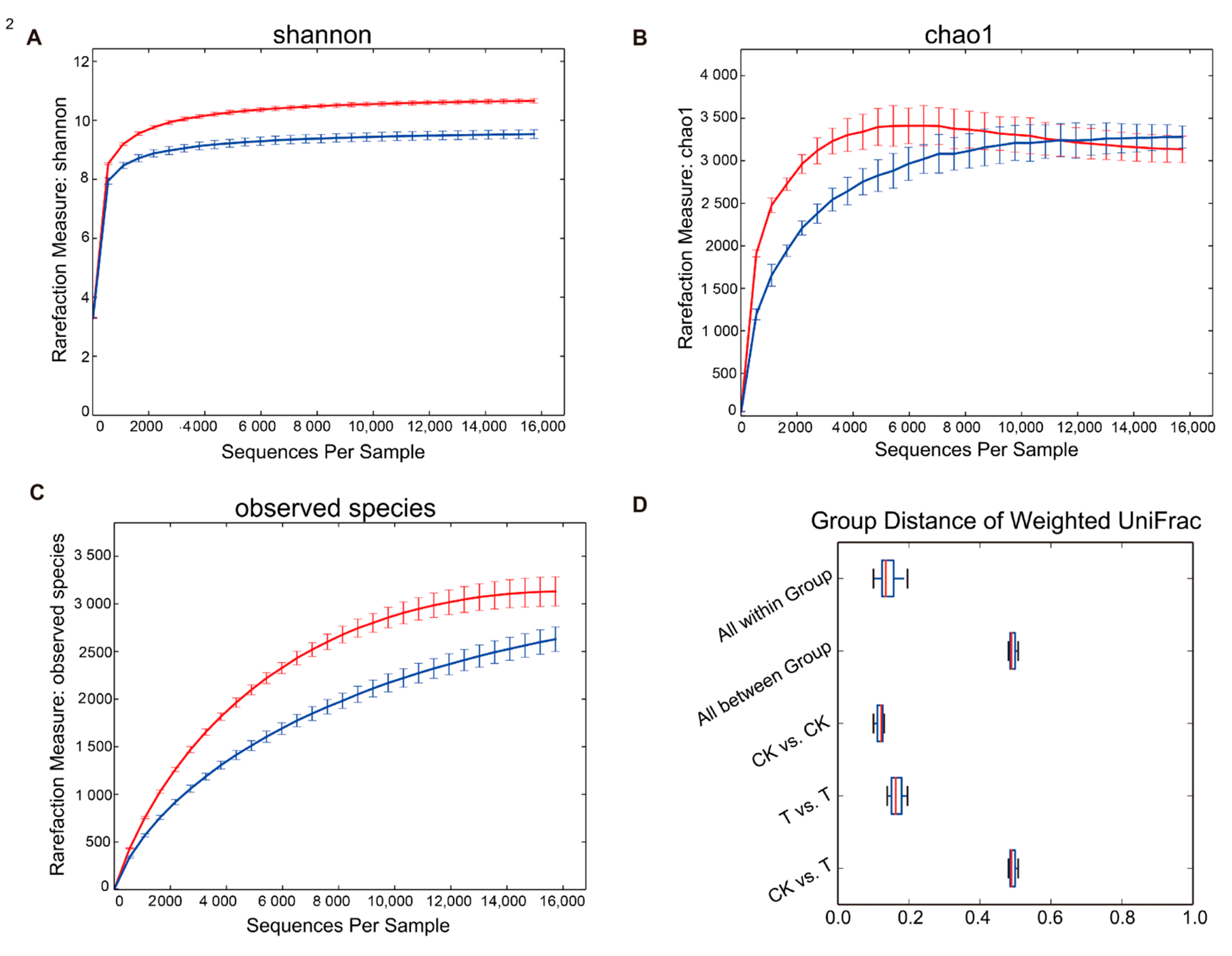
3.3. Diversity of the Microbial Community
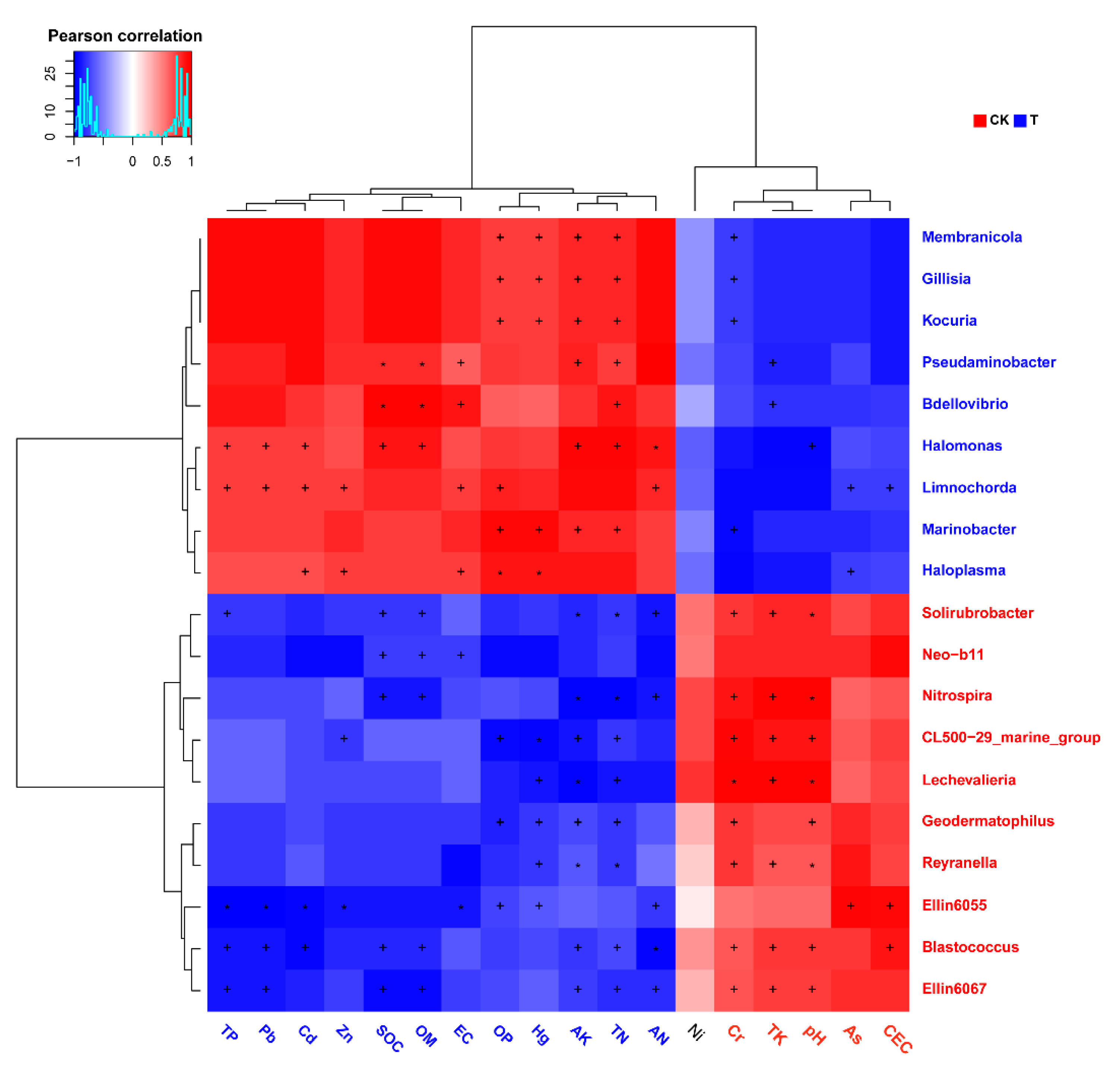
3.4. The Interactions between the Microorganisms and Physical or Chemical Indicators
3.5. The Predictive Function Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| MSW | municipal solid waste |
| SOC | soil organic carbon |
| TK | total potassium |
| TP | total phosphorus |
| TN | total nitrogen |
| Hg | mercury |
| As | arsenic |
| Pb | lead |
| Cr | chromium |
| Ni | nickel |
| Cd | cadmium |
| Zn | zinc |
| CSPSA | control standards of pollutants in sludge for agricultural use |
| SCAL | soil contamination risk of agricultural land |
Appendix A
| Metal (mg/kg) | Aged Refuse | Control | CSPSA Class A Upper Limit | CSPSAU Class B Upper Limit | SCAL Upper Limit |
|---|---|---|---|---|---|
| Pb | 38.48 ± 4.1 | 16.92 ± 0.92 | 300 | 1000 | 170 |
| Cr | 46.36 ± 4.53 | 65.89 ± 4.32 | 500 | 1000 | 250 |
| Ni | 29.12 ± 3.28 | 32.19 ± 2.01 | 100 | 200 | 190 |
| Cd | 0.42 ± 0.06 | 0.21 ± 0.01 | 3 | 15 | 0.6 |
| Zn | 163.83 ± 9.67 | 58.67 ± 1.81 | 1200 | 3000 | 300 |
| As | 8.45 ± 0.33 | 13.18 ± 0.38 | 30 | 75 | 25 |
| Hg | 0.15 ± 0.02 | 0.06 ± 0.01 | 3 | 15 | 3.4 |
References
- Zhu, Y.; Zhang, Y.; Luo, D.; Chong, Z.; Li, E.; Kong, X. A review of municipal solid waste in China: Characteristics, compositions, influential factors and treatment technologies. Environ. Dev. Sustain. 2020. [Google Scholar] [CrossRef]
- Chengdu, Q.; Jun, H.; Bin, W.; Shubo, D.; Yujue, W.; Gang, Y. Contaminants of emerging concern in landfill leachate in China: Areview. Emerg. Contam. 2018, 4, 1–10. [Google Scholar]
- Zink, T.; Geyer, R. Material Recycling and the Myth of Landfill Diversion. J. Ind. Ecol. 2018, 23, 541–548. [Google Scholar] [CrossRef]
- Makhatova, A.; Mazhit, B.; Sarbassov, Y.; Meiramkulova, K.; Inglezakis, V.J.; Poulopoulos, S.G. Effective photochemical treatment of a municipal solid waste landfill leachate. PLoS ONE 2020, 15, e0239433. [Google Scholar] [CrossRef] [PubMed]
- Pecorini, I.; Iannelli, R. Characterization of Excavated Waste of Different Ages in View of Multiple Resource Recovery in Landfill Mining. Sustainability 2020, 12, 1780. [Google Scholar] [CrossRef]
- Anijiofor Chinenyenwa, N.N.; Idrus, S.; Ahsan, A. Aged Refuse Characterization as Resource for Wastewater Treatment and Landfill Remediation. Int. J. Waste Resour. 2017, 7, 1000269. [Google Scholar]
- Li, G.; Hou, F.; Guo, Z.; Yao, G.; Sang, N. Analyzing nutrient distribution in different particle-size municipal aged refuse. Waste Manag. 2011, 31, 2203–2207. [Google Scholar] [CrossRef]
- Cerqueda-García, D.; Améndola-Pimenta, M.; Zamora-Briseño, J.A.; González-Penagos, C.E.; Árcega-Cabrera, F.; Ceja-Moreno, V.; Rodríguez-Canul, R. Effects of chronic exposure to water accommodated fraction (WAF) of light crude oil on gut microbiota composition of the lined sole (Achirus lineatus). Mar. Environ. Res. 2020, 161, 105116. [Google Scholar] [CrossRef]
- Chen, F.; Li, X.; Zhu, Q.; Ma, J.; Hou, H.; Zhang, S. Bioremediation of petroleum-contaminated soil enhanced by aged refuse. Chemosphere 2019, 222, 98–105. [Google Scholar] [CrossRef]
- Zhao, J.; Jing, Y.; Zhang, J.; Sun, Y.; Wang, Y.; Wang, H.; Bi, X. Aged refuse enhances anaerobic fermentation of food waste to produce short-chain fatty acids. Bioresour. Technol. 2019, 289, 121547. [Google Scholar] [CrossRef]
- Zgavarogea, R.; Niculescu, V.; Miricioiu, M.; Paun, N.; Constantinescu, M. Wastewater treatment: Application and future perspective of anaerobic membrane bioreactors. In Proceedings of the 16th International Multidisciplinary Scientific GeoConference SGEM 2016, Albena, Bulgaria, 28 June–6 July 2016. [Google Scholar]
- Niculescu, V.; Miricoiu, M.; Zgavarogea, R.; Paun, N. Advances in membranes technology for gas separation and wastewater treatment. In Proceedings of the International Multidisciplinary Scientific GeoConference-SGEM2016, Albena, Bulgaria, 28 June–6 July 2016. [Google Scholar]
- Sekhohola-Dlamini, L.; Tekere, M. Microbiology of municipal solid waste landfills: A review of microbial dynamics and ecological influences in waste bioprocessing. Biodegradation 2020, 31, 1–21. [Google Scholar] [CrossRef]
- Zainun, M.Y.; Simarani, K. Metagenomics profiling for assessing microbial diversity in both active and closed landfills. Sci. Total Environ. 2018, 616–617, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.; Huang, Y.; Qiu, Z.; Li, Q. Microbial response during treatment of different types of landfill leachate in a semi-aerobic aged refuse biofilter. Chemosphere 2021, 262, 127822. [Google Scholar] [CrossRef]
- Gupta, J.; Rathour, R.; Kumar, M.; Thakur, I.S. Metagenomic Analysis of Microbial Diversity in Landfill Lysimeter Soil of Ghazipur Landfill Site, New Delhi, India. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Yadav, B.; Johri, A.K.; Dua, M. Metagenomic Analysis of the Microbial Diversity in Solid Waste from Okhla Landfill, New Delhi, India. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Dombard, D.R.; Bogner, J.E.; Malas, J. A Review of Landfill Microbiology and Ecology: A Call for Modernization With ‘Next Generation’ Technology. Front. Microbiol. 2020, 11, 1127. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, Z.G.; Wu, H.T.; Lü, X.G. Soil Microorganism Characteristics and Soil Nutrients of Different Wetlands in Sanjinag Plain, Northeast China. Huan Jing Ke Xue = Huanjing Kexue 2015, 36, 1842–1848. [Google Scholar] [PubMed]
- Oldroyd, G.E.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Ann. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Zhuang, X.; Gao, J.; Ma, A.; Fu, S.; Zhuang, G. Bioactive molecules in soil ecosystems: Masters of the underground. Int. J. Mol. Sci. 2013, 14, 8841–8868. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.D. Agricultural and Chemistry Analysis of Soil. Agric. Press China 2005, 30–34. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Pop, M.; Bravo, H.C.J.G.B. Metastats: An improved statistical method for analysis of metagenomic data. Genome Biol. 2011, 12, 1–27. [Google Scholar] [CrossRef]
- Douglas, G.M.; Beiko, R.G.; Langille, M.G. Predicting the functional potential of the microbiome from marker genes using PICRUSt. In Microbiome Analysis; Springer: Berlin, Germany, 2018; pp. 169–177. [Google Scholar]
- Jani, Y.; Kaczala, F.; Marchand, C.; Hogland, M.; Kriipsalu, M.; Hogland, W.; Kihl, A. Characterisation of excavated fine fraction and waste composition from a Swedish landfill. Waste Manag. Res. J. Int. Solid Wastes Public Cleans. Assoc. 2016, 34, 1292–1299. [Google Scholar] [CrossRef]
- Weber, R.; Gaus, C.; Tysklind, M.; Johnston, P.; Forter, M.; Hollert, H.; Heinisch, E.; Holoubek, I.; Lloyd-Smith, M.; Masunaga, S.; et al. Dioxin- and POP-contaminated sites—Contemporary and future relevance and challenges: Overview on background, aims and scope of the series. Environ. Sci. Pollut. Res. Int. 2008, 15, 363–393. [Google Scholar] [CrossRef] [PubMed]
- Gajski, G.; Oreščanin, V.; Garaj-Vrhovac, V. Chemical composition and genotoxicity assessment of sanitary landfill leachate from Rovinj, Croatia. Ecotoxicol. Environ. Saf. 2012, 78, 253–259. [Google Scholar] [CrossRef]
- Rikta, S.Y.; Tareq, S.M.; Uddin, M.K. Toxic metals (Ni2+, Pb2+, Hg2+) binding affinity of dissolved organic matter (DOM) derived from different ages municipal landfill leachate. Appl. Water Sci. 2018, 8, 5. [Google Scholar] [CrossRef]
- Khaledian, Y.; Brevik, E.C.; Pereira, P.; Cerdà, A.; Fattah, M.A.; Tazikeh, H. Modeling soil cation exchange capacity in multiple countries. Catena 2017, 158, 194–200. [Google Scholar] [CrossRef]
- Stadler, A.; Rudolph, S.; Kupisch, M.; Langensiepen, M.; Ewert, F. Quantifying the effect of soil variability on crop growth using apparent soil electrical conductivity measurements. Eur. J. Agron. 2015, 64, 8–20. [Google Scholar] [CrossRef]
- Feng, S.; Hou, S.; Huang, X.; Fang, Z.; Yang, H. Insights into the microbial community structure of anaerobic digestion of municipal solid waste landfill leachate for methane production by adaptive thermophilic granular sludge. Electr. J. Biotechnol. 2019, 39, 98–106. [Google Scholar] [CrossRef]
- Rajasekar, A.; Sekar, R.; Medina-Roldán, E.; Bridge, J.; Moy, C.K.S.; Wilkinson, S. Next-generation sequencing showing potential leachate influence on bacterial communities around a landfill in China. Can. J. Microbiol. 2018, 64, 537–549. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial Exopolysaccharide mediated heavy metal removal: A Review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Handley, K.M.; Lloyd, J.R. Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 2013, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.; Webb, E.A.; Nelson, W.C.; Heidelberg, J.F.; Ivanova, N.; Pati, A.; Edwards, K.J. Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph”. Appl. Environ. Microbiol. 2011, 77, 2763–2771. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, H.; Shao, Z.; Liao, S.; Johnstone, L.; Rensing, C.; Wang, G. Genome sequence of deep-sea manganese-oxidizing bacterium Marinobacter manganoxydans MnI7-9. J. Bacteriol. 2012, 194, 899–900. [Google Scholar] [CrossRef]
- Cui, H.; Liu, L.L.; Dai, J.R.; Yu, X.N.; Guo, X.; Yi, S.J.; Zhou, D.Y.; Guo, W.H.; Du, N. Bacterial community shaped by heavy metals and contributing to health risks in cornfields. Ecotoxicol. Environ. Saf. 2018, 166, 259–269. [Google Scholar] [CrossRef]
- Viesser, J.A.; Sugai-Guerios, M.; Malucelli, L.C.; Pincerati, M.R.; Karp, S.G.; Maranho, L.T. Petroleum-Tolerant Rhizospheric Bacteria: Isolation, Characterization and Bioremediation Potential. Sci. Rep. 2020, 10, 2060. [Google Scholar] [CrossRef]
- Luo, Y.J.; Xie, B.S.; Lv, X.L.; Cai, M.; Wang, Y.N.; Cui, H.L.; Cai, H.; Wu, X.L. Marinobacter shengliensis sp. nov. a moderately halophilic bacterium isolated from oil-contaminated saline soil. Antonie Leeuwenhoek 2015, 107, 1085–1094. [Google Scholar] [CrossRef]
- Martín, S.; Márquez, M.C.; Sánchez-Porro, C.; Mellado, E.; Arahal, D.R.; Ventosa, A. Marinobacter lipolyticus sp. nov. a novel moderate halophile with lipolytic activity. Int. J. Syst. Evolut. Microbiol. 2003, 53, 1383–1387. [Google Scholar] [CrossRef]
- Bonin, P.; Vieira, C.; Grimaud, R.; Militon, C.; Cuny, P.; Lima, O.; Guasco, S.; Brussaard, C.P.; Michotey, V. Substrates specialization in lipid compounds and hydrocarbons of Marinobacter genus. Environ. Sci. Pollut. Res. Int. 2015, 22, 15347–15359. [Google Scholar] [CrossRef] [PubMed]


| Samples | Simpson | Chao1 | ACE | Shannon |
|---|---|---|---|---|
| T1 | 0.993826 | 3118.17 | 3207.86 | 9.33 |
| T2 | 0.996125 | 3321.49 | 3708.06 | 9.69 |
| T3 | 0.995019 | 3411.14 | 3652.43 | 9.59 |
| CK1 | 0.998292 | 3049 | 3049 | 10.73 |
| CK2 | 0.997839 | 3000 | 3000 | 10.56 |
| CK3 | 0.997648 | 3349.22 | 3359.57 | 10.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, F.; Du, J.; Yuan, Y.; Wu, X.; Zhao, S. Analysis of Microbial Communities in Aged Refuse Based on 16S Sequencing. Sustainability 2021, 13, 4111. https://doi.org/10.3390/su13084111
Hou F, Du J, Yuan Y, Wu X, Zhao S. Analysis of Microbial Communities in Aged Refuse Based on 16S Sequencing. Sustainability. 2021; 13(8):4111. https://doi.org/10.3390/su13084111
Chicago/Turabian StyleHou, Fen, Junjie Du, Ye Yuan, Xihui Wu, and Sai Zhao. 2021. "Analysis of Microbial Communities in Aged Refuse Based on 16S Sequencing" Sustainability 13, no. 8: 4111. https://doi.org/10.3390/su13084111
APA StyleHou, F., Du, J., Yuan, Y., Wu, X., & Zhao, S. (2021). Analysis of Microbial Communities in Aged Refuse Based on 16S Sequencing. Sustainability, 13(8), 4111. https://doi.org/10.3390/su13084111





