Topographic Effects on the Spatial Species Associations in Diverse Heterogeneous Tropical Evergreen Forests
Abstract
1. Introduction
2. Materials and Methods
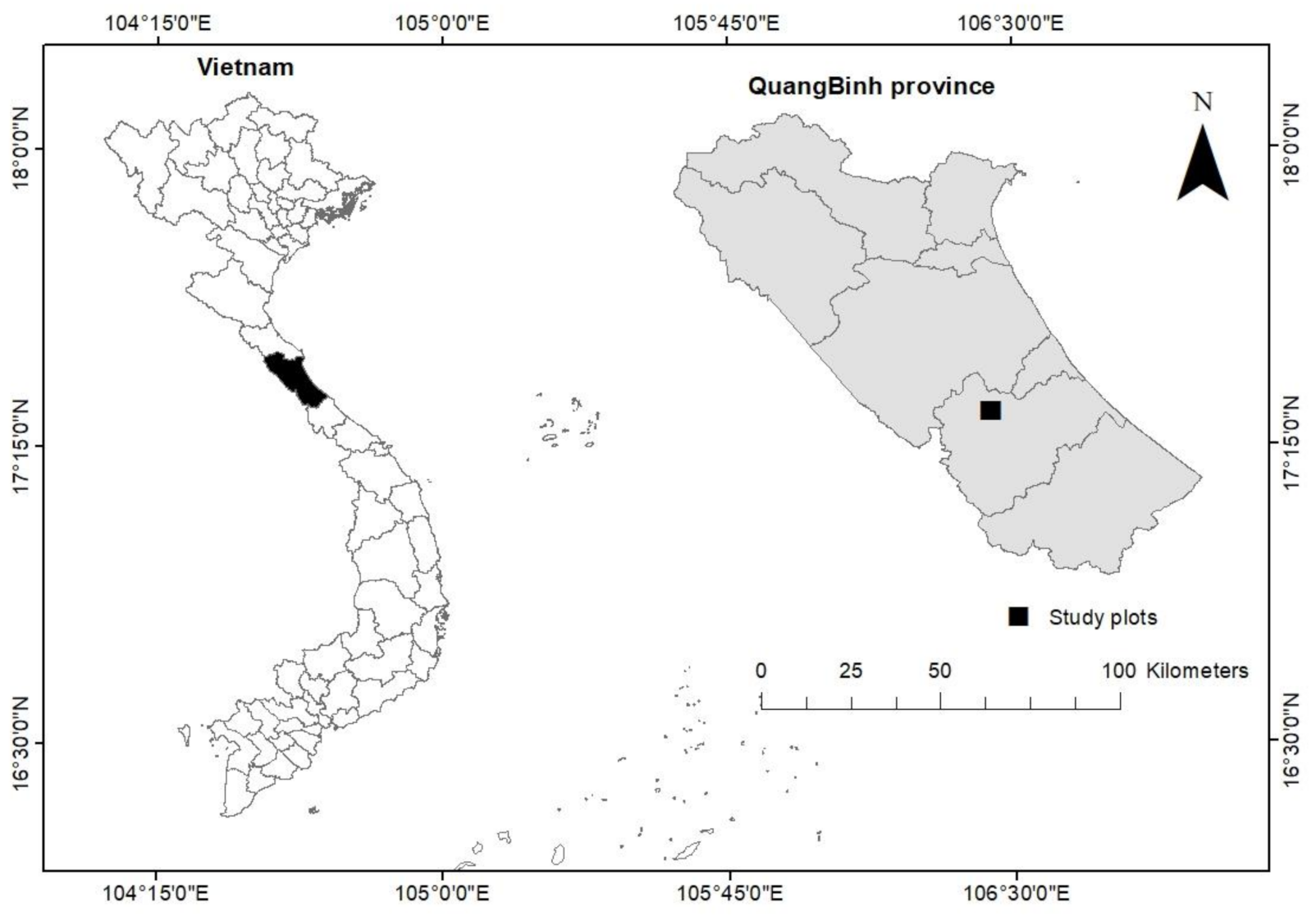
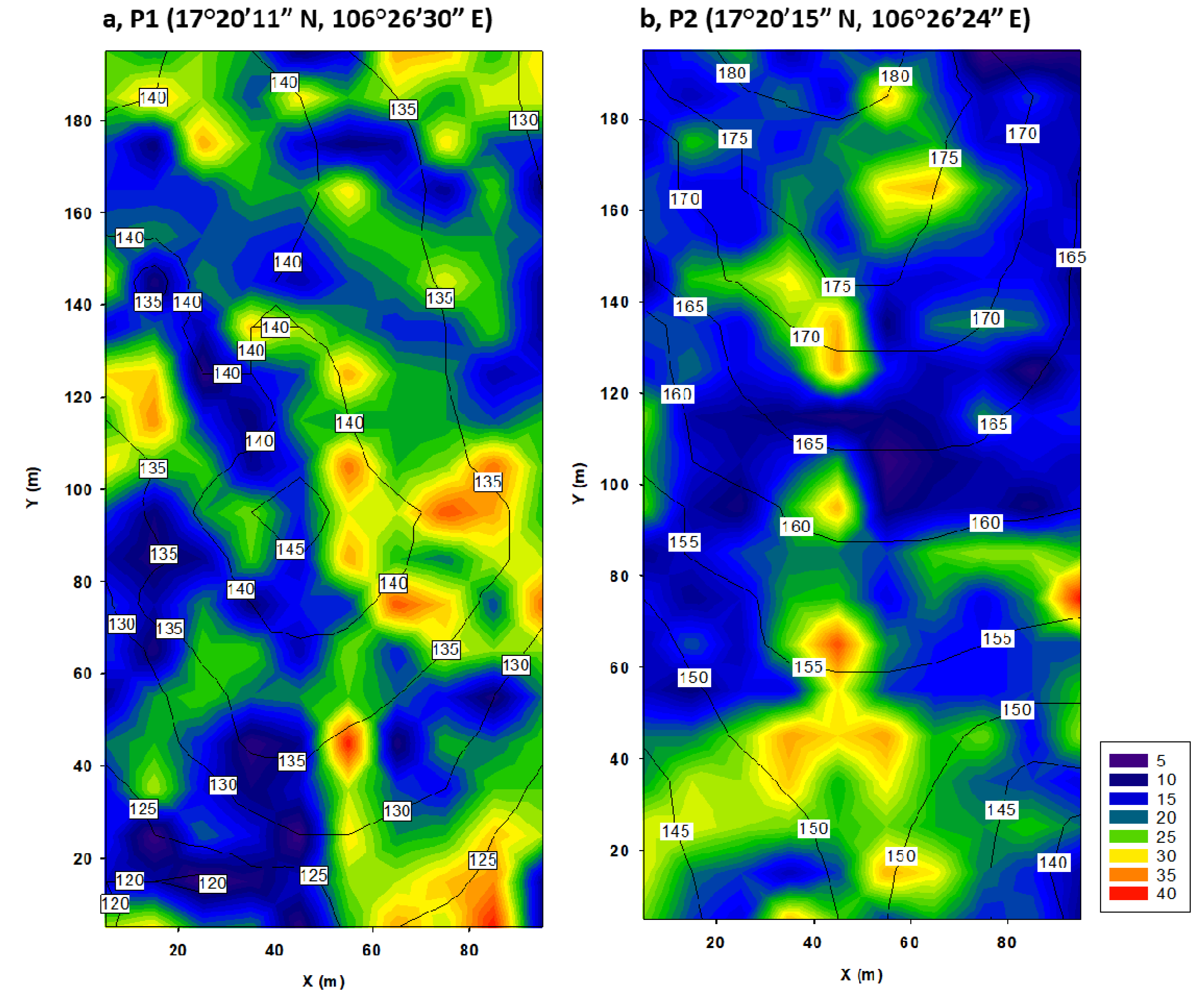
2.1. Study Site and Data Collection
2.2. Data Analysis
2.2.1. Community Properties
2.2.2. Topographical Effects on Community Properties
2.2.3. Overall Species–Species Associations
3. Results
3.1. Community Properties
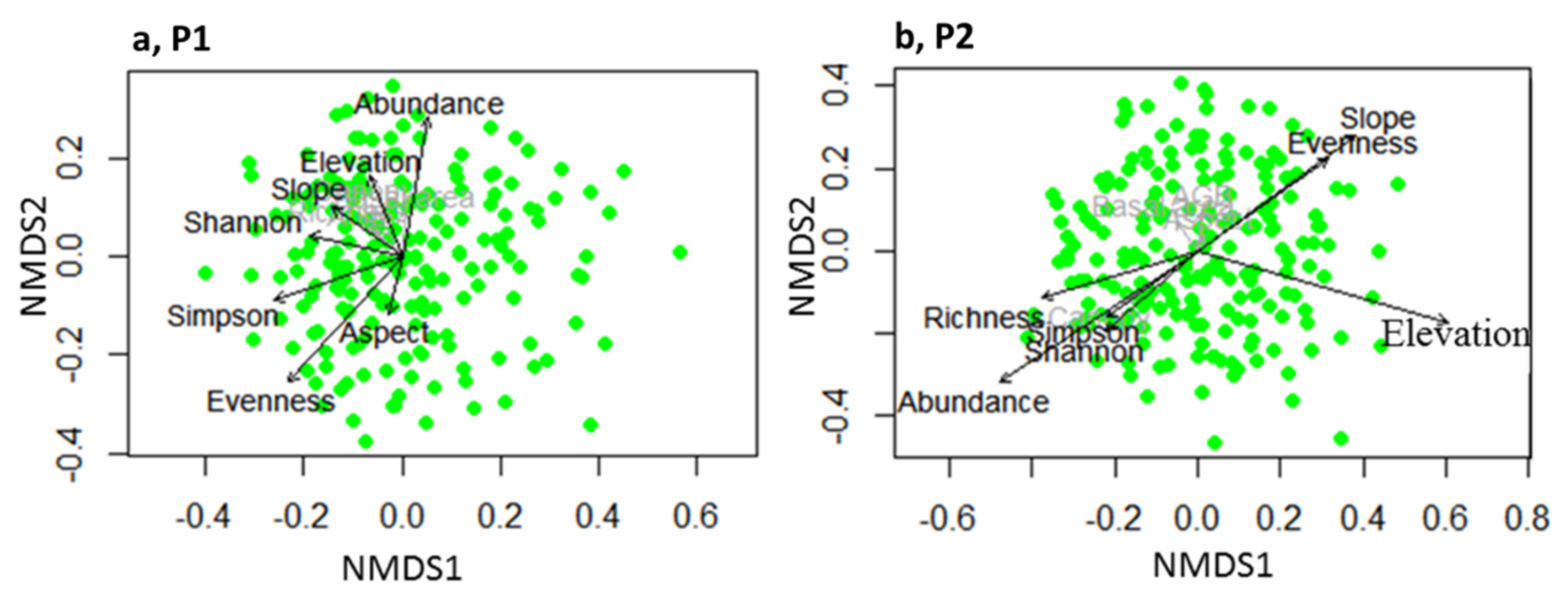
3.2. Effects of Topography on Community Properties
3.3. Overall Species–Species Associations
4. Discussion
4.1. Topographical Effects on Community Properties
4.2. Overall Species–Species Associations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| No | Tree Species | P1 | P2 | Shade Tolerance | ||||
|---|---|---|---|---|---|---|---|---|
| N | DBH (cm) | IVI (%) | N | DBH (cm) | IVI (%) | |||
| 1 | Garuga pierrei | 282 | 10.08 ± 10.89 | 8.98 | 232 | 11.30 ± 13.26 | 7.72 | Tolerant |
| 2 | Tarrietia javanica | 383 | 5.62 ± 6.39 | 7.28 | 330 | 4.52 ± 3.58 | 5.14 | Intolerant |
| 3 | Ormosia balansae | 138 | 17.05 ± 12.97 | 7.26 | 187 | 14.75 ± 10.81 | 6.60 | Intolerant |
| 4 | Bursera tonkinensis | 384 | 6.15 ± 4.16 | 6.72 | 253 | 6.67 ± 4.12 | 4.41 | Medium |
| 5 | Paviesia annamensis | 240 | 9.18 ± 7.64 | 6.02 | 239 | 6.94 ± 4.86 | 4.32 | Intolerant |
| 6 | Litsea glutinosa | 229 | 8.06 ± 6.21 | 4.96 | 264 | 8.26 ± 6.70 | 5.49 | Intolerant |
| 7 | Castanopsis indica | 168 | 10.21 ± 8.27 | 4.65 | - | - | Intolerant | |
| 8 | Polyalthia nemoralis | 303 | 5.02 ± 1.77 | 4.58 | 244 | 5.53 ± 1.88 | 3.78 | Intolerant |
| 9 | Syzygium wightianum | 179 | 9.36 ± 7.04 | 4.40 | 81 | 11.56 ± 8.17 | 1.54 | Intolerant |
| 10 | Erythrophfloeum fordii | 63 | 18.52 ± 15.35 | 3.96 | - | - | - | Medium |
| 11 | Mallotus kurzii | 265 | 4.01 ± 0.98 | 3.76 | 114 | 3.71 ± 0.73 | 1.63 | Intolerant |
| 12 | Amoora dasyclada | 148 | 7.99 ± 6.73 | 3.28 | 96 | 8.89 ± 6.93 | 2.08 | Medium |
| 13 | Cinnamomun bejolghota | 100 | 10.71 ± 9.25 | 3.00 | 267 | 13.01 ± 10.59 | 8.51 | Intolerant |
| 14 | Gironniera Subaequalis | 92 | 9.71 ± 6.65 | 2.27 | 137 | 11.19 ± 9.28 | 3.73 | Medium |
| 15 | Endosperrmun sinensis | 54 | 11.77 ± 13.18 | 2.14 | 83 | 21.67 ± 13.33 | 4.63 | Intolerant |
| 16 | Garcinia oblongifolia | 121 | 6.23 ± 4.08 | 2.11 | 67 | 6.22 ± 3.48 | 1.11 | Tolerant |
| 17 | Canarium album | - | - | - | 155 | 11.03 ± 6.04 | 3.68 | Intolerant |
| 18 | Koilodepas hainanense | 104 | 5.83 ± 2.61 | 1.68 | 80 | 8.41 ± 4.52 | 1.54 | Tolerant |
| 19 | Cassine glauca | 74 | 8.41 ± 5.51 | 1.59 | 89 | 8.69 ± 7.66 | 1.97 | Tolerant |
| 20 | Litsea vang | 71 | 6.54 ± 3.30 | 1.27 | 76 | 8.72 ± 4.67 | 1.5 | Intolerant |
| 21 | Symplocos laurina | 55 | 9.31 ± 5.61 | 1.25 | 145 | 11.81 ± 6.86 | 3.71 | Intolerant |
| 22 | Engelhardtia roxburghiana | - | - | 63 | 28.78 ± 11.91 | 4.84 | Tolerant | |
Appendix B
| Properties | Plot P1 | Plot P2 | ||||||
|---|---|---|---|---|---|---|---|---|
| NMDS1 | NMDS2 | r2 | Pr (>r) | NMDS1 | NMDS2 | r2 | Pr (>r) | |
| Richness | −0.84966 | −0.52733 | 0.0412 | 0.016 | −0.95625 | −0.29256 | 0.0775 | 0.001 |
| Abundance | 0.08552 | −0.99634 | 0.2051 | 0.001 | −0.82451 | −0.56584 | 0.1677 | 0.001 |
| Canopy cover | −0.57273 | −0.81974 | 0.0292 | 0.051 | −0.8942 | −0.44766 | 0.017 | 0.171 |
| Basal area | 0.01327 | −0.99991 | 0.0204 | 0.159 | −0.33053 | 0.9438 | 0.0047 | 0.678 |
| Simpson | −0.95727 | 0.28919 | 0.1998 | 0.001 | −0.82216 | −0.56926 | 0.0379 | 0.025 |
| Shannon | −0.97059 | −0.24074 | 0.1194 | 0.001 | −0.76235 | −0.64716 | 0.0444 | 0.009 |
| Evenness | −0.65468 | 0.7559 | 0.2773 | 0.001 | 0.80735 | 0.59007 | 0.0731 | 0.001 |
| AGB | −0.76702 | −0.64163 | 0.0121 | 0.315 | 0.21548 | 0.97651 | 0.0085 | 0.457 |
| Elevation | −0.25379 | −0.96726 | 0.0634 | 0.004 | 0.96418 | −0.26524 | 0.2172 | 0.001 |
| Slope | −0.81517 | −0.57922 | 0.0572 | 0.006 | 0.80928 | 0.58743 | 0.1188 | 0.001 |
| Aspect | −0.24281 | 0.97007 | 0.033 | 0.04 | −0.10431 | 0.99454 | 0.0007 | 0.927 |
References
- Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef]
- Wright, J.S. Plant diversity in tropical forests: A review of mechanisms of species coexistence. Oecologia 2002, 130, 1–14. [Google Scholar] [CrossRef]
- May, F.; Huth, A.; Wiegand, T. Moving beyond abundance distributions: Neutral theory and spatial patterns in a tropical forest. Proc. R. Soc. B Biol. Sci. 2015, 282, 20141657. [Google Scholar] [CrossRef]
- Wiegand, T.; Huth, A.; Getzin, S.; Wang, X.; Hao, Z.; Gunatilleke, C.S.; Gunatilleke, I.N. Testing the independent species’ arrangement assertion made by theories of stochastic geometry of biodiversity. Proc. R. Soc. B. 2012, 279, 3312–3320. [Google Scholar] [CrossRef]
- Volkov, I.; Banavar, J.R.; Hubbell, S.P.; Maritan, A. Inferring species interactions in tropical forests. Proc. Natl. Acad. Sci. USA 2009, 106, 13854–13859. [Google Scholar] [CrossRef] [PubMed]
- Harms, K.E.; Condit, R.; Hubbell, S.P.; Foster, R.B. Habitat associations of trees and shrubs in a 50-ha neotropical forest plot. J. Ecol. 2001, 89, 947–959. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Uria-Diez, J.; Wiegand, K. Spatial distribution and association patterns in a tropical evergreen broad-leaved forest of north-central Vietnam. J. Veg. Sci. 2016, 27, 318–327. [Google Scholar] [CrossRef]
- May, F.; Giladi, I.; Ziv, Y.; Jeltsch, F. Dispersal and diversity–unifying scale-dependent relationships within the neutral theory. Oikos 2012, 121, 942–951. [Google Scholar] [CrossRef]
- Adler, P.B.; Fajardo, A.; Kleinhesselink, A.R.; Kraft, N.J. Trait-based tests of coexistence mechanisms. Ecol. Lett. 2013, 16, 1294–1306. [Google Scholar] [CrossRef]
- Shipley, B.; Paine, C.T.; Baraloto, C. Quantifying the importance of local niche-based and stochastic processes to tropical tree community assembly. Ecology 2012, 93, 760–769. [Google Scholar] [CrossRef]
- Kraft, N.J.; Ackerly, D.D. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 2010, 80, 401–422. [Google Scholar] [CrossRef]
- Hubbell, S.P.; Ahumada, J.A.; Condit, R.; Foster, R.B. Local neighborhood effects on long-term survival of individual trees in a neotropical forest. Ecol. Res. 2001, 16, 859–875. [Google Scholar] [CrossRef]
- Uriarte, M.; Condit, R.; Canham, C.D.; Hubbell, S.P. A spatially explicit model of sapling growth in a tropical forest: Does the identity of neighbours matter? J. Ecol. 2004, 92, 348–360. [Google Scholar] [CrossRef]
- Wiegand, T.; Gunatilleke, S.; Gunatilleke, N. Species associations in a heterogeneous Sri lankan dipterocarp forest. Am. Nat. 2007, 170, E77–E95. [Google Scholar] [CrossRef]
- Lan, G.; Getzin, S.; Wiegand, T.; Hu, Y.; Xie, G.; Zhu, H.; Cao, M. Spatial Distribution and Interspecific Associations of Tree Species in a Tropical Seasonal Rain Forest of China. PLoS ONE 2012, 7, e46074. [Google Scholar] [CrossRef]
- Aiba, S.; Kitayama, K.; Takyu, M. Habitat associations with topography and canopy structure of tree species in a tropical montane forest on Mount Kinabalu, Borneo. Plant Ecol. 2004, 174, 147–161. [Google Scholar] [CrossRef]
- Punchi-Manage, R.; Wiegand, T.; Wiegand, K.; Getzin, S.; Gunatilleke, C.S.; Gunatilleke, I.N. Effect of spatial processes and topography on structuring species assemblages in a Sri Lankan dipterocarp forest. Ecology 2014, 95, 376–386. [Google Scholar] [CrossRef]
- Lai, J.; Mi, X.; Ren, H.; Ma, K. Species-habitat associations change in a subtropical forest of China. J. Veg. Sci. 2009, 20, 415–423. [Google Scholar] [CrossRef]
- Illian, J.; Penttinen, A.; Stoyan, H.; Stoyan, D. Statistical Analysis and Modelling of Spatial Point Patterns; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 70. [Google Scholar]
- Wiegand, T.; Moloney, K.A. Handbook of Spatial Point-Pattern Analysis in Ecology; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Law, R.; Illian, J.; Burslem, D.F.; Gratzer, G.; Gunatilleke, C.V.S.; Gunatilleke, I.A.U.N. Ecological information from spatial patterns of plants: Insights from point process theory. J. Ecol. 2009, 97, 616–628. [Google Scholar] [CrossRef]
- Pacala, S.W.; Levin, S.A. Biologically generated spatial pattern and the coexistence of competing species. In Spatial Ecology: The Role of Space in Population Dynamics and Interspecific Interactions; Princeton University Press: Princeton, NJ, USA, 1997; pp. 204–232. [Google Scholar]
- Stoll, P.; Prati, D. Intraspecific aggregation alters competitive interactions in experimental plant communities. Ecology 2001, 82, 319–327. [Google Scholar] [CrossRef]
- McGill, B.J. Towards a unification of unified theories of biodiversity. Ecol. Lett. 2010, 13, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Getzin, S.; Wiegand, T.; Wiegand, K.; He, F. Heterogeneity influences spatial patterns and demographics in forest stands. J. Ecol. 2008, 96, 807–820. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Erfanifard, Y.; Pham, V.D.; Le, X.T.; Petritan, I.C. Spatial Association and Diversity of Dominant Tree Species in Tropical Rainforest, Vietnam. Forests 2018, 9, 615. [Google Scholar] [CrossRef]
- Tichý, L. Field test of canopy cover estimation by hemispherical photographs taken with a smartphone. J. Veg. Sci. 2016, 27, 427–435. [Google Scholar] [CrossRef]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: Princeton, NJ, USA, 1988. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Heip, C.H.; Herman, P.M.; Soetaert, K. Indices of diversity and evenness. Oceanis 1998, 24, 61–88. [Google Scholar]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; Ver. 3.5. 1; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Oksanen, J. Vegan: An Introduction to Ordination. Available online: http://cran.r-project.org/web/packages/vegan/vignettes/introvegan.pdf (accessed on 1 December 2020).
- Ripley, B.D. Modelling spatial patterns. J. R. Stat. Soc. Ser. B (Methodol.) 1977, 39, 172–212. [Google Scholar] [CrossRef]
- Wiegand, T.; Moloney, K. Rings, circles, and null-models for point pattern analysis in ecology. Oikos 2004, 104, 209–229. [Google Scholar] [CrossRef]
- Comita, L.S.; Condit, R.; Hubbell, S.P. Developmental changes in habitat associations of tropical trees. J. Ecol. 2007, 95, 482–492. [Google Scholar] [CrossRef]
- Wang, Z.; Ye, W.; Cao, H.; Huang, Z.; Lian, J.; Li, L.; Wei, S.; Sun, I.F. Species–topography association in a species-rich subtropical forest of China. Basic Appl. Ecol. 2009, 10, 648–655. [Google Scholar] [CrossRef]
- Gentry, A.H. Changes in plant community diversity and floristic composition on environmental and geographical gradients. Ann. Mo. Bot. Gard. 1988, 1–34. [Google Scholar] [CrossRef]
- Paoli, G.D.; Curran, L.M.; Zak, D.R. Soil nutrients and beta diversity in the Bornean Dipterocarpaceae: Evidence for niche partitioning by tropical rain forest trees. J. Ecol. 2006, 94, 157–170. [Google Scholar] [CrossRef]
- Prada, C.M.; Stevenson, P.R. Plant composition associated with environmental gradients in tropical montane forests (Cueva de Los Guacharos National Park, Huila, Colombia). Biotropica 2016, 48, 568–576. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, B.; Li, D.; Mallik, A.U.; Xiang, W.; Ding, T.; Wen, S.; Lu, S.; Huang, F.; He, Y.; et al. Effects of topography and spatial processes on structuring tree species composition in a diverse heterogeneous tropical karst seasonal rainforest. Flora 2017, 231, 21–28. [Google Scholar] [CrossRef]
- Lasky, J.R.; Sun, I.F.; Su, S.H.; Chen, Z.S.; Keitt, T.H. Trait-mediated effects of environmental filtering on tree community dynamics. J. Ecol. 2013, 101, 722–733. [Google Scholar] [CrossRef]
- Lan, G.; Hu, Y.; Cao, M.; Zhu, H. Topography related spatial distribution of dominant tree species in a tropical seasonal rain forest in China. For. Ecol. Manag. 2011, 262, 1507–1513. [Google Scholar] [CrossRef]
- Liu, J.; Yunhong, T.; Slik, J.F. Topography related habitat associations of tree species traits, composition and diversity in a Chinese tropical forest. For. Ecol. Manag. 2014, 330, 75–81. [Google Scholar] [CrossRef]
- Raventós, J.; Wiegand, T.; Luis, M.D. Evidence for the spatial segregation hypothesis: A test with nine-year survivorship data in a Mediterranean shrubland. Ecology 2010, 91, 2110–2120. [Google Scholar] [CrossRef]
- Wang, X.; Wiegand, T.; Hao, Z.; Li, B.; Ye, J.; Lin, F. Species associations in an old-growth temperate forest in north-eastern China. J. Ecol. 2010, 98, 674–686. [Google Scholar] [CrossRef]
- Velázquez, E.; Paine, C.T.; May, F.; Wiegand, T. Linking trait similarity to interspecific spatial associations in a moist tropical forest. J. Veg. Sci. 2015, 26, 1068–1079. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, H.; Shu, X.; Xie, F.; Zhang, K.; Zhang, Q.; Dang, H. Spatial distribution and interspecific associations in a deciduous broad-leaved forest in north-central China. J. Veg. Sci. 2019, 30, 1153–1163. [Google Scholar] [CrossRef]
- Chesson, P. General theory of competitive coexistence in spatially-varying environments. Theor. Popul. Biol. 2000, 58, 211–237. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.A. Neighbour-regulated mortality: The influence of positive and negative density dependence on tree populations in species-rich tropical forests. Ecol. Lett. 2003, 6, 757–765. [Google Scholar] [CrossRef]
- Pacala, S.W. Dynamics of plant communities. Plant Ecol. 1997, 532–555. [Google Scholar]


| Type of Association | Function |
|---|---|
| Segregation (i.e., less individuals of species 2 on average around species 1 than expected by chance) | |
| Partial overlap (i.e., frequent individuals of species 2 around some individuals of species 1, despite others) | |
| Mixing (i.e., frequent individuals of species 2 around individuals of species 1) | |
| Type IV (i.e., overlapping of individuals of species 2 on dense cluster of individuals of species 1 and 2) |
| Characteristics (Mean ± SD) | P1 | P2 | p-Value for the Test | ||
|---|---|---|---|---|---|
| Levene’s | Post Hoc Scheffe | Mann–Whitney | |||
| Elevation (m) | 134.30 ± 6.32 a | 160.67 ± 11.09 b | <0.001 | <0.001 | |
| Slope (°) | 20.22 ± 6.61 a | 26.6 ± 7.54 b | 0.203 | <0.001 | |
| Aspect (°) | 87.51 ± 55.46 a | 91.08 ± 47.96 a | <0.001 | 0.413 | |
| Species richness | 11.39 ± 2.38 a | 11.48 ± 2.68 a | 0.013 | 0.533 | |
| Abundance | 19.67 ± 5.19 a | 18.69 ± 6.35 b | 0.018 | 0.025 | |
| Canopy cover (%) | 83.38 ± 4.52 a | 81.01 ± 7.37 b | <0.001 | <0.001 | |
| Shannon index | 2.22 ± 0.36 a | 2.29 ± 0.29 b | 0.605 | 0.035 | |
| Simpson index | 0.87 ± 0.04 a | 0.88 ± 0.03 b | 0.258 | 0.024 | |
| Evenness index | 0.85 ± 0.07 a | 0.88 ± 0.06 b | 0.391 | 0.002 | |
| Total basal area (m2) | 0.24 ± 0.15 a | 0.32 ± 0.18 b | <0.001 | <0.001 | |
| AGB (Mg) | 1559.27 ± 1406.18 a | 2152.42 ± 1545.60 b | 0.005 | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hai, N.H.; Erfanifard, Y.; Bui, V.B.; Mai, T.H.; Petritan, A.M.; Petritan, I.C. Topographic Effects on the Spatial Species Associations in Diverse Heterogeneous Tropical Evergreen Forests. Sustainability 2021, 13, 2468. https://doi.org/10.3390/su13052468
Hai NH, Erfanifard Y, Bui VB, Mai TH, Petritan AM, Petritan IC. Topographic Effects on the Spatial Species Associations in Diverse Heterogeneous Tropical Evergreen Forests. Sustainability. 2021; 13(5):2468. https://doi.org/10.3390/su13052468
Chicago/Turabian StyleHai, Nguyen Hong, Yousef Erfanifard, Van Bac Bui, Trinh Hien Mai, Any Mary Petritan, and Ion Catalin Petritan. 2021. "Topographic Effects on the Spatial Species Associations in Diverse Heterogeneous Tropical Evergreen Forests" Sustainability 13, no. 5: 2468. https://doi.org/10.3390/su13052468
APA StyleHai, N. H., Erfanifard, Y., Bui, V. B., Mai, T. H., Petritan, A. M., & Petritan, I. C. (2021). Topographic Effects on the Spatial Species Associations in Diverse Heterogeneous Tropical Evergreen Forests. Sustainability, 13(5), 2468. https://doi.org/10.3390/su13052468









