Microbiological Attributes and Performance of the Bacterial Community in Brazilian Cerrado Soil with Different Cover Crops
Abstract
:1. Introduction
2. Materials and Methods
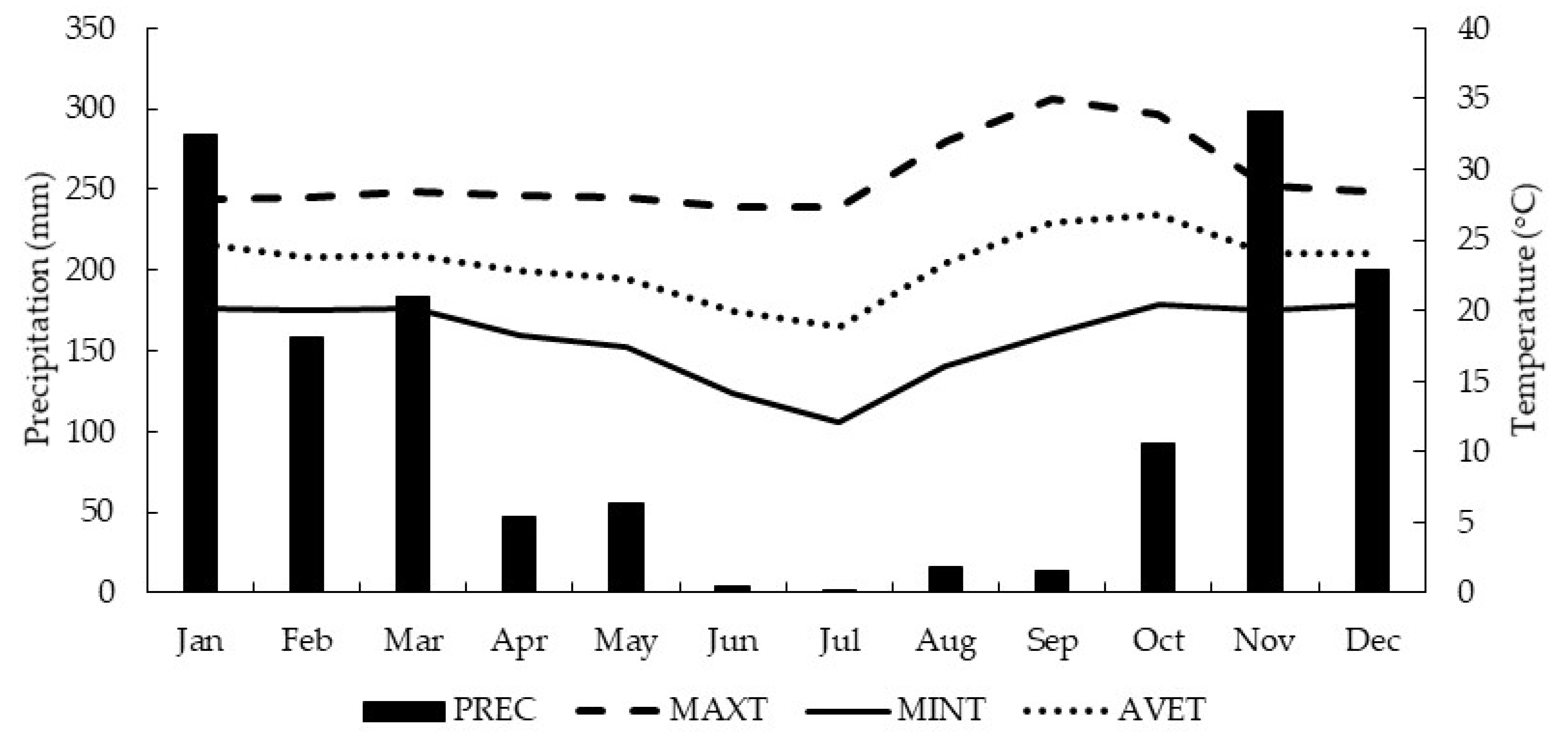
2.1. Experimental Characterization
2.2. Experimental Design
2.3. Soil Sampling for Microbiological Determination and Plant Biomass
2.4. Determination of Microbial Biomass Carbon (MBC), Basal Soil Respiration (BR), and Metabolic Quotient (qCO2)
2.5. Soil Total DNA Extraction
2.6. Abundance of the Bacterial Community by Quantitative PCR (qPCR)
2.7. Determination of the Structure of the Bacterial Community by the Terminal Restriction Fragment Length Polymorphism (T-RFLP)
2.8. Statistical Analysis
3. Results and Discussion
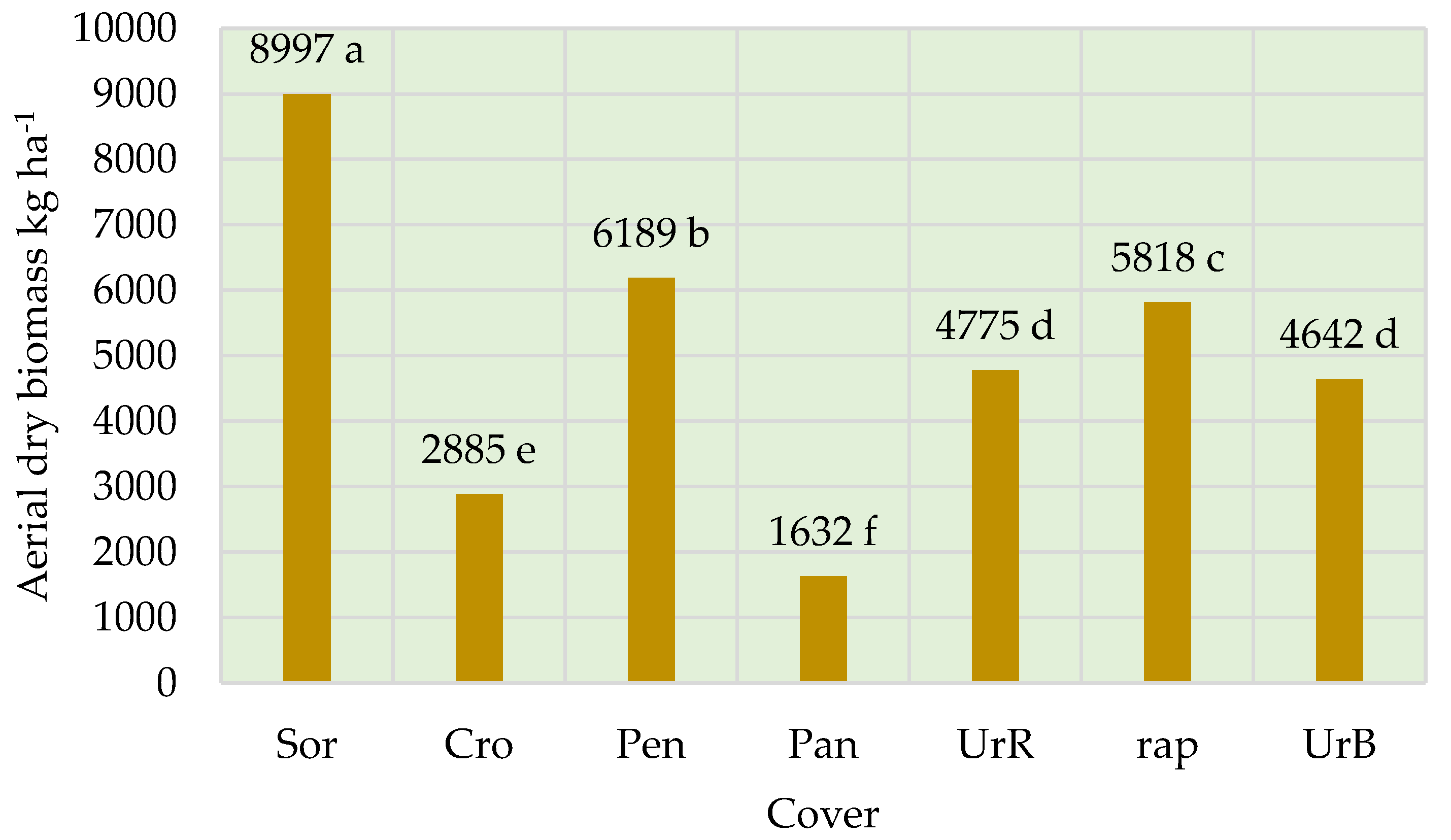
3.1. Cover Crops Biomass
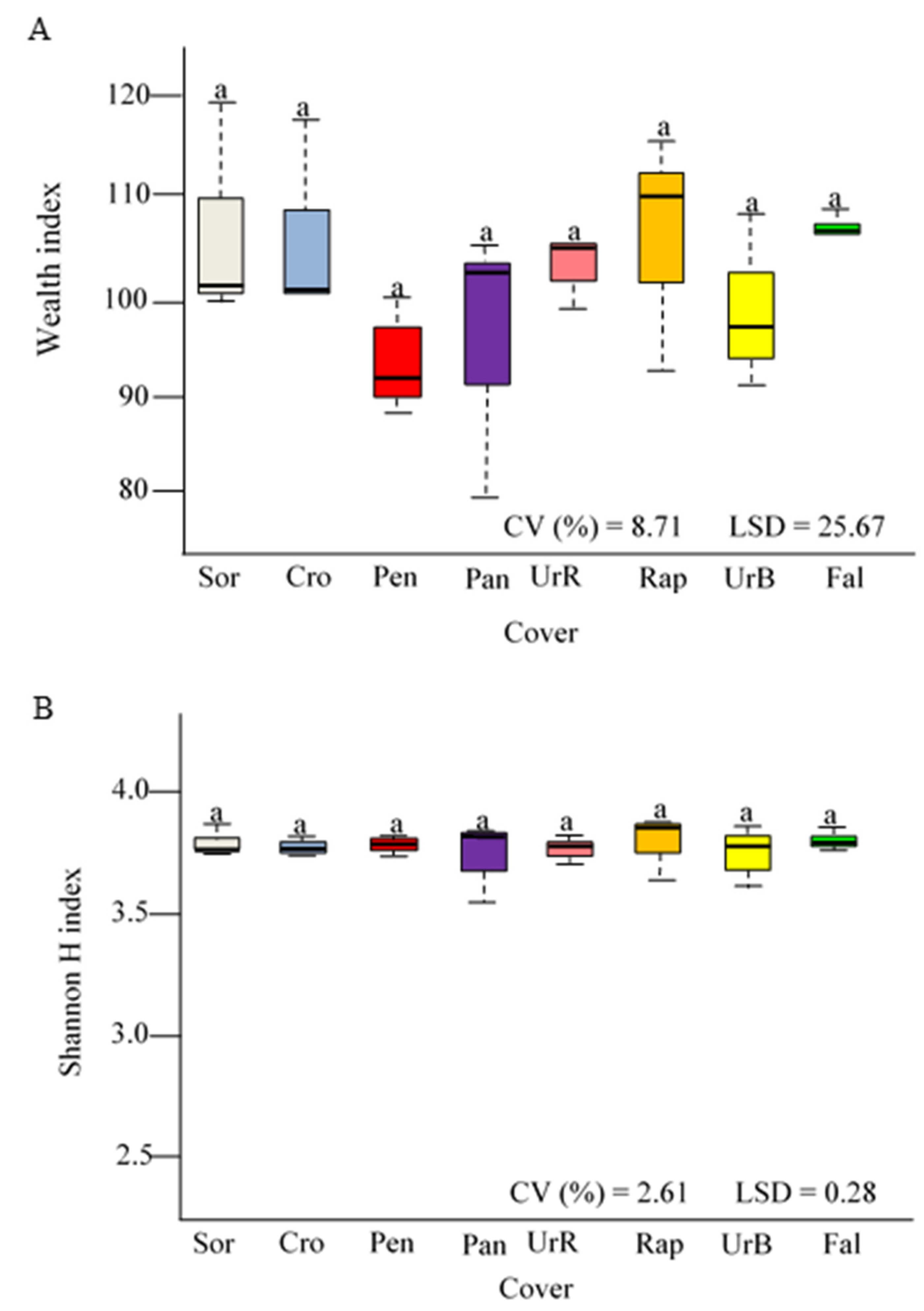
3.2. Microbial Biomass Carbon, Basal Soil Respiration, Metabolic Quotient, and Abundance of the Rrs Gene (16S rRNA)
3.3. Abundance of the Bacterial Community by Quantitative PCR (qPCR)
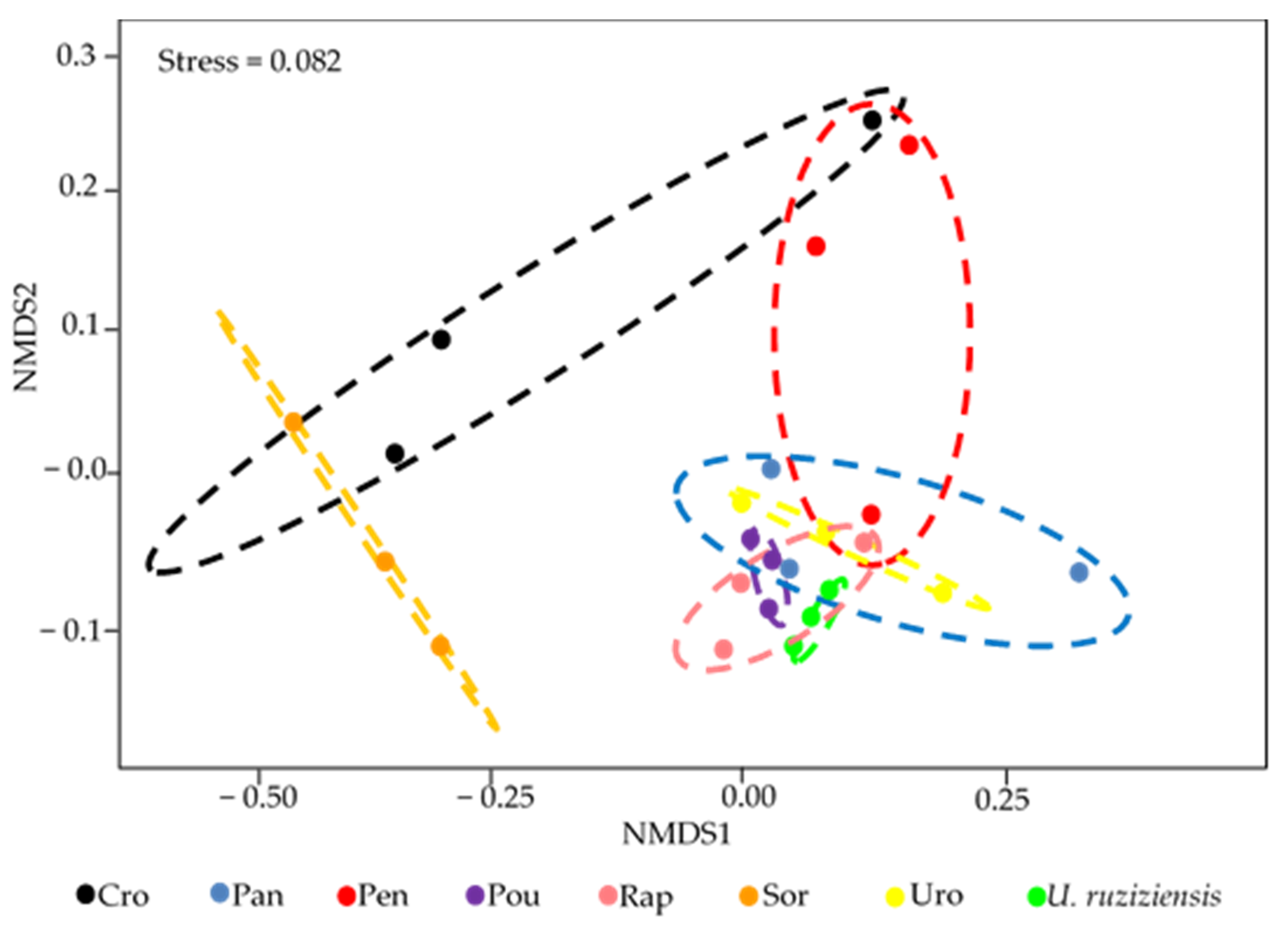
3.4. Changes in the Structure of the Bacterial Community
3.5. Correlation of the Structure of the Bacterial Community with the Microbiological Attributes Evaluated
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, X.; Fu, X.; Yang, Y.; Cai, P.; Peng, S.; Chen, W.; Huang, Q. Microbial communities play important roles in modulating paddy soil fertility. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Cardoso, E.J.B.N.; Vasconcellos, R.L.F.; Bini, D.; Miyauchi, M.Y.H.; Santos, C.A.D.; Alves, P.R.L.; De Paula, A.M.; Nakatani, A.S.; Pereira, J.M.I.; Nogueira, M.A. Soil health: Looking for suitable indicators. What should be considered to assess the effects of use and management on soil health? Sci. Agric. 2013, 70, 274–289. [Google Scholar] [CrossRef] [Green Version]
- Quiquampoix, H.; Servagent-Noiville, S.; Baron, M. Enzyme adsorption on soil mineral surfaces and consequences for the catalytic activity. In Enzymes in the Environment; Burns, R.G., Dick, R.P., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 285–306. [Google Scholar]
- Epelde, L.; Burges, A.; Mijangos, I.; Garbisu, C. Microbial properties and attributes of ecological relevance for soil quality monitoring during a chemical stabilization field study. Appl. Soil Ecol. 2014, 75, 1–12. [Google Scholar] [CrossRef]
- Ferreira, E.P.B.; Santos, H.P.; Costa, J.R.; De-Polli, H.; Rumjanek, N.G. Microbial soil quality indicators under different crop rotations and tillage management. Cienc. Agron. 2010, 41, 177–183. [Google Scholar] [CrossRef]
- Ferreira, E.P.B.; Wendland, A.; Didonet, A.D. Microbial biomass and enzyme activity of a Cerrado Oxisol under agroecological production system. Bragantia 2011, 70, 1–9. [Google Scholar] [CrossRef]
- Blaud, A.; Herrmann, A.M.; Diouf, F.; Lerch, T. Analysing the effect of soil organic matter on bacterial communities using T-RFLP fingerprinting: Different methods, different stories? Biol. Fertil. Soils 2015, 51, 959–971. [Google Scholar] [CrossRef] [Green Version]
- Bunemann, E.K.; Bongiorno, G.; Baic, Z.; Creamer, R.E.; De Deyn, G.; Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mader, P.; et al. Soil quality–A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Simon, C.A.; Lima, S.F.; Cordeiro, M.S.; Secco, V.A.; Nacata, G.; Silva, A.M.M.; Simon, C.C.; Brasil, M.S. Cover crops as modifying agents of microbiological soil attribute. Aust. J. Crop. Sci. 2019, 13, 1578–1585. [Google Scholar] [CrossRef]
- Soares, D.S.; Gerosa, M.L.; Marchão, R.L.; Maciel, G.A.; Oliveira, A.D.; Malaquias, J.V.; Carvalho, A.M.S. How diversity of crop residues in long-term no-tillage systems affect chemical and microbiological soil properties. Soil Tillage Res. 2019, 194, 1–12. [Google Scholar]
- Carvalho, A.M.; Coelho, M.C.; Dantas, R.A.; Fonseca, O.P.; Guimarães Júnior, R.; Figueiredo, C.C. Chemical composition of cover plants and its effect on maize yield in no-tillage systems in the Brazilian savanna. Crop. Pasture Sci. 2012, 63, 1075–1081. [Google Scholar] [CrossRef]
- Andreote, F.D.; Silva, M.C.P. Microbial communities associated with plants: Learning from nature to apply it in agriculture. Curr. Opin. Microbiol. 2017, 37, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, R.C.; Waldo, A.L.C.; Cruz, J.C.; Santana, D.P. Plantas de cobertura de solo para sistema plantio direto. Inf. Agropecu. 2001, 22, 25–36. [Google Scholar]
- Chenu, C.; Angers, D.A.; Barré, P.; Derrien, D.; Arrouays, D.; Balesdent, J. Increasing organic stocks in agricultural soils: Knowledge gaps and potential innovations. Soil Tillage Res. 2018, 167, 1–12. [Google Scholar] [CrossRef]
- Santos, H.G.; Jacomine, P.K.T.; Anjos, L.H.C.; Oliveira, V.A.; Lumbreras, J.F.; Coelho, M.R.; Almeida, J.A.; Araujo Filho, J.C.; Oliveira, J.B.; Cunha, T.J.F. Sistema Brasileiro de Classificação de Solos, 5th ed.; Embrapa: Brasilia, Brazil, 2018; pp. 195–212. [Google Scholar]
- Silva, F.C.D.S. (Ed.) Manual de Análises Químicas de Solos, Plantas e Fertilizantes; Embrapa: Rio de Janeiro, Brazil, 2009; pp. 107–190. [Google Scholar]
- Donagemma, G.K.; Campos, D.V.B.; Calderano, S.B.; Teixeira, W.G.; Viana, J.H.M. Manual de Métodos de Análise de Solo, 2nd ed.; Embrapa: Rio de Janeiro, Brazil, 2011; pp. 13–94. [Google Scholar]
- Vance, E.D.; Bookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 9, 703–707. [Google Scholar] [CrossRef]
- Ohlinger, R. Bestimmung der bodenatmung im laborversuch. In Bodenbiologische Arbeitsmethoden; Schinner, F., Ohlinger, R., Kandeler, E., Margesin, R., Eds.; Springer: Berlin, Germany, 1993; pp. 86–90. [Google Scholar]
- Anderson, T.H.; Domsch, K.H. The metabolic quotient for CO2 (qCO2) as a specific activit parameter to assess the effects of environmental conditions. such as pH. on the microbial biomass of forest soils. Soil Biol. Biochem. 1993, 25, 393–395. [Google Scholar] [CrossRef]
- Muyzer, G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, D.F. Sisvar: A computer statistical analysis system. Cinc. Agrotecnol. 2011, 35, 1039–1042. [Google Scholar] [CrossRef] [Green Version]
- Ling, N.; Chen, D.; Guo, H.; Wei, J.; Bai, Y.; Shen, Q.; Hu, S. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 2017, 292, 25–33. [Google Scholar] [CrossRef]
- Yang, L.; Wang, N.; Chen, Y.; Yang, W.; Tian, D.; Zhang, C.; Niu, S. Carbon management practices regulate soil bacterial communities in response to nitrogen addition in a pine forest. Plant Soil 2020, 452, 137–151. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O´Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Package ‘vegan’. Community Ecol. 2013, 2, 1–295. [Google Scholar]
- Doneda, A.; Aita, C.; Giacomini, S.J.; Miola, E.C.C.; Giacomini, D.A.; Schirmann, J.; Gonzato, R. Biomass and decomposition of cover crop residues in monoculture and intercropping. Rev. Bras. Cienc. Solo 2012, 36, 1714–1723. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, D.M.P.; Spehar, C.R. Decomposição da Palhada de Culturas Para Plantio no Período de Safrinha em Solos de Tabuleiros Costeiros; Embrapa Meio-Norte: Teresina, Brazil, 2002; pp. 1–4. [Google Scholar]
- Silva, P.C.G.; Foloni, J.S.S.; Fabris, L.B.; Tiritan, C.S. Fitomassa e relação C/N em consórcios de sorgo e milho com espécies de cobertura. Pesqui. Agropecu. Bras. 2009, 44, 1504–1512. [Google Scholar] [CrossRef]
- Roscoe, R.; Mercante, F.M.; Salton, J.C. Dinâmica da Matéria Orgânica do Solo em Sistemas Conservacionistas: Modelagem Matemática e Métodos Auxiliares; Embrapa Agropecuária Oeste: Dourados, Brazil, 2006; pp. 17–41. [Google Scholar]
- Hungria, M.; Franchini, J.C.; Brandão-Júnior, O.; Kaschuk, G.; Souza, R.A. Soil microbial activity and crop sustainability in a long-term experiment with three soil-tillage and two crop-rotation systems. Appl. Soil Ecol. 2009, 42, 288–296. [Google Scholar] [CrossRef]
- Teixeira, C.M.; Carvalho, G.J.; Andrade, M.J.B.; Silva, C.A.; Pereira, J.M. Decomposição e liberação de nutrientes das palhadas de milheto e milheto + crotalária no plantio direto do feijoeiro. Acta Sci. Agron. 2009, 31, 647–653. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, N.F.; Gallo, A.S.; Fontanetti, A.; Meneghin, S.P.; Souza, M.D.B.; Morinigo, K.P.G.; Silva, R.F. Biomassa e atividade microbiana do solo em diferentes sistemas de cultivo do cafeeiro. Rev. Ciênc. Agrárias 2017, 40, 34–44. [Google Scholar] [CrossRef]
- Silva, J.M.; Alburquerque, L.S.D.; Santos, T.M.C.D.; Oliveira, J.U.L.D.; Guedes, E.L.F. Mineralização de vermicompostos estimada pela respiração microbiana. Rev. Verde Agroecol. Desenv. Sustent. 2013, 8, 132–135. [Google Scholar]
- Souza, E.D.; Costa, S.E.V.G.A.; Anghinoni, I.; Lima, C.V.S.; Carvalho, P.C.F.V.; Martins, A.P. Biomassa microbiana do solo em sistema de integração lavoura-pecuária em plantio direto. submetido a intensidades de pastejo. Rev. Bras. Cienc. Solo 2010, 34, 79–88. [Google Scholar] [CrossRef]
- Carneiro, M.A.C.; Souza, E.D.; Reis, E.F.; Pereira, H.S.; Azevedo, W.R. Atributos físicos, químicos e biológicos de solo de Cerrado sob diferentes sistemas de uso e manejo. Rev. Bras. Cienc. Solo 2009, 33, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Alves, T.S.; Campos, A.L.L.; Elias Neto, N.; Matsuoka, M.; Loureiro, M.F. Biomassa e atividade microbiana de solo sob vegetação nativa e diferentes sistemas de manejos. Acta Sci. Agron. 2011, 33, 341–347. [Google Scholar] [CrossRef]
- Schmidt, R.O.; Sana, R.S.; Leal, F.K.; Andreazza, R.; Camargo, F.A.O.; Meurer, E.J. Biomassa e atividade microbiana do solo em sistemas de produção olerícola orgânica e convencional. Cienc. Rural. 2013, 43, 270–276. [Google Scholar] [CrossRef]
- Schmidt, R.G.K.; Bossange, A.V.; Mitchell, J.; Scow, K. Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS ONE 2018, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analysis of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Clarke, K.R. Detecting change in benthic community structure. In Proceedings of the 14th International Biometric Conference, Namour, Belgium, 1988; Oger, R., Ed.; Société Adolphe Quiételet: Gembloux, Belgium; pp. 131–142.
- Clarke, K.R.; Gorley, R.N. Primer V5: User Manual/Tutorial; Primer-E Ltd.: Plymouth, New Zealand, 2001. [Google Scholar]
- Schlemper, T.R.; Leite, M.F.A.; Lucheta, A.R.; Shimels, M.; Bouwmeester, H.J.; Veen, J.A.; Kuramae, E.E. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 2017, 93, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romdhane, S.; Spor, A.; Busset, H.; Falchetto, L.; Martin, J.; Bizouard, F.; Bru, D.; Breuil, M.; Philippot, L.; Cordeau, S. Cover crop management practices rather than composition of cover crop mixtures affect bacterial communities in no-till agroecosystems. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Madari, B.E.; Coutinho, H.L.C.; Machado, P. Soil aggregation and bacterial community structure as affected by tillage and cover cropping in the Brazilian Cerrados. Soil Tillage Res. 2006, 90, 16–28. [Google Scholar] [CrossRef]
- Lannes, L.S.; Bustamante, M.M.C.; Edwards, P.J.; Venterink, H.O. Native and alien herbaceous plants in the Brazilian Cerrado are (co-) limited by different nutrients. Plant Soil 2016, 400, 231–243. [Google Scholar] [CrossRef] [Green Version]
| Cover Crop | Clay | Silt | Sand | OM | pH | P | K | Zn | Ca | Mg | H + Al | CEC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | CaCl2 | mg·dm−3 | cmolc·dm−3 | |||||||||
| Sorghum bicolor | 41.0 | 19.0 | 40.0 | 2.9 | 5.1 | 14.8 | 53.0 | 9.5 | 2.3 | 1.16 | 3.5 | 7.1 |
| Crotalaria ochroleuca | 40.0 | 20.0 | 40.0 | 3.0 | 6.0 | 17.4 | 71.0 | 6.5 | 4.0 | 1.42 | 3.0 | 7.5 |
| Pennisetum americanum | 41.0 | 17.0 | 42.0 | 3.2 | 6.2 | 12.4 | 78.0 | 5.5 | 4.8 | 1.65 | 1.7 | 8.3 |
| Panicum miliaceum | 42.0 | 20.0 | 38.0 | 3.0 | 5.8 | 21.2 | 70.0 | 5.7 | 3.1 | 1.47 | 2.3 | 7.0 |
| Urochloa ruziziensis | 38.0 | 11.0 | 51.0 | 2.7 | 5.6 | 20.6 | 86.0 | 8.1 | 3.2 | 1.63 | 1.9 | 6.9 |
| Raphanus sativus | 39.0 | 19.0 | 42.0 | 2.3 | 5.4 | 17.3 | 53.0 | 6.7 | 2.9 | 1.21 | 2.8 | 7.0 |
| Urochloabrizantha | 38.0 | 20.0 | 42.0 | 2.9 | 5.4 | 25.1 | 71.0 | 7.4 | 3.4 | 1.54 | 2.3 | 7.4 |
| Fallow | 44.0 | 18.0 | 38.0 | 2.6 | 5.8 | 24.4 | 65.0 | 7.2 | 3.9 | 1.9 | 2.1 | 8.1 |
| Cover Crop | MBC µg Microbial C G−1 Soil | C-CO2 100 G Soil−1 | Qco2 | 16S Rrna N° of 16S Rrna Gene Copies G−1 Soil |
|---|---|---|---|---|
| Sorghum bicolor | 199.52a | 120.40c | 6.04d | 1.61 × 108b |
| Crotalaria ochroleuca | 187.98b | 123.20c | 6.56d | 1.18 × 108c |
| Pennisetum americanum | 152.93c | 84.30e | 5.52d | 2.38 × 108a |
| Panicum miliaceum | 90.09e | 67.41f | 7.50c | 9.83 × 107c |
| Urochloa ruziziensis | 71.93f | 142.27a | 19.79a | 1.42 × 108b |
| Raphanus sativus | 83.46e | 129.47b | 15.57b | 1.07 × 108c |
| Urochloabrizantha | 123.06d | 102.30d | 8.33c | 1.15 × 108c |
| Fallow | 180.85b | 119.17c | 6.59d | 3.95 × 107d |
| S. bicolor | C. ochroleuca | P. americanum | P. miliaceum | U. ruziziensis | R. sativus | U. brizantha | Fallow | |
|---|---|---|---|---|---|---|---|---|
| S. bicolor | 0 | |||||||
| C. ochroleuca | 0.22 ns | 0 | ||||||
| P. americanum | 1 ns | 0.25 ns | 0 | |||||
| P. miliaceum | 1 ns | 0.37 ns | 0.44 ns | 0 | ||||
| U. ruziziensis | 1 ns | 0.33 ns | 0.70 ns | 0.18 ns | 0 | |||
| R. sativus | 0.96 ns | 0.33 ns | 0.51 ns | −0.33 ns | 0.22 ns | 0 | ||
| U. brizantha | 1 ns | 0.33 ns | 0.48 ns | −0.33 ns | 0.40 ns | −0.40 ns | 0 | |
| Fallow | 1 ns | 0.33 ns | 0.77 ns | 0.18 ns | 1 ns | 0.11 ns | −0.07 ns | 0 |
| MBC | BR | qCO2 | qPCR | ADB | |
|---|---|---|---|---|---|
| Sorghum bicolor | 0.50 ns | 1.00 ** | −0.50 ns | 0.50 ns | −0.50 ns |
| Pennisetum americanum | 0.87 ns | −0.87 ns | −0.87 ns | −0.87 ns | 0.00 ns |
| Panicum miliaceum | 0.50 ns | 1.00 ** | −0.50 ns | 1.00 ** | −1.00 ** |
| Urochloa Ruziziensis | 1.00 ** | −0.50 ns | −1.00 ** | 0.50 ns | 0.50 ns |
| Raphanus sativus | 0.50 ns | 0.50 ns | −0.50 ns | −0.50 ns | −1.00 ** |
| Urochloa brizantha | 1.00 ** | 0.00 ns | −1.00 ** | −0.50 ns | −1.00 ** |
| Fallow | −1.00 ** | −0.50 ns | 1.00 ** | 0.50 ns | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Lima, S.F.; Secco, V.A.; Simon, C.A.; Silva, A.M.M.; Vendruscolo, E.P.; Andrade, M.G.d.O.; Contardi, L.M.; de Lima, A.P.L.; Cordeiro, M.A.S.; Abreu, M.S. Microbiological Attributes and Performance of the Bacterial Community in Brazilian Cerrado Soil with Different Cover Crops. Sustainability 2021, 13, 8318. https://doi.org/10.3390/su13158318
de Lima SF, Secco VA, Simon CA, Silva AMM, Vendruscolo EP, Andrade MGdO, Contardi LM, de Lima APL, Cordeiro MAS, Abreu MS. Microbiological Attributes and Performance of the Bacterial Community in Brazilian Cerrado Soil with Different Cover Crops. Sustainability. 2021; 13(15):8318. https://doi.org/10.3390/su13158318
Chicago/Turabian Stylede Lima, Sebastião Ferreira, Vinicius Andrade Secco, Cátia Aparecida Simon, Antônio Marcos Miranda Silva, Eduardo Pradi Vendruscolo, Maria Gabriela de Oliveira Andrade, Lucymara Merquides Contardi, Ana Paula Leite de Lima, Meire Aparecida Silvestrini Cordeiro, and Mariele Silva Abreu. 2021. "Microbiological Attributes and Performance of the Bacterial Community in Brazilian Cerrado Soil with Different Cover Crops" Sustainability 13, no. 15: 8318. https://doi.org/10.3390/su13158318
APA Stylede Lima, S. F., Secco, V. A., Simon, C. A., Silva, A. M. M., Vendruscolo, E. P., Andrade, M. G. d. O., Contardi, L. M., de Lima, A. P. L., Cordeiro, M. A. S., & Abreu, M. S. (2021). Microbiological Attributes and Performance of the Bacterial Community in Brazilian Cerrado Soil with Different Cover Crops. Sustainability, 13(15), 8318. https://doi.org/10.3390/su13158318






