Addition of Activated Carbon into a Cattle Diet to Mitigate GHG Emissions and Improve Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Measure Animal Responses on Farm
2.1.1. Location and Animals
2.1.2. Powdered Activated Carbon (PAC)
2.1.3. Experimental Design
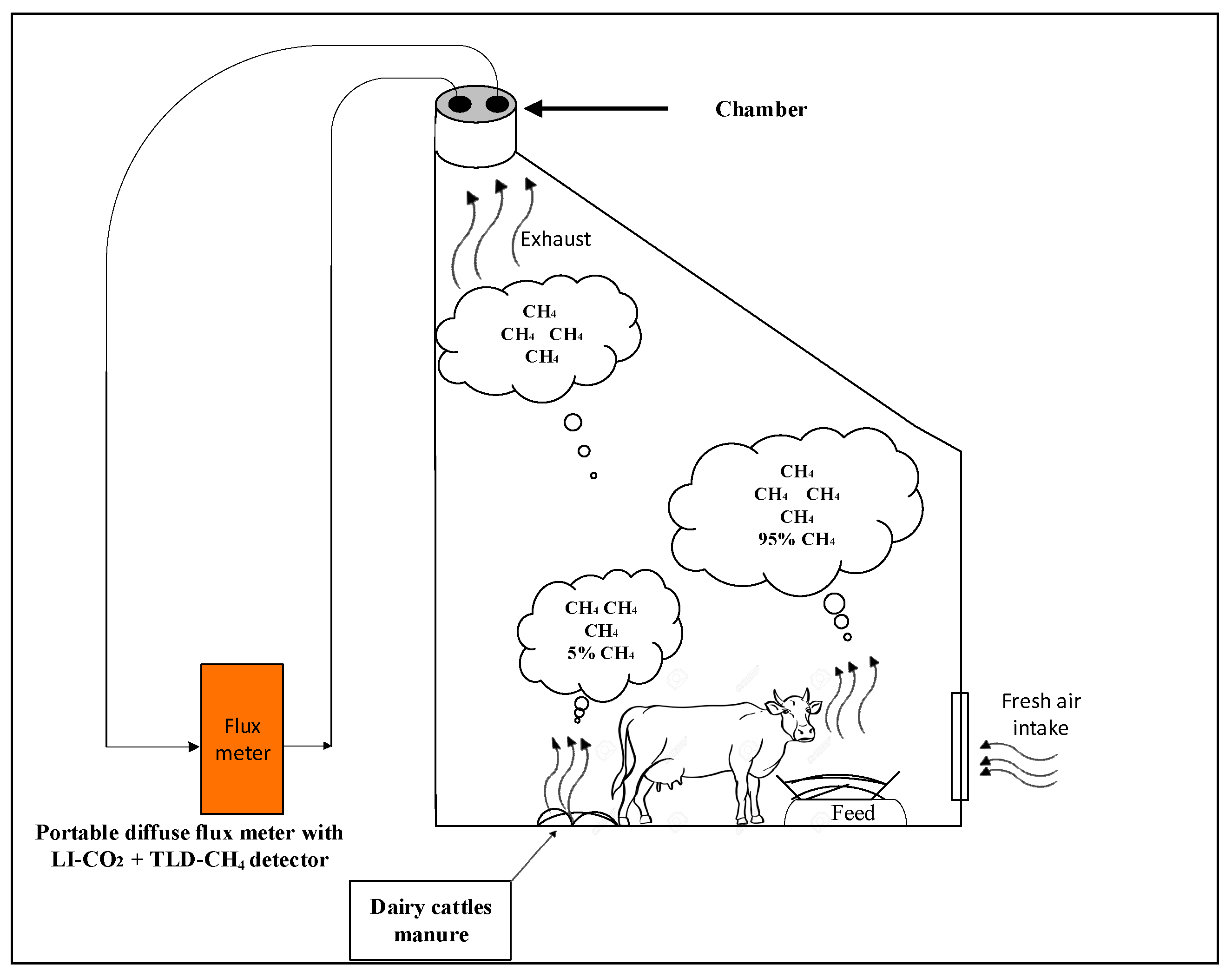
2.1.4. Measuring GHG Emissions before, during and after Milking Dairy Cattle
2.2. Measuring GHG Emissions from the Manure
2.2.1. Measurement of CO2 and CH4 Emissions
2.2.2. Manure Collection
2.2.3. Archaeal and Bacterial 16S rRNA Gene Sequencing
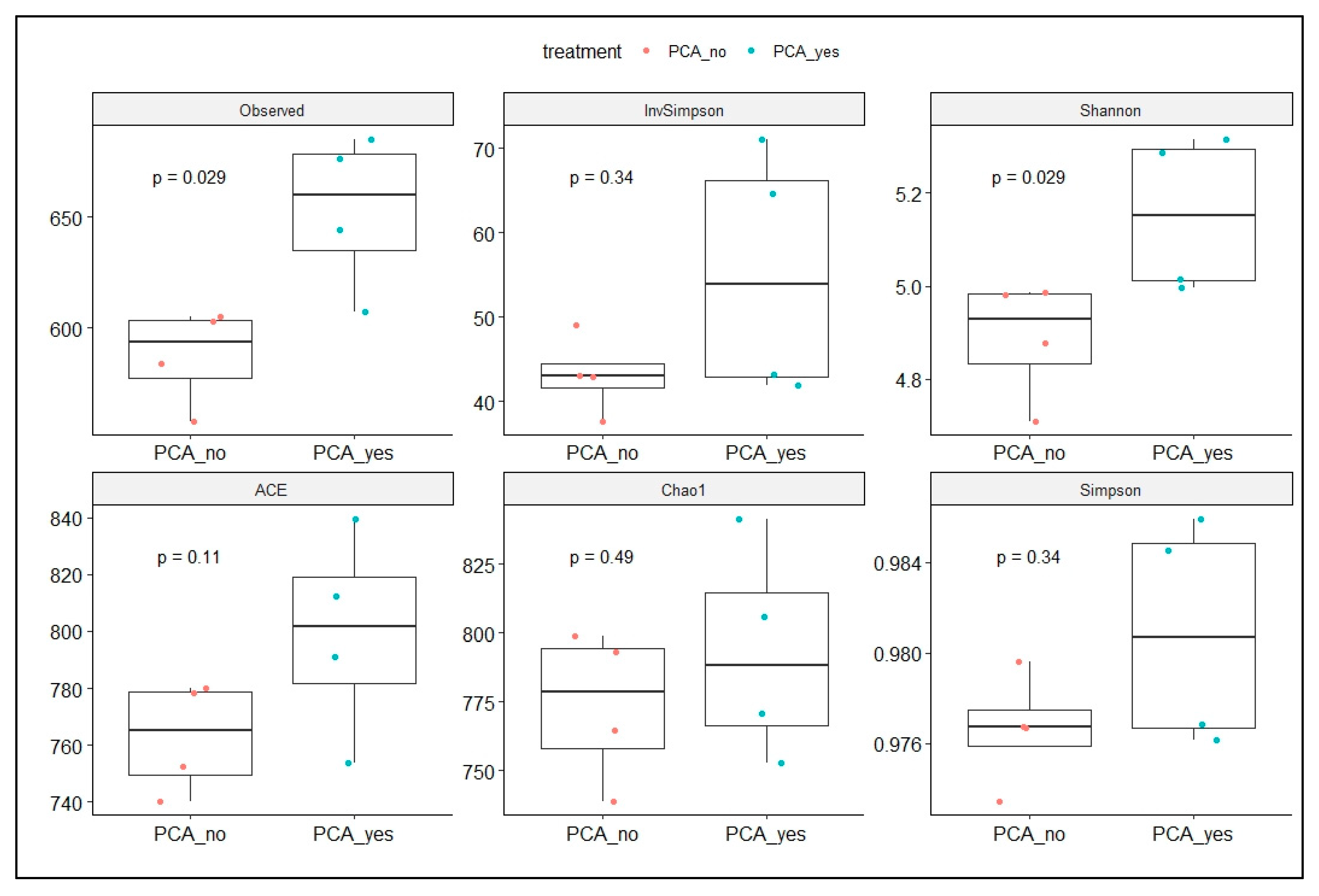
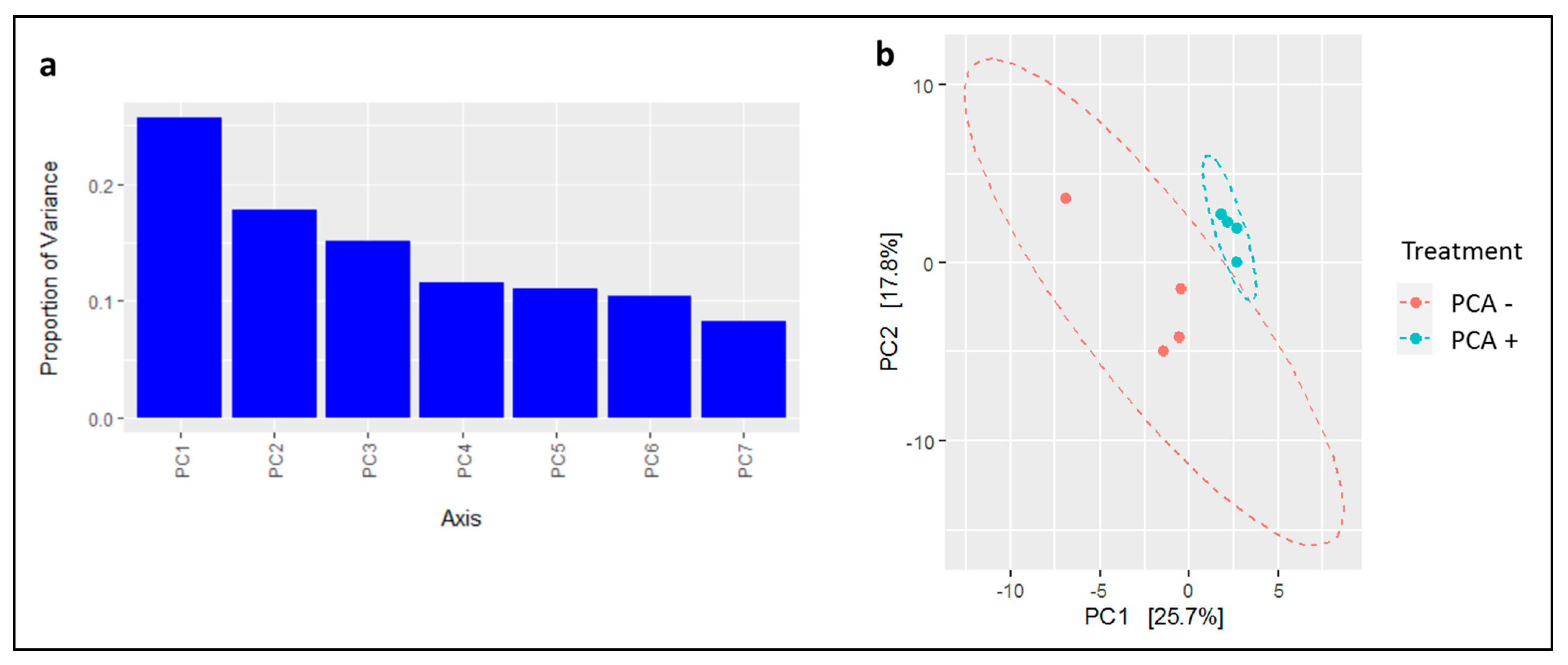
2.2.4. Microbiome Beta and Alpha Diversity Analysis
2.3. Statistical Analysis
3. Experimental Results
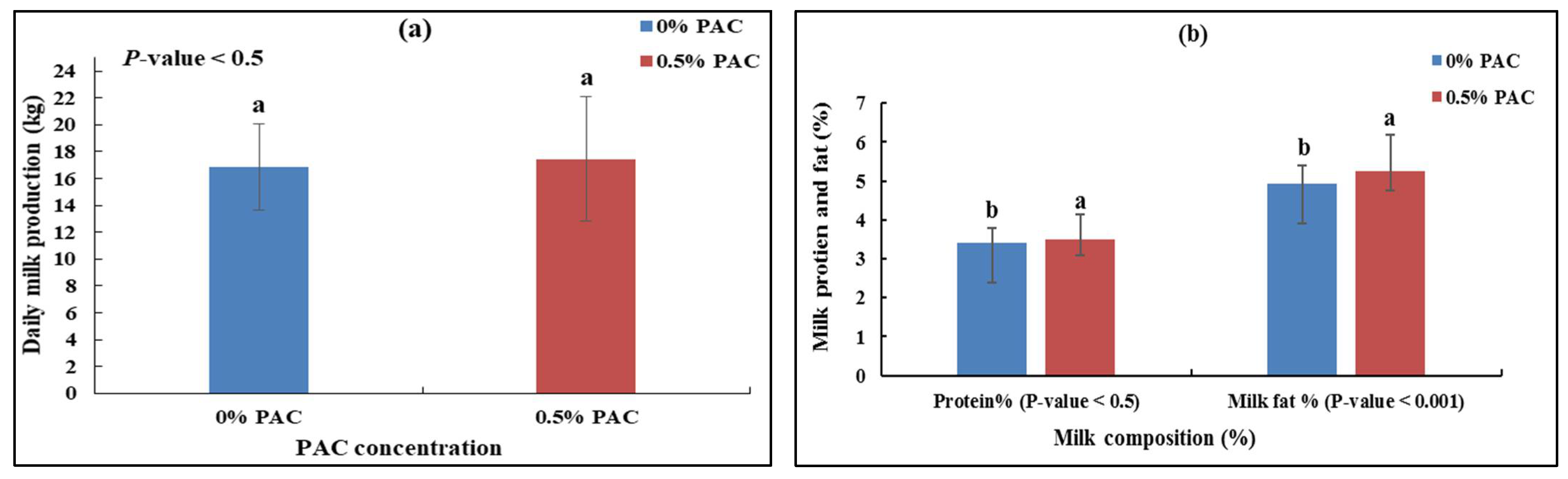
3.1. The Effect of PAC on Dairy Milk Production
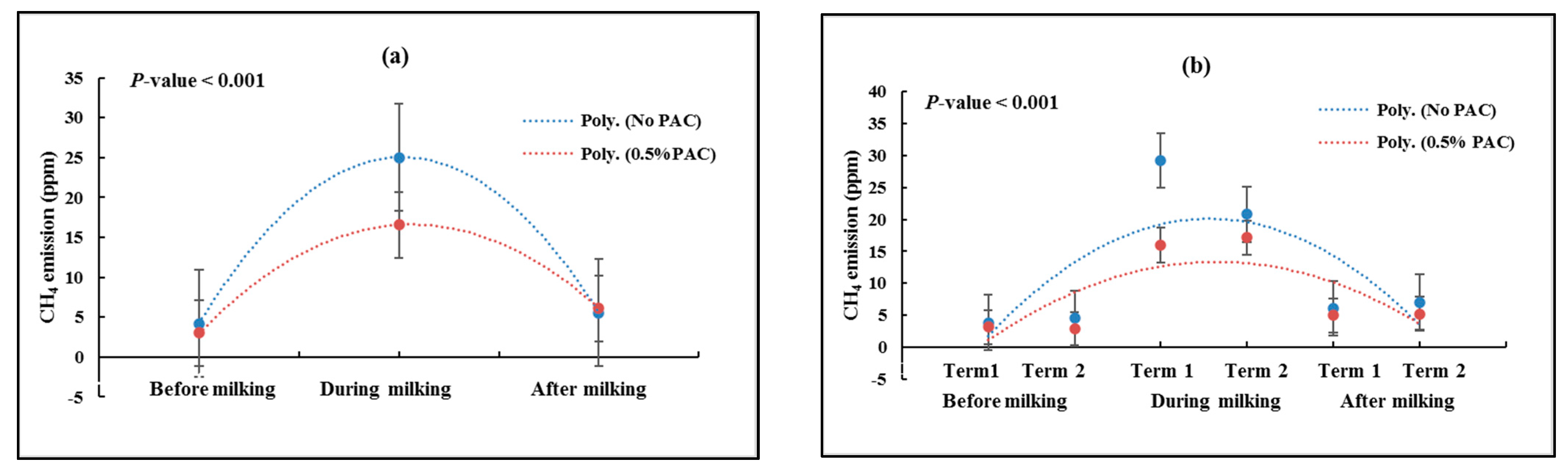
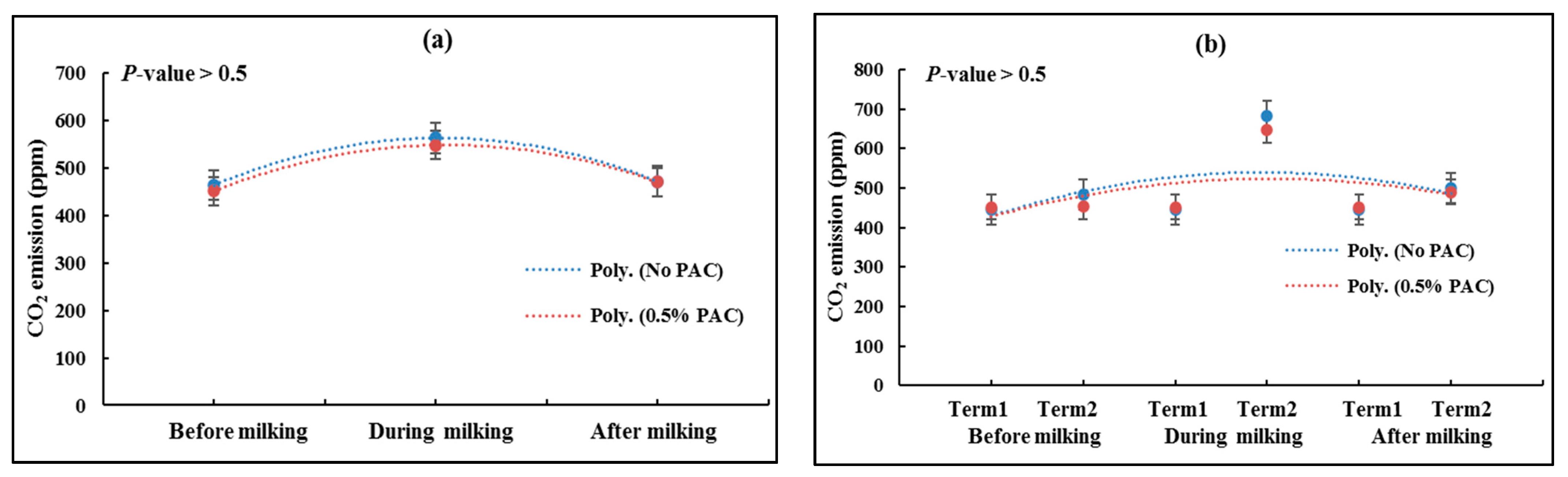
3.2. Measuring Total GHG Emissions before, during and after Milking
3.2.1. Methane Emissions (CH4)
3.2.2. CO2 Emissions
3.3. Measuring Total GHG Emissions and Chemical Compounds in Dairy Cattle’s Manure
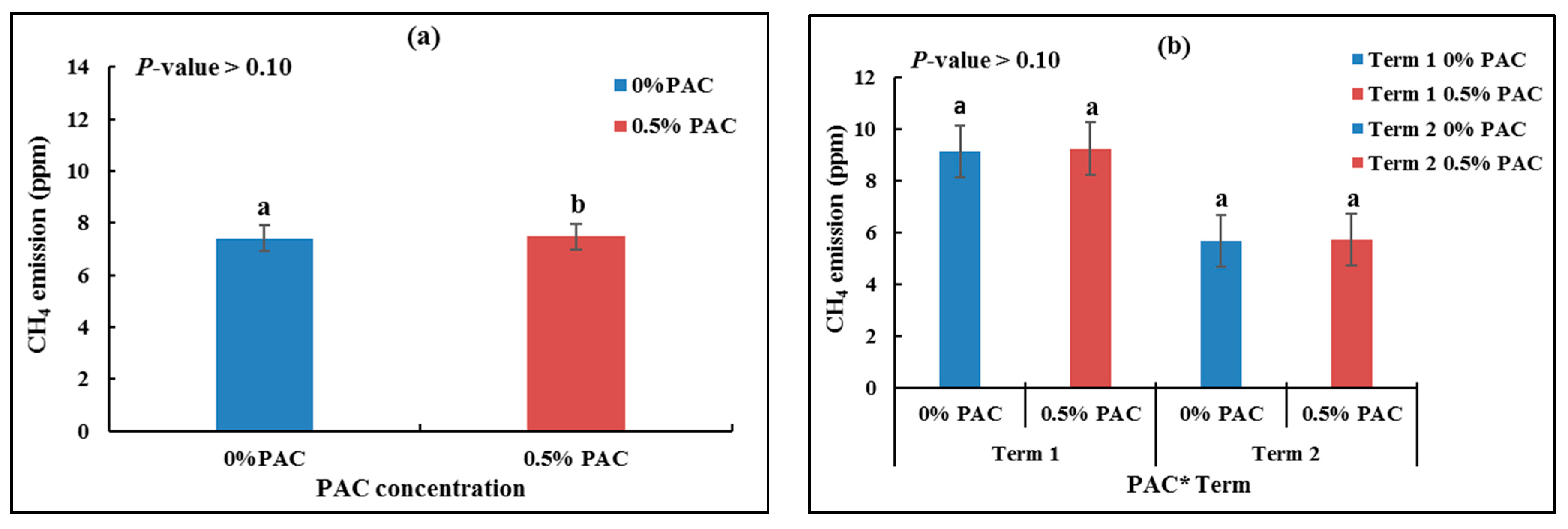
3.3.1. CH4 Emissions
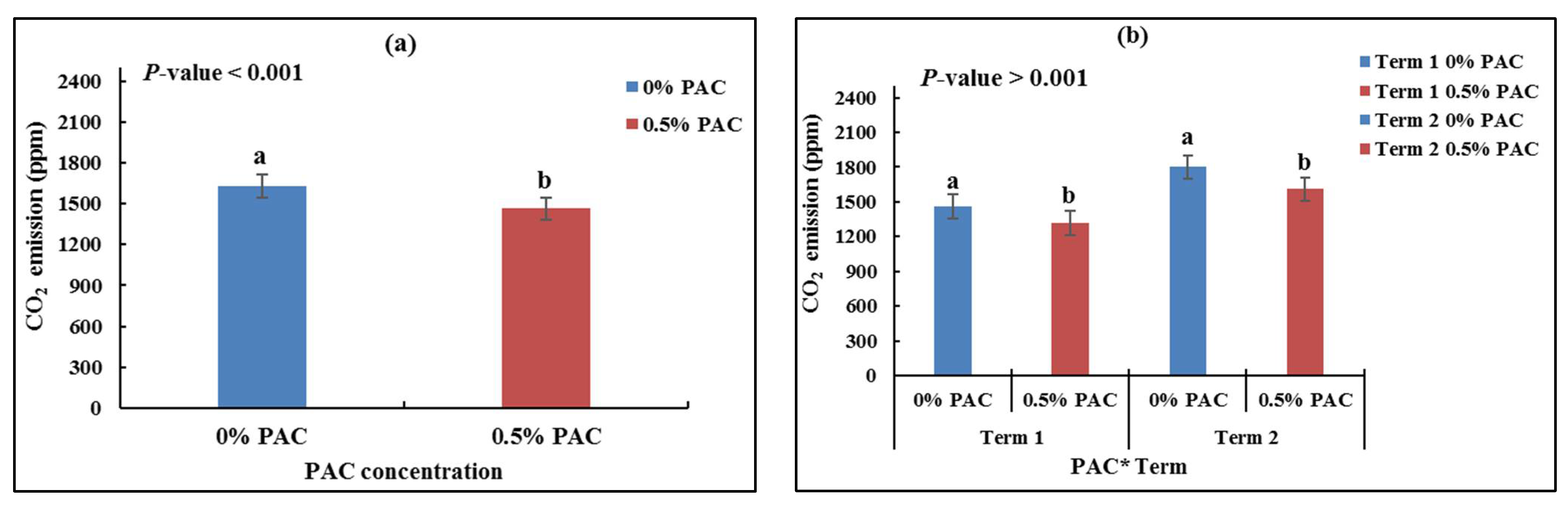
3.3.2. CO2 Emissions
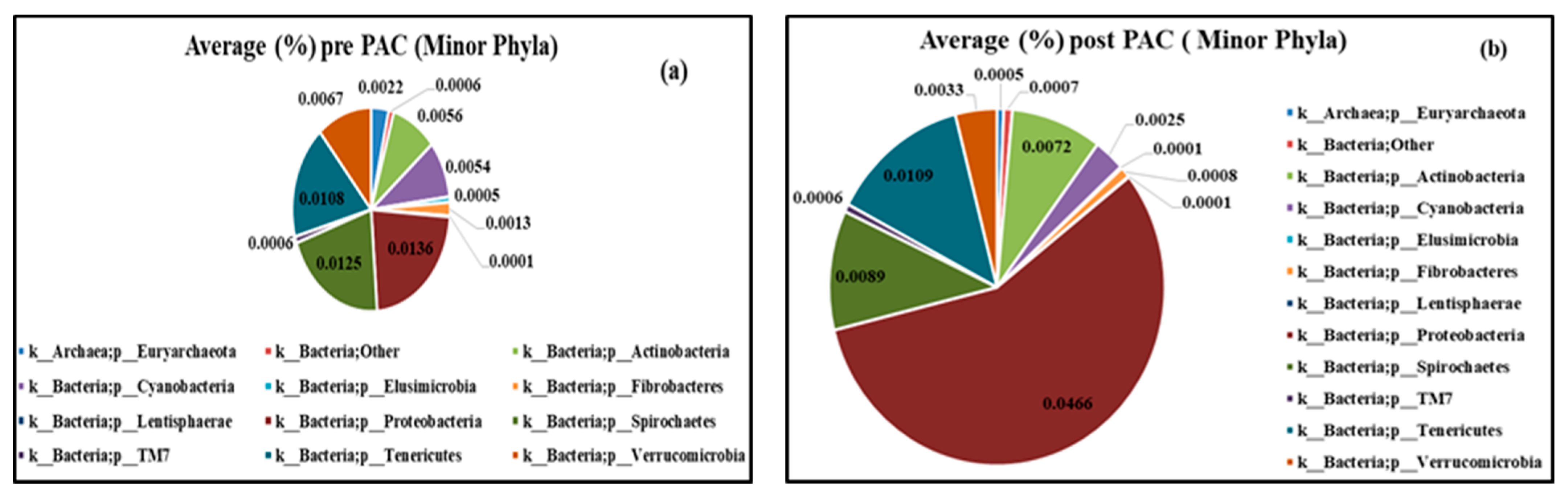
3.4. The Effect of PAC on the Bacterial and Archaeal Microbial in Dairy Cattle’s Manure
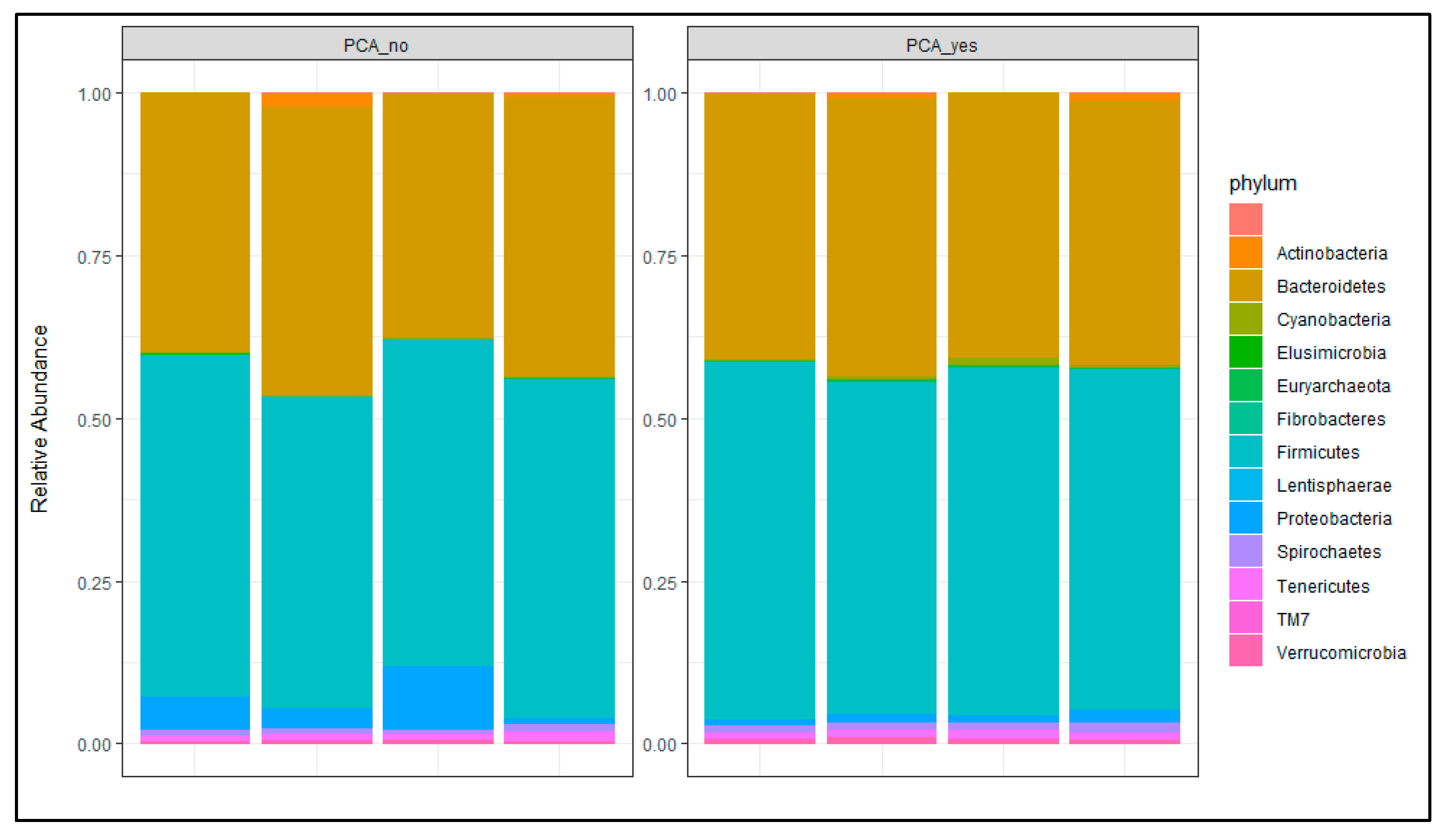
3.4.1. Microbial Community Analysis
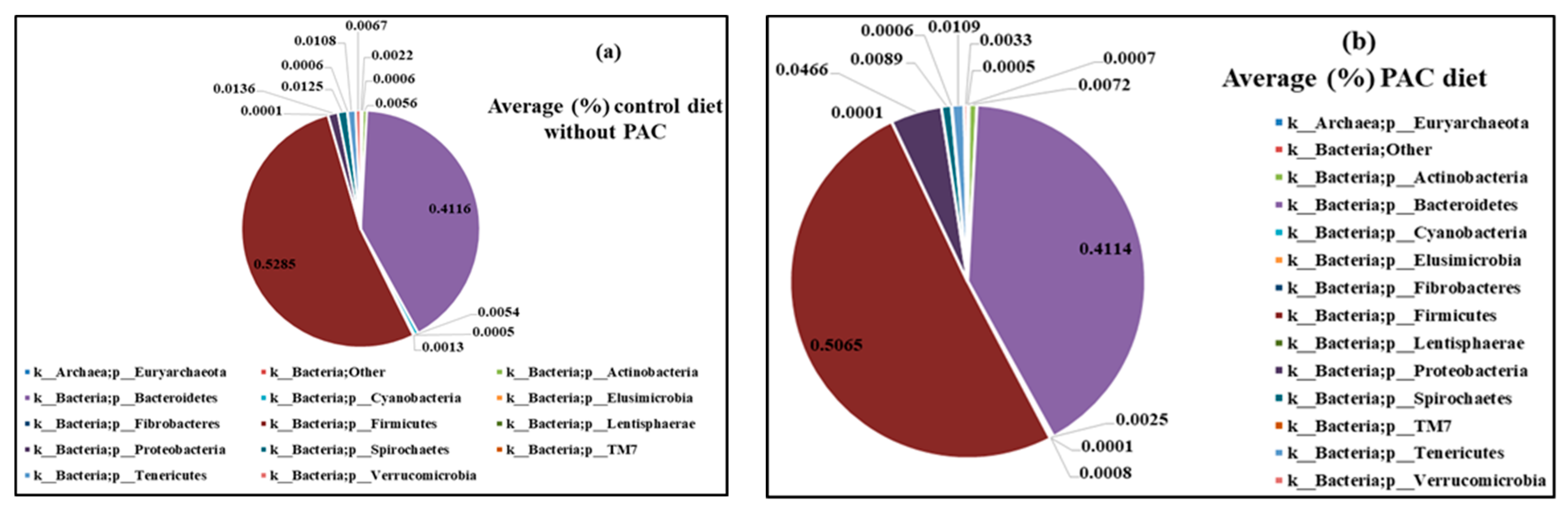
3.4.2. Chemical Compounds of Manure
4. Discussion
4.1. Milking Production and GHG Emissions
4.2. Microbial Community
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eggleston, H.; Buendia, L.; Miwa, K.; Ngara, T.; Tanabe, K. 2006 IPCC Guidelines for National Greenhouse Gas Inventories; Institute for Global Environmental Strategies: Kanagawa, Japan, 2006. [Google Scholar]
- Bellarby, J.; Tirado, R.; Leip, A.; Weiss, F.; Lesschen, J.P.; Smith, P. Livestock greenhouse gas emissions and mitigation potential in Europe. Glob. Chang. Biol. 2012, 19, 3–18. [Google Scholar] [CrossRef]
- Vergé, X.; De Kimpe, C.; Desjardins, R. Agricultural production, greenhouse gas emissions and mitigation potential. Agric. For. Meteorol. 2007, 142, 255–269. [Google Scholar] [CrossRef]
- Rotz, C.A.; Montes, F.; Chianese, D.S. The carbon footprint of dairy production systems through partial life cycle assessment. J. Dairy Sci. 2010, 93, 1266–1282. [Google Scholar] [CrossRef] [PubMed]
- Larney, F.J.; Hao, X. A review of composting as a management alternative for beef cattle feedlot manure in southern Alberta, Canada. Bioresour. Technol. 2007, 98, 3221–3227. [Google Scholar] [CrossRef]
- Holman, D.B.; Hao, X.; Topp, E.; Yang, H.E.; Alexander, T.W. Effect of Co-Composting Cattle Manure with Construction and Demolition Waste on the Archaeal, Bacterial, and Fungal Microbiota, and on Antimicrobial Resistance Determinants. PLoS ONE 2016, 11, e0157539. [Google Scholar] [CrossRef] [Green Version]
- Pandey, P.K.; Biswas, S.; Vaddella, V.K.; Soupir, M.L. Escherichia coli persistence kinetics in dairy manure at moderate, mesophilic, and thermophilic temperatures under aerobic and anaerobic environments. Bioprocess Biosyst. Eng. 2015, 38, 457–467. [Google Scholar] [CrossRef]
- Pandey, P.; Chiu, C.; Miao, M.; Wang, Y.; Settles, M.; Del Rio, N.S.; Castillo, A.; Souza, A.; Pereira, R.; Jeannotte, R. 16S rRNA analysis of diversity of manure microbial community in dairy farm environment. PLoS ONE 2018, 13, e0190126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Chen, S. Ignored sediment fungal populations in water supply reservoirs are revealed by quantitative PCR and 454 pyrosequencing. BMC Microbiol. 2015, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zaheer, R.; Cook, S.; Klima, C.; Stanford, K.; Alexander, T.; Topp, E.; Read, R.; Mcallister, T. Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 2013, 4, 133. [Google Scholar] [CrossRef] [Green Version]
- Vahjen, W.; Pietruszyńska, D.; Starke, I.C.; Zentek, J. High dietary zinc supplementation increases the occurrence of tetracycline and sulfonamide resistance genes in the intestine of weaned pigs. Gut Pathog. 2015, 7, 23. [Google Scholar] [CrossRef] [Green Version]
- Joseph, S.; Pow, D.; Dawson, K.; Mitchell, D.R.; Rawal, A.; Hook, J.; Taherymoosavi, S.; Van Zwieten, L.; Rust, J.; Donne, S.; et al. Feeding Biochar to Cows: An Innovative Solution for Improving Soil Fertility and Farm Productivity. Pedosphere 2015, 25, 666–679. [Google Scholar] [CrossRef]
- Winders, T.M.; Jolly-Breithaupt, M.L.; Wilson, H.C.; MacDonald, J.C.; Erickson, G.E.; Watson, A.K. Evaluation of the effects of biochar on diet digestibility and methane production from growing and finishing steers. Transl. Anim. Sci. 2019, 3, 775–783. [Google Scholar] [CrossRef] [Green Version]
- McHenry, M.P. Carbon-based stock feed additives: A research methodology that explores ecologically delivered C biosequestration, alongside live weights, feed use efficiency, soil nutrient retention, and perennial fodder plantations. J. Sci. Food Agric. 2010, 90, 183–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banner, R.E.; Rogosic, J.; Burritt, E.A.; Provenza, F.D. Supplemental barley and charcoaI increase intake of sagebrush by lambs. J. Range Manag. 2000, 53, 415–420. [Google Scholar] [CrossRef]
- Knutson, H.J.; Carr, M.A.; Branham, L.A.; Scott, C.B.; Callaway, T.R. Effects of activated charcoal on binding E. coli O157:H7 and Salmonella typhimurium in sheep. Small Rumin. Res. 2006, 65, 101–105. [Google Scholar] [CrossRef]
- Rogosic, J.; Pfister, J.A.; Provenza, F.D.; Grbesa, D. The effect of activated charcoal and number of species offered on intake of Mediterranean shrubs by sheep and goats. Appl. Anim. Behav. Sci. 2006, 101, 305–317. [Google Scholar] [CrossRef]
- Al-Kindi, A.; Schiborra, A.; Buerkert, A.; Schlecht, E. Effects of quebracho tannin extract and activated charcoal on nutrient digestibility, digesta passage and faeces composition in goats. J. Anim. Physiol. Anim. Nutr. 2017, 101, 576–588. [Google Scholar] [CrossRef]
- Poage, G.W.; Scott, C.B.; Bisson, M.G.; Hartmann, S.F. Activated Charcoal Attenuates Bitterweed Toxicosis in Sheep. J. Range Manag. Arch. 2000, 53, 73–78. [Google Scholar] [CrossRef]
- Bisson, M.G.; Scott, C.B.; Taylor, C.A. Activated Charcoal and Experience Affect Intake of Juniper by Goats. J. Range Manag. Arch. 2001, 54, 274–278. [Google Scholar] [CrossRef]
- Leng, R.A.; Preston, T.R.; Inthapanya, S. Biochar reduces enteric methane and improves growth and feed conversion in local yellow cattle fed cassava root chips and fresh cassava foliage. Livest. Res. Rural. Dev. 2012, 24, 11. [Google Scholar]
- Lockyer, D.; Jarvis, S. The measurement of methane losses from grazing animals. Environ. Pollut. 1995, 90, 383–390. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Community ecology package. R Package Version 2018, 2, 2–5. [Google Scholar]
- Chatonnet, F.; Pignarre, A.; Sérandour, A.A.; Caron, G.; Avner, S.; Robert, N.; Kassambara, A.; Laurent, A.; Bizot, M.; Agirre, X.; et al. The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation. Haematologica 2020, 105, 774–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [Green Version]
- Swan, A.R.; Sandilands, M. Introduction to geological data analysis. In International Journal of Rock Mechanics and Mining Sciences and Geomechanics Abstracts; Elsevier: Amsterdam, The Netherlands, 1995; p. 387A. [Google Scholar]
- Saleem, A.M.; Ribeiro, G.O., Jr.; Yang, W.Z.; Ran, T.; Beauchemin, K.A.; McGeough, E.J.; Ominski, K.H.; Okine, E.K.; McAllister, T.A. Effect of engineered biocarbon on rumen fermentation, microbial protein synthesis, and methane production in an artificial rumen (RUSITEC) fed a high forage diet1. J. Anim. Sci. 2018, 96, 3121–3130. [Google Scholar] [CrossRef] [Green Version]
- Hansen, H.H.; Storm, I.M.L.D.; Sell, A.M. Effect of biochar on in vitro rumen methane production. Acta Agric. Scand. Sect. A Anim. Sci. 2012, 62, 305–309. [Google Scholar] [CrossRef]
- Cabeza, I.; Waterhouse, T.; Sohi, S.; Rooke, J. Effect of biochar produced from different biomass sources and at different process temperatures on methane production and ammonia concentrations in vitro. Anim. Feed. Sci. Technol. 2018, 237, 1–7. [Google Scholar] [CrossRef]
- Leng, R.A.; Inthapanya, S.; Preston, T.R. Methane production is reduced in an in vitro incubation when the rumen fluid is taken from cattle that previously received biochar in their diet. Gas 2012, 1050, 1367. [Google Scholar]
- McFarlane, Z.D.; Myer, P.R.; Cope, E.R.; Evans, N.D.; Bone, T.C.; Biss, B.E.; Mulliniks, J.T. Effect of Biochar Type and Size on in Vitro Rumen Fermentation of Orchard Grass Hay. Agric. Sci. 2017, 08, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Baumann, E.; Chouinard, P.Y.; Lebeuf, Y.; Rico, D.E.; Gervais, R. Effect of lipid supplementation on milk odd- and branched-chain fatty acids in dairy cows. J. Dairy Sci. 2016, 99, 6311–6323. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; McSweeney, C.; Wright, A.D.G.; Bishop-Hurley, G.; Kalantar-Zadeh, K. Measuring Methane Production from Ruminants. Trends Biotechnol. 2016, 34, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Kempton, T.; Murray, R.; Leng, R. Methane Production and Digestibility Measurements in the Grey Kangaroo and Sheep. Aust. J. Biol. Sci. 1976, 29, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Torrent, J.; Johnson, D.E.; Kujawa, M.A. Co-product fiber digestibility: Kinetic and in vivo assessment. J. Anim. Sci. 1994, 72, 790–795. [Google Scholar] [CrossRef]
- Xie, Z.; Xu, Y.; Liu, G.; Liu, Q.; Zhu, J.; Tu, C.; Amonette, J.E.; Cadisch, G.; Yong, J.W.; Hu, S. Impact of biochar application on nitrogen nutrition of rice, greenhouse-gas emissions and soil organic carbon dynamics in two paddy soils of China. Plant Soil 2013, 370, 527–540. [Google Scholar] [CrossRef]
- Martin, S.L.; Clarke, M.L.; Othman, M.; Ramsden, S.J.; West, H.M. Biochar-mediated reductions in greenhouse gas emissions from soil amended with anaerobic digestates. Biomass Bioenergy 2015, 79, 39–49. [Google Scholar] [CrossRef]
- Lai, W.Y.; Lai, C.M.; Ke, G.R.; Chung, R.S.; Chen, C.T.; Cheng, C.H.; Pai, C.W.; Chen, S.Y.; Chen, C.C. The effects of woodchip biochar application on crop yield, carbon sequestration and greenhouse gas emissions from soils planted with rice or leaf beet. J. Taiwan Inst. Chem. Eng. 2013, 44, 1039–1044. [Google Scholar] [CrossRef]
- Wang, J.; Pan, X.; Liu, Y.; Zhang, X.; Xiong, Z. Effects of biochar amendment in two soils on greenhouse gas emissions and crop production. Plant Soil 2012, 360, 287–298. [Google Scholar] [CrossRef]
- Angst, T.E.; Six, J.; Reay, D.S.; Sohi, S.P. Impact of pine chip biochar on trace greenhouse gas emissions and soil nutrient dynamics in an annual ryegrass system in California. Agric. Ecosyst. Environ. 2014, 191, 17–26. [Google Scholar] [CrossRef]
- DeVore, D.W. In-vitro digestibility and gas production of by-product feedstuffs and the effects of monesin on in-vitro digestibility and gas production. Master’s Thesis, Missouri State University, Springfield, MO, USA, 2018. [Google Scholar]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Durso, L.M.; Harhay, G.P.; Smith, T.P.; Bono, J.L.; DeSantis, T.Z.; Clawson, M.L. Bacterial Community Analysis of Beef Cattle Feedlots Reveals That Pen Surface Is Distinct from Feces. Foodborne Pathog. Dis. 2011, 8, 647–649. [Google Scholar] [CrossRef] [Green Version]
- Callaway, T.R.; Dowd, S.E.; Edrington, T.S.; Anderson, R.C.; Krueger, N.; Bauer, N.; Kononoff, P.J.; Nisbet, D.J. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 2010, 88, 3977–3983. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D. Connections Between the Gut Microbiome and Metabolic Hormones in Early Pregnancy in Overweight and Obese Women. Diabetes 2016, 65, 2214–2223. [Google Scholar] [CrossRef] [Green Version]
- Tobita, T.; Manzari, M.T.; Ozutsumi, O.; Ueda, K.; Uzuoka, R.; Iai, S. Benchmark Centrifuge Tests and Analyses of Liquefaction-Induced Lateral Spreading during Earthquake; Taylor and Francis: London, UK, 2014. [Google Scholar]
- Bhatt, K.; Maheshwari, D.K. Decoding multifarious role of cow dung bacteria in mobilization of zinc fractions along with growth promotion of C. annuum L. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Poulsen, M.M.; Vestergaard, P.F.; Clasen, B.F.; Radko, Y.; Christensen, L.P.; Stødkilde-Jørgensen, H.; Møller, N.; Jessen, N.; Pedersen, S.B.; Jørgensen, J.O.L. High-Dose Resveratrol Supplementation in Obese Men: An Investigator-Initiated, Randomized, Placebo-Controlled Clinical Trial of Substrate Metabolism, Insulin Sensitivity, and Body Composition. Diabetes 2013, 62, 1186–1195. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Park, C.Y.; Theesfeld, C.L.; Wong, A.K.; Yuan, Y.; Scheckel, C.; Fak, J.J.; Funk, J.; Yao, K.; Tajima, Y.; et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat. Genet. 2019, 51, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Indugu, N.; Vecchiarelli, B.; Pitta, D.W. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front. Microbiol. 2015, 6, 781. [Google Scholar] [CrossRef] [Green Version]
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997, 63, 2802–2813. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Pandya, P.; Parnerkar, S.; Tripathi, A.; Rank, D.; Kothari, R.; Joshi, C. Molecular Identification of Methanogenic Archaea From Surti Buffaloes (Bubalus Bubalis), Reveals More Hydrogenotrophic Methanogens Phylotypes. Braz. J. Microbiol. 2011, 42, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Singh, K.; Tripathi, A.; Pandya, P.; Parnerkar, S.; Rank, D.; Kothari, R.; Joshi, C. Methanogen diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. Res. Veter. Sci. 2012, 92, 451–455. [Google Scholar] [CrossRef]
- Zhang, M.; Li, J.; Wang, Y.; Yang, C. Impacts of different biochar types on the anaerobic digestion of sewage sludge. RSC Adv. 2019, 9, 42375–42386. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wang, C.; Lin, C.; Zhang, Y.; Chen, X.; Tang, L.; Liu, C.; Chen, Q.; Onwuka, M.I.; Song, T. Methane and Nitrous Oxide Flux after Biochar Application in Subtropical Acidic Paddy Soils under Tobacco-Rice Rotation. Sci. Rep. 2019, 9, 17277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Wu, D.; Feng, X.; Müllen, K. Nitrogen-Doped Ordered Mesoporous Graphitic Arrays with High Electrocatalytic Activity for Oxygen Reduction. Angew. Chem. 2010, 122, 2619–2623. [Google Scholar] [CrossRef]


| Ingredient | As Fed | Dry Matter at 89% |
|---|---|---|
| Total Crude Protein (minimum) | 16.0% | 18.0% |
| Crude Protein (minimum) | 13.1% | 14.7% |
| Equivalent Crude Protein (maximum) | 2.9% | 3.2% |
| Urea (maximum) | 1.0% | 1.1% |
| Crude Fibre (maximum) | 12.0% | 13.5% |
| Crude Fat (minimum) | 1.5% | 1.7% |
| Calcium (minimum) | 1.0% | 1.1% |
| Phosphorus (minimum) | 0.5% | 0.56% |
| Magnesium (minimum) | 0.4% | 0.45% |
| Salt (maximum added) | 0.5% | 0.56% |
| Copper (added) | 45 mg/kg | 50 mg/kg |
| Zinc (added) | 150 mg/kg | 170 mg/kg |
| Items | 0% PAC | 0.5% PAC | p-Value |
|---|---|---|---|
| Dry matter | 88.56 | 89.11 | 0.001 |
| Organic matter | 77.7 | 78.62 | 0.02 |
| Ash | 22.3 | 21.37 | 0.02 |
| Curd protein | 13.11 | 10.37 | 0.001 |
| NDF% | 43.15 | 43.06 | 0.10 |
| ADF% | 26.56 | 28.34 | 0.001 |
| Lignin% | 9.63 | 8.99 | 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Azzawi, M.; Bowtell, L.; Hancock, K.; Preston, S. Addition of Activated Carbon into a Cattle Diet to Mitigate GHG Emissions and Improve Production. Sustainability 2021, 13, 8254. https://doi.org/10.3390/su13158254
Al-Azzawi M, Bowtell L, Hancock K, Preston S. Addition of Activated Carbon into a Cattle Diet to Mitigate GHG Emissions and Improve Production. Sustainability. 2021; 13(15):8254. https://doi.org/10.3390/su13158254
Chicago/Turabian StyleAl-Azzawi, Mohammed, Les Bowtell, Kerry Hancock, and Sarah Preston. 2021. "Addition of Activated Carbon into a Cattle Diet to Mitigate GHG Emissions and Improve Production" Sustainability 13, no. 15: 8254. https://doi.org/10.3390/su13158254
APA StyleAl-Azzawi, M., Bowtell, L., Hancock, K., & Preston, S. (2021). Addition of Activated Carbon into a Cattle Diet to Mitigate GHG Emissions and Improve Production. Sustainability, 13(15), 8254. https://doi.org/10.3390/su13158254







