Rodent Virus Diversity and Differentiation across Post-Katrina New Orleans
Abstract
1. Introduction
2. Materials and Methods
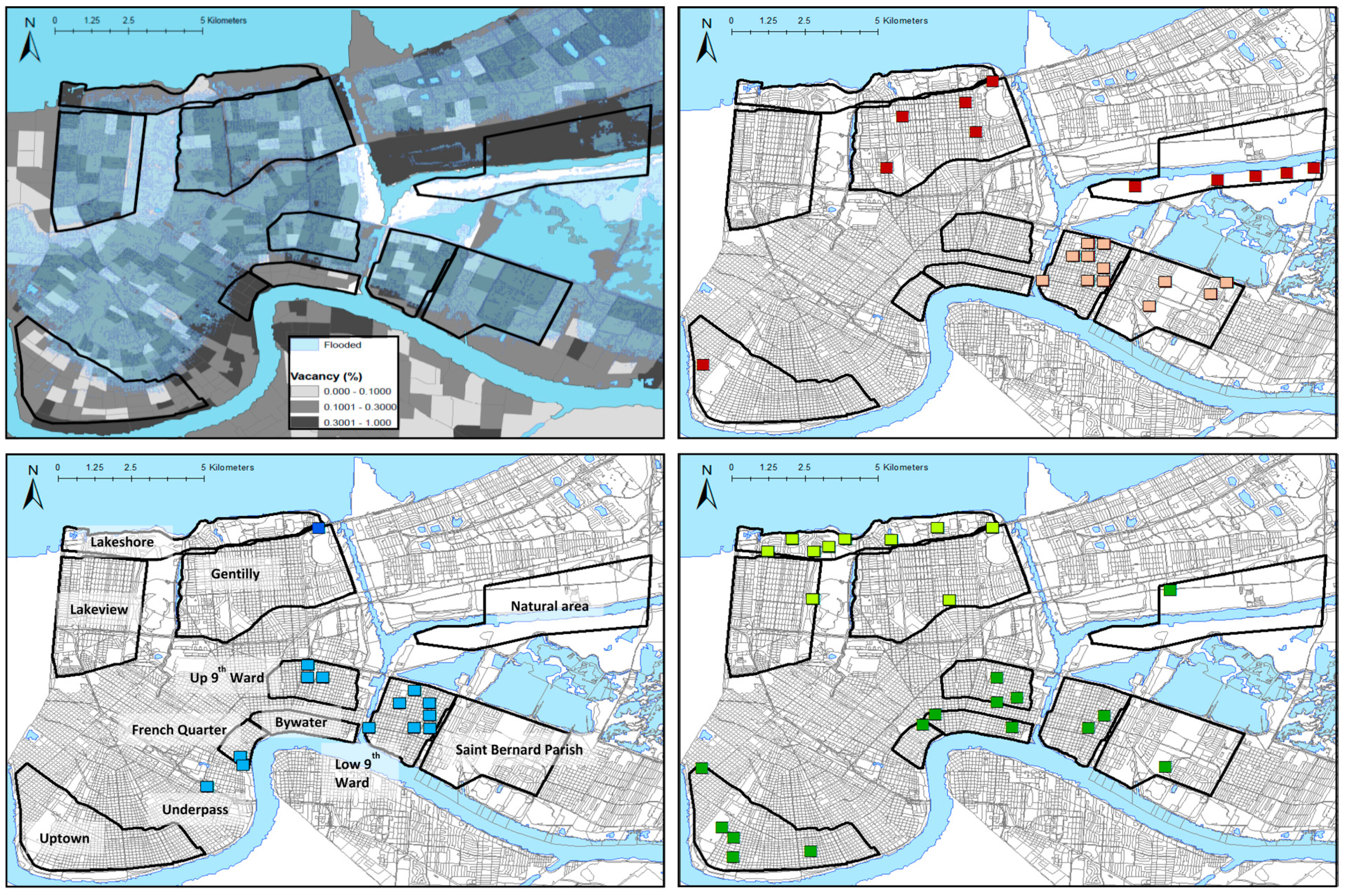
2.1. Study Areas and Rodent Trapping
2.2. Tissue Collection and Specimen Measurements
2.3. Metagenomic Sequencing
2.4. Bioinformatics
2.5. Statistical Analyses
3. Results
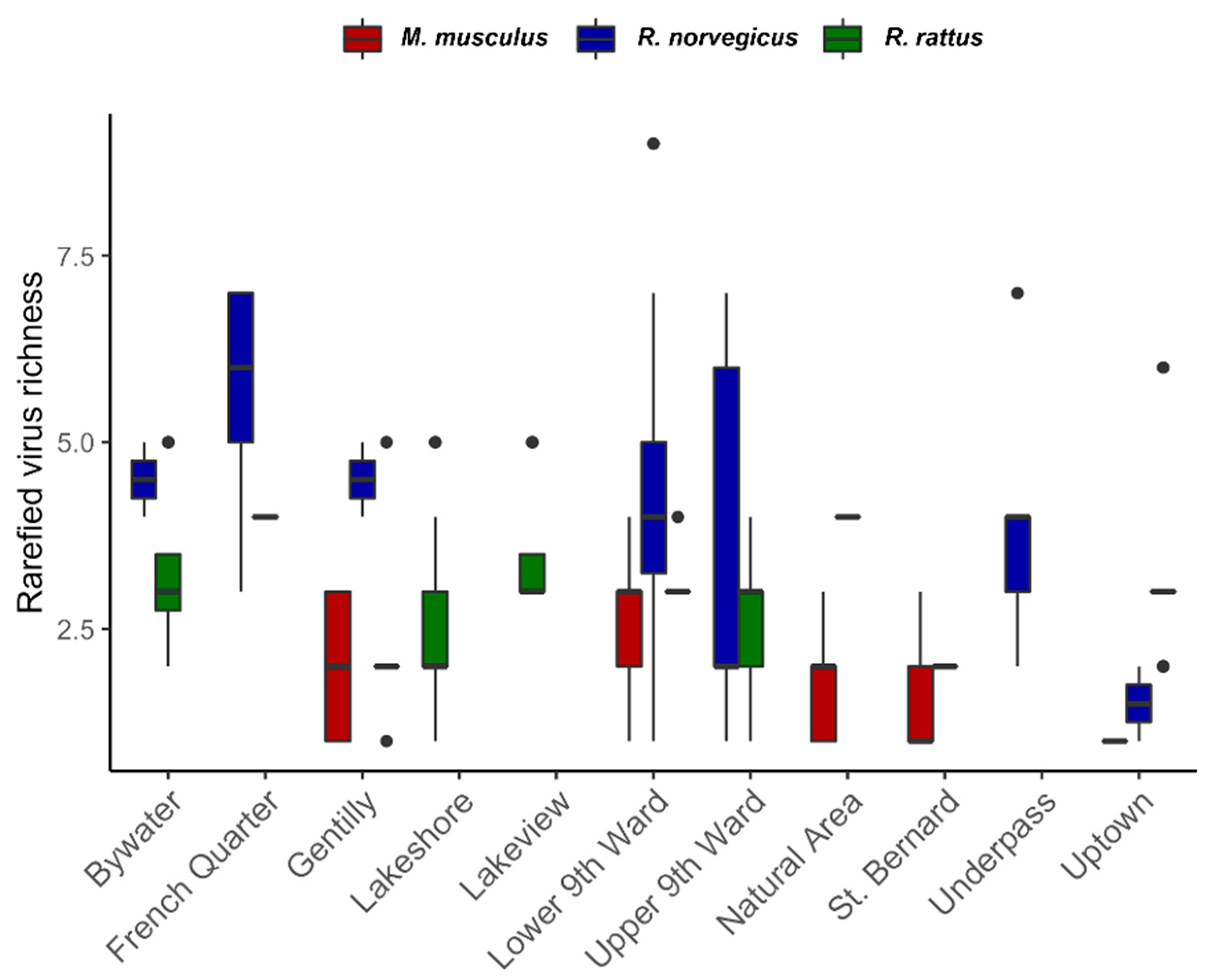
3.1. Viral Richness
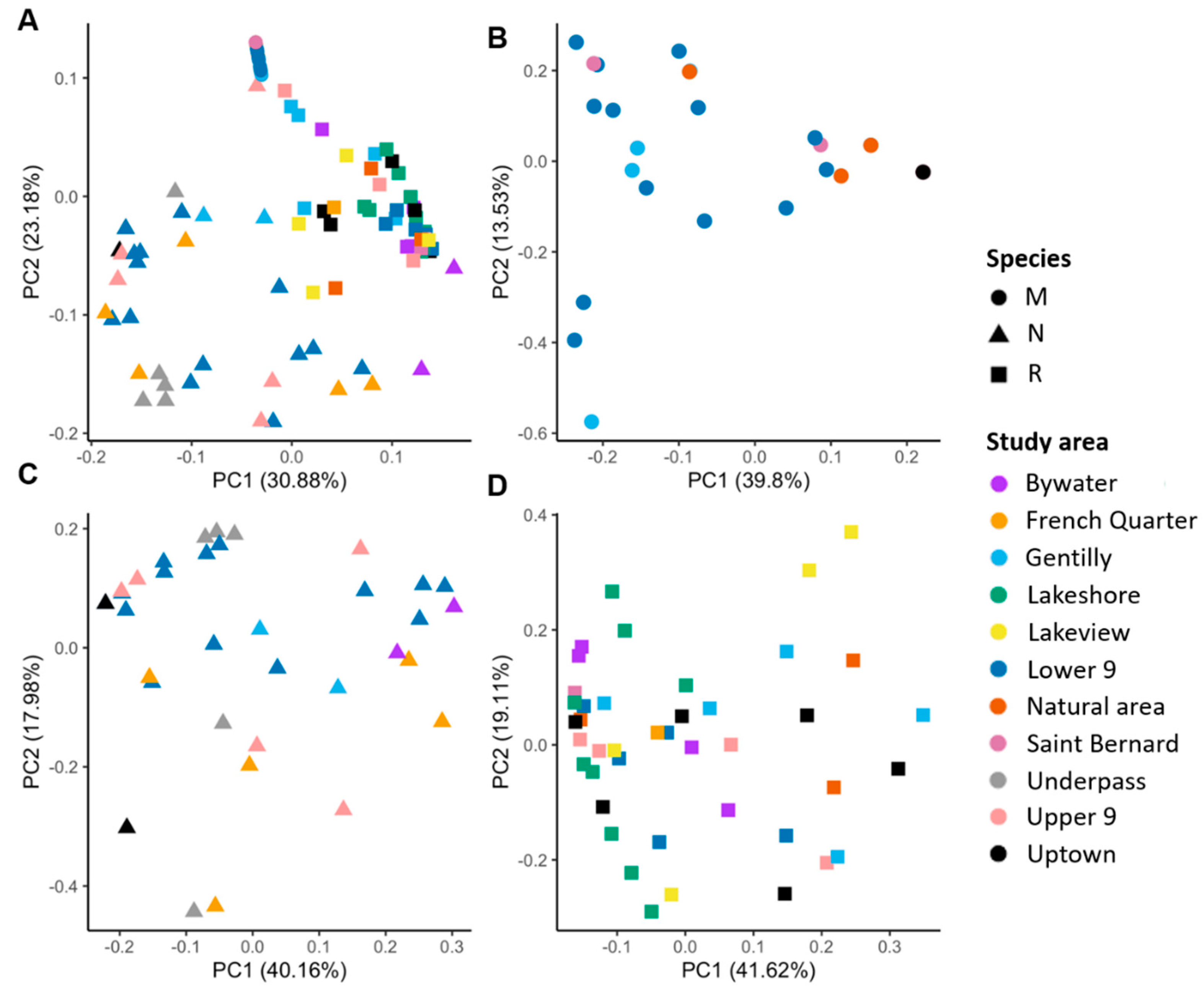
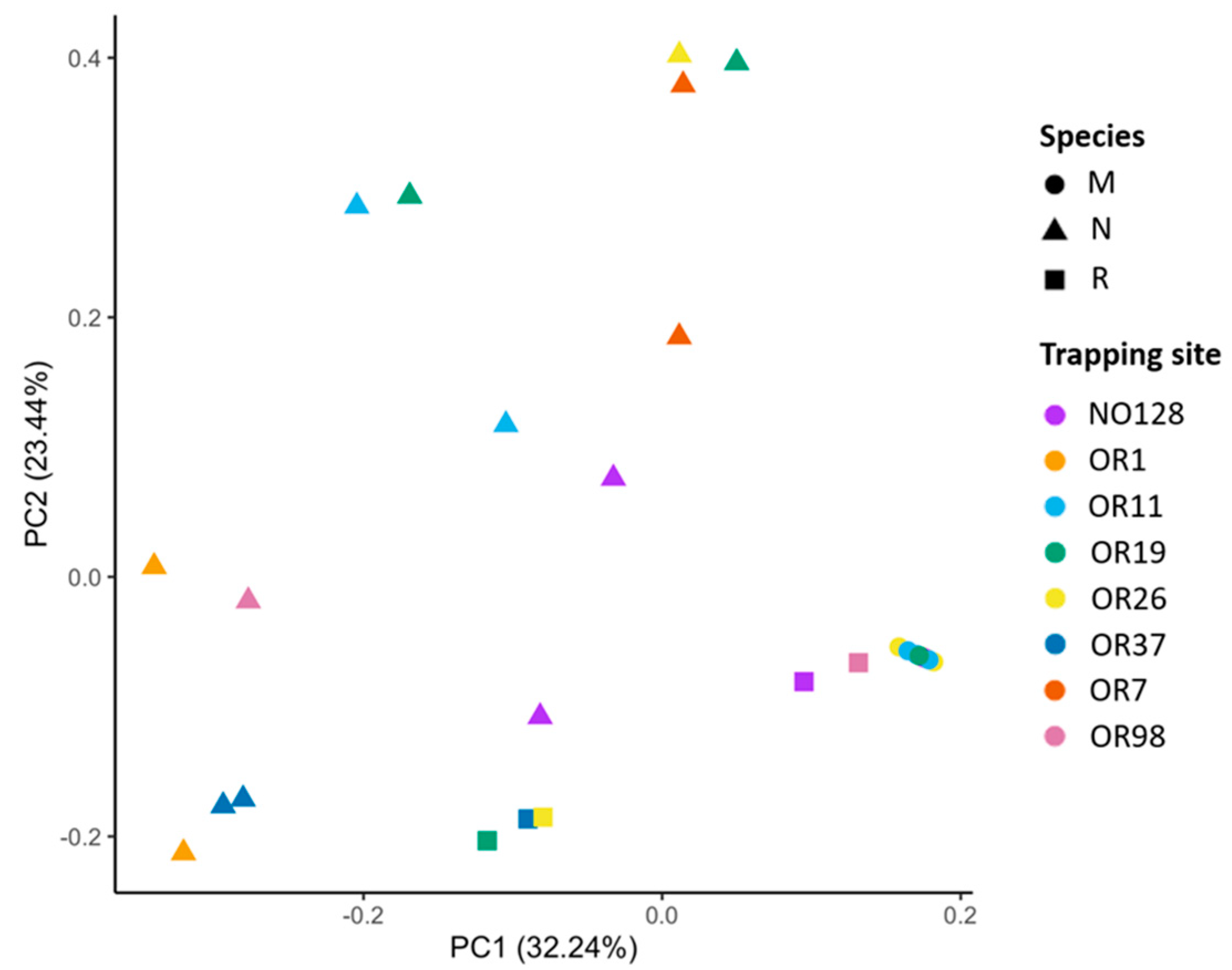
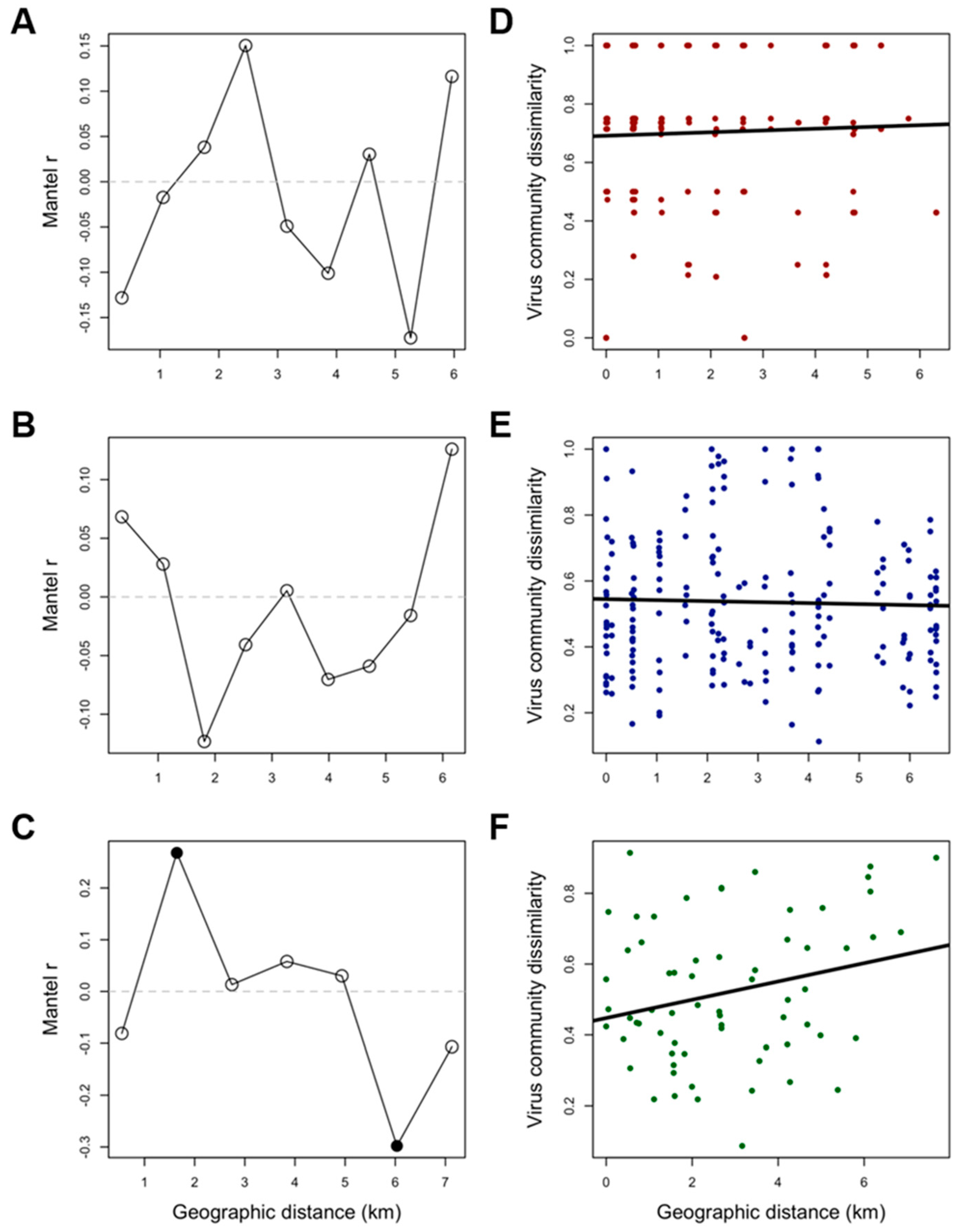
3.2. Correlates of Viral Richness and Differentiation
4. Discussion
4.1. Patterns of Viral Richness
4.2. Patterns of Viral Assemblage Structure
4.3. Predictors of Viral Diversity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Small, C.; Nicholls, R.J. A global analysis of human settlement in coastal zones. J. Coast Res. 2003, 19, 584–599. [Google Scholar]
- Elsner, J.B.; Kossin, J.P.; Jagger, T.H. The increasing intensity of the strongest tropical cyclones. Nature 2008, 455, 92–95. [Google Scholar] [CrossRef]
- Leaning, J.; Guha-Sapir, D. Natural disasters, armed conflict, and public health. N. Engl. J. Med. 2013, 369, 1836–1842. [Google Scholar] [CrossRef]
- Rael, R.C.; Peterson, A.C.; Ghersi, B.M.; Childs, J.; Blum, M.J. Disturbance, reassembly, and disease risk in socioecological systems. Ecohealth 2016, 13, 450–455. [Google Scholar] [CrossRef]
- Ostfeld, R.S.; Holt, R.D. Are predators good for your health? Evaluating evidence for top-down regulation of zoonotic disease reservoirs. Front. Ecol. Environ. 2004, 2, 13–20. [Google Scholar] [CrossRef]
- Watson, J.T.; Gayer, M.; Connolly, M.A. Epidemics after natural disasters. Emerg. Infect. Dis. 2007, 13, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, B.A.; Gubler, D.J. Disease ecology and the global emergence of zoonotic pathogens. Environ. Health Prev. Med. 2005, 10, 263. [Google Scholar] [CrossRef]
- Peterson, A.C.; Ghersi, B.M.; Campanella, R.; Riegel, C.; Lewis, J.A.; Blum, M.J. Rodent assemblage structure reflects socioecological mosaics of counter-urbanization across post-Hurricane Katrina New Orleans. Landsc. Urban Plan. 2020, 195, 103710. [Google Scholar] [CrossRef]
- Eskew, E.A.; Olival, K.J. De-urbanization and zoonotic disease risk. Ecohealth 2018, 15, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Himsworth, C.G.; Parsons, K.L.; Jardine, C.; Patrick, D.M. Rats, cities, people, and pathogens: A systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 2013, 13, 349–359. [Google Scholar] [CrossRef]
- Himsworth, C.G.; Bidulka, J.; Parsons, K.L.; Feng, A.Y.T.; Tang, P.; Jardine, C.M.; Kerr, T.; Mak, S.; Robinson, J.; Patrick, D.M. Ecology of Leptospira interrogans in Norway rats (Rattus norvegicus) in an inner-city neighborhood of Vancouver, Canada. PLoS Negl. Trop Dis. 2013, 7, e2270. [Google Scholar] [CrossRef]
- Peterson, A.C.; Ghersi, B.M.; Alda, F.; Firth, C.; Frye, M.J.; Bai, Y.; Osikowicz, L.M.; Riegel, C.; Lipkin, W.I.; Kosoy, M.Y.; et al. Rodent-borne Bartonella infection varies according to host species within and among cities. Ecohealth 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rothenburger, J.L.; Himsworth, C.H.; Nemeth, N.M.; Pearl, D.L.; Jardine, C.M. Environmental factors and zoonotic pathogen ecology in urban exploiter species. Ecohealth 2017, 14, 630–641. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Himsworth, C.G.; Bai, Y.; Kosoy, M.Y.; Wood, H.; DiBernardo, A.; Lindsay, R.; Bidulka, J.; Tang, P.; Jardine, C.; Patrick, D. An investigation of Bartonella spp., Rickettsia typhi, and Seoul hantavirus in rats (Rattus spp.) from an inner-city neighborhood of Vancouver, Canada: Is pathogen presence a reflection of global and local rat population structure? Vector Borne Zoonotic Dis. 2015, 15, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Childs, J.E.; Glass, G.E.; Korch, G.W.; Ksiazek, T.G.; Leduc, J.W. Lymphocytic choriomeningitis virus infection and house mouse (Mus Musculus) distribution in urban Baltimore. Am. J. Trop. Med. Hyg. 1992, 47, 27–31. [Google Scholar] [CrossRef]
- Easterbrook, J.D.; Ka, J.B.; Vanasco, N.B.; Reeves, W.K.; Pu, R.H.; Kosoy, M.Y.; Glass, G.E.; Watson, J.; Klein, S.L. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. J. Epidemiol. Infect. 2007, 135, 1192–1199. [Google Scholar] [CrossRef]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Peterson, A.C.; Ghersi, B.M.; Riegel, C.; Wunder Jr, E.A.; Childs, J.E.; Blum, M.J. Amplification of pathogenic Leptospira infection with greater abundance and co-occurrence of rodent hosts across a counter-urbanizing landscape. Mol. Ecol. 2021, 30, 2145–2161. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Bhat, M.; Firth, M.A.; Williams, S.H.; Frye, M.J.; Simmonds, P.; Conte, J.M.; Ng, J.; Garcia, J.; Bhuva, N.P.; et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio 2014, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.H.; Che, X.; Garcia, J.A.; Klena, J.D.; Lee, B.; Muller, D.; Ulrich, W.; Corrigan, R.M.; Nichol, S.; Jain, K.; et al. Viral diversity of house mice in New York City. MBio 2018, 9, e01354-17. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.H.; Che, X.; Paulick, A.; Guo, C.; Lee, B.; Muller, D.; Uhlemann, A.-C.; Lowy, F.D.; Corrigan, R.M.; Lipkin, W.I. New York City house mice (Mus musculus) as potential reservoirs for pathogenic bacteria and antimicrobial resistance determinants. MBio 2018, 9, e00624-18. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, L.; Du, J.; Yang, L.; Ren, X.; Liu, B.; Jiang, J.; Yang, J.; Dong, J.; Sun, L.; et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 2018, 6, 178. [Google Scholar] [CrossRef]
- Morse, S.S.; Mazet, J.A.; Woolhouse, M.; Parrish, C.R.; Carroll, D.; Karesh, W.B.; Zambrana-Torrelio, C.; Lipkin, W.I.; Daszak, P. Prediction and prevention of the next pandemic zoonosis. Lancet 2012, 380, 1956–1965. [Google Scholar] [CrossRef]
- Han, B.A.; Schmidt, J.P.; Bowden, S.E.; Drake, J.M. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. USA 2015, 112, 7039–7044. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 546, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Bergner, L.M.; Orton, R.J.; Benavides, J.A.; Becker, D.J.; Tello, C.; Biek, R.; Streicker, D.G. Demographic and environmental drivers of metagenomic viral diversity in vampire bats. Mol. Ecol. 2020, 29, 26–39. [Google Scholar] [CrossRef]
- Wille, M. Unravelling virus community ecology in bats through the integration of metagenomics and community ecology. Mol. Ecol. 2020, 29, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Kapusinszky, B.; Wang, C.; Rose, R.K.; Lipton, H.L.; Delwart, E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011, 7, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Rabiela, F.; Suzán, G.; Wiratsudakul, A.; Rico-Chávez, O. Viral metacommunities associated to bats and rodents at different spatial scales. Community Ecol. 2018, 19, 168–175. [Google Scholar] [CrossRef]
- Shaw, L.P.; Wang, A.D.; Dylus, D.; Meier, M.; Pogacnik, G.; Dessimoz, C.; Balloux, F. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol. Ecol. 2020, 29, 3361–3379. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.N. Biodiversity loss and emerging infectious disease: An example from the rodent-borne hemorrhagic fevers. Biodiversity 2006, 7, 9–17. [Google Scholar] [CrossRef]
- Dizney, L.J.; Ruedas, L.A. Increased host species diversity and decreased prevalence of sin nombre virus. Emerg. Infect Dis. 2009, 15, 1012–1018. [Google Scholar] [CrossRef]
- Dearing, M.D.; Clay, C.; Lehmer, E.; Dizney, L. The roles of community diversity and contact rates on pathogen prevalence. J. Mammal. 2015, 96, 29–36. [Google Scholar] [CrossRef]
- Krasnov, B.R.; Shenbrot, G.I.; Mouillot, D.; Khokhlova, I.S.; Poulin, R. Spatial variation in species diversity and composition of flea assemblages in small mammalian hosts: Geographical distance or faunal similarity? J. Biogeogr. 2005, 32, 633–644. [Google Scholar] [CrossRef]
- Krasnov, B.R.; Mouillot, D.; Shenbrot, G.I.; Khokhlova, I.S.; Vinarski, M.V.; Korallo-Vinarskaya, N.P.; Poulin, R. Similarity in ectoparasite faunas of Palaearctic rodents as a function of host phylogenetic, geographic or environmental distances: Which matters the most? Int. J. Parasitol. 2010, 40, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.R.; Altizer, S.; Smith, K.F.; Alonso Aguirre, A.; Brown, J.H.; Budischak, S.A.; Byers, J.E.; Dallas, T.A.; Jonathan Davies, T.; Drake, J.M.; et al. The macroecology of infectious diseases: A new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 2016, 19, 1159–1171. [Google Scholar] [CrossRef]
- Kendig, A.E.; Borer, E.T.; Mitchell, C.E.; Power, A.G.; Seabloom, E.W. Characteristics and drivers of plant virus community spatial patterns in US west coast grasslands. Oikos 2017, 126, 1281–1290. [Google Scholar] [CrossRef]
- Robertson, C. Settlement Is Reached in Suit Over Katrina Grants. New York Times, 6 July 2011. Available online: https://www.nytimes.com/2011/07/07/us/07orleans.html (accessed on 29 July 2020).
- Green, T.F.; Olshansky, R.B. Rebuilding housing in New Orleans: The road home program after the Hurricane Katrina disaster. Hous. Policy Debate 2012, 22, 75–99. [Google Scholar] [CrossRef]
- Green, R.D.; Kouassi, M.; Mambo, B. Housing, race, and recovery from Hurricane Katrina. Rev. Black Political. Econ. 2013, 40, 145–163. [Google Scholar] [CrossRef]
- Gotham, K.F. Reinforcing inequalities: The impact of the CDBG program on post-Katrina rebuilding. Hous. Policy Debate 2014, 24, 192–212. [Google Scholar] [CrossRef]
- Lewis, J.A.; Zipperer, W.C.; Ernstson, H.; Bernik, B.; Hazen, R.; Elmqvist, T.; Blum, M.J. Socioecological disparities in New Orleans following Hurricane Katrina. Ecosphere 2017, 8, e01922. [Google Scholar] [CrossRef]
- Gulachenski, A.; Ghersi, B.M.; Lesen, A.E.; Blum, M.J. Abandonment, ecological assembly and public health risks in counter-urbanizing cities. Sustainability 2016, 8, 491. [Google Scholar] [CrossRef]
- Ghersi, B.M. Rat Demography and Rodent-Borne Pathogens across Post-Katrina New Orleans. Ph.D. Thesis, University of Tennessee, Knoxville, TN, USA, 2020. [Google Scholar]
- Bergner, L.M.; Orton, R.J.; da Silva Filipe, A.; Shaw, A.E.; Becker, D.J.; Tello, C.; Biek, R.; Streicker, D.G. Using noninvasive metagenomics to characterize viral communities from wildlife. Mol. Ecol. Resourc. 2019, 19, 128–143. [Google Scholar] [CrossRef]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Mcardle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Combs, M.; Byers, K.A.; Ghersi, B.M.; Blum, M.J.; Caccone, A.; Costa, F.; Himsworth, C.G.; Richardson, J.L.; Munshi-South, J. Urban rat races: Spatial population genomics of brown rats (Rattus norvegicus) compared across multiple cities. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180245. [Google Scholar] [CrossRef]
- Oksanen, J.; Guillaume, F.B.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-5. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 August 2020).
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Maechler, M.; Bolker, B.M. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 2017, 9, 378–400. [Google Scholar] [CrossRef]
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 2007, 22, 1–19. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Childs, J.E.; Glass, G.E.; Ksiazek, T.G.; Rossi, C.A.; Oro, J.G.B.; Leduc, J.W. Human-rodent contact and infection with lymphocytic choriomeningitis and Seoul viruses in an inner-city population. Am. J. Trop. Med. Hyg. 1991, 44, 117–121. [Google Scholar] [CrossRef]
- Knowles, S.; Eccles, R.; Baltrūnaitė, L. Species identity dominates over environment in shaping the microbiota of small mammals. Ecol. Lett. 2019, 22, 826–837. [Google Scholar] [CrossRef]
- Patz, J.A.; Daszak, P.; Tabor, G.M.; Aguirre, A.A.; Pearl, M.; Epstein, J.; Wolfe, N.D.; Kilpatrick, A.M.; Foufopoulos, J.; Molyneux, D.; et al. Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004, 112, 1092–1098. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, P.R.; Mills, J.N.; Prieur-Richard, A.-H.; Ezenwa, V.O.; Bailly, X.; Rizzoli, A.; Suzán, G.; Vittecoq, M.; García-Peña, G.E.; Daszak, P.; et al. Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. Philos. Trans. R Soc. B Biol. Sci. 2017, 372, 20160129. [Google Scholar] [CrossRef]
- Little, T.J.; Shuker, D.M.; Colegrave, N.; Day, T.; Graham, A.L. The coevolution of virulence: Tolerance in perspective. PLoS Pathog. 2010, 6, e1001006. [Google Scholar] [CrossRef] [PubMed]
- Faust, C.L.; Dobson, A.P.; Gottdenker, N.; Bloomfield, L.S.P.; McCallum, H.I.; Gillespie, T.R.; Diuk-Wasser, M.; Plowright, R.K. Null expectations for disease dynamics in shrinking habitat: Dilution or amplification? Philos. Trans. R. Soc. B 2017, 372, 20160173. [Google Scholar] [CrossRef] [PubMed]
- Mori, E.; Ferretti, F.; Fattorini, N. Alien war: Ectoparasite load, diet and temporal niche partitioning in a multi-species assembly of small rodents. Biol. Invasions. 2019, 21, 3305–3318. [Google Scholar] [CrossRef]
- Puckett, E.E.; Park, J.; Combs, M.; Blum, M.J.; Bryant, J.E.; Caccone, A.; Costa, F.; Deinum, E.E.; Esther, A.; Himsworth, C.G.; et al. Global population divergence and admixture of the brown rat (Rattus norvegicus). Proc. R. Soc. B Biol. Sci. 2016, 283, 20161762. [Google Scholar] [CrossRef]
- Torchin, M.E.; Lafferty, K.D.; Dobson, A.P.; McKenzie, V.J.; Kuris, A.M. Introduced species and their missing parasites. Nature 2003, 421, 628–630. [Google Scholar] [CrossRef]
- van Dijk, J.G.B.; Hoye, B.J.; Verhagen, J.H.; Nolet, B.A.; Fouchier, R.A.M.; Klaassen, M. Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J. Anim. Ecol. 2013, 83, 266–275. [Google Scholar] [CrossRef]
- Nishiyama, S.; Dutia, B.M.; Stewart, J.P.; Meredith, A.L.; Shaw, D.J.; Simmonds, P.; Sharp, C.P. Identification of novel anelloviruses with broad diversity in UK rodents. J. Gen. Virol. 2014, 95, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Takahashi, M.; Nishizawa, T.; Tawara, A.; Fukai, K.; Muramatsu, U.; Naito, Y.; Yoshikawa, A. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 2002, 83, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Tagliapietra, V.; Rosà, R.; Hauffe, H.C.; Laakkonen, J.; Voutilainen, L.; Vapalahti, O.; Vaheri, A.; Henttonen, H.; Rizzoli, A. Spatial and temporal dynamics of lymphocytic choriomeningitis virus in wild rodents, northern Italy. Emerg. Infect. Dis. 2009, 15, 1019–1025. [Google Scholar] [CrossRef][Green Version]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Mihaljevic, J.R. Linking metacommunity theory and symbiont evolutionary ecology. Trends Ecol. Evol. 2012, 27, 323–329. [Google Scholar] [CrossRef]
- Poulin, R. The decay of similarity with geographical distance in parasite communities of vertebrate hosts. J. Biogeogr. 2003, 30, 1609–1615. [Google Scholar] [CrossRef]
- Munshi-South, J.; Kharchenko, K. Rapid, pervasive genetic differentiation of urban white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 2010, 19, 4242–4254. [Google Scholar] [CrossRef]
- Combs, M.; Puckett, E.E.; Richardson, J.; Mims, D.; Munshi-South, J. Spatial population genomics of the brown rat (Rattus norvegicus) in New York City. Mol. Ecol. 2018, 27, 83–98. [Google Scholar] [CrossRef]
- Hechinger, R.F.; Lafferty, K.D. Host diversity begets parasite diversity: Bird final hosts and trematodes in snail intermediate hosts. Proc. R. Soc. B Biol. Sci. 2005, 272, 1059–1066. [Google Scholar] [CrossRef]
- Kamiya, T.; O’Dwyer, K.; Nakagawa, S.; Poulin, R. Host diversity drives parasite diversity: Meta-analytical insights into patterns and causal mechanisms. Ecography 2014, 37, 689–697. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; Ostfeld, R.S.; Keesing, F. Frontiers in research on biodiversity and disease. Ecol. Lett. 2015, 18, 1119–1133. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.L.; Zgliczynski, B.J.; Haupt, A.J.; Guerra, A.S.; Micheli, F.; Sandin, S.A. Human impacts decouple a fundamental ecological relationship—The positive association between host diversity and parasite diversity. Glob. Chang. Biol. 2018, 24, 3666–3679. [Google Scholar] [CrossRef] [PubMed]
- Streicker, D.G.; Allgeier, J.E. Foraging choices of vampire bats in diverse landscapes: Potential implications for land-use change and disease transmission. J. Appl. Ecol. 2016, 53, 1280–1288. [Google Scholar] [CrossRef]
- Reaser, J.K.; Witt, A.; Tabor, G.M.; Hudson, P.J.; Plowright, R.K. Ecological countermeasures for preventing zoonotic disease outbreaks: When ecological restoration is a human health imperative. Restor. Ecol. 2021, 29, e13357. [Google Scholar] [CrossRef]
- Plowright, R.K.; Reaser, J.K.; Locke, H.; Woodley, S.J.; Patz, J.A.; Becker, D.J.; Oppler, G.; Hudson, P.J.; Tabor, G.M. Land use-induced spillover: A call to action to safeguard environmental, animal, and human health. Lancet Planet. Health 2021, 5, e237–e245. [Google Scholar] [CrossRef]
| Study Area | M. musculus | R. norvegicus | R. rattus | Total |
|---|---|---|---|---|
| Bywater | 0, 0 | 2, 6 | 4, 12 | 6, 18 |
| French Quarter | 0, 0 | 5, 25 | 1, 4 | 6, 29 |
| Gentilly | 5, 25 | 2, 10 | 5, 21 | 12, 56 |
| Lakeshore | 0, 0 | 0, 0 | 9, 28 | 9, 28 |
| Lakeview | 0, 0 | 0, 0 | 4, 23 | 4, 23 |
| Lower 9th Ward | 14, 71 | 14, 60 | 5, 21 | 33, 152 |
| Natural Area | 5, 19 | 0, 0 | 3, 17 | 8, 36 |
| St. Bernard Parish | 7, 32 | 0, 0 | 1, 4 | 8, 36 |
| Underpass | 0, 0 | 5, 26 | 0, 0 | 5, 26 |
| Upper 9th Ward | 0, 0 | 5, 26 | 5, 20 | 10, 46 |
| Uptown | 1, 2 | 2, 7 | 6, 23 | 9, 32 |
| Total | 32, 149 | 35, 160 | 47, 173 | 110, 482 |
| Virus | M. musculus (n = 32) | R. norvegicus (n = 35) | R. rattus (n = 43) |
|---|---|---|---|
| Anelloviridae sp. | 0 (0) | 14 (40) | 40 (93) |
| Beihai picobirna-like virus 8 | 3 (9) | 0 (0) | 0 (0) |
| H1 parvovirus | 1 (3) | 1 (3) | 1 (2) |
| Lymphocytic choriomeningitis virus | 2 (6) | 0 (0) | 0 (0) |
| Mouse kidney parvovirus | 5 (16) | 0 (0) | 0 (0) |
| Mouse parvovirus 1 | 10 (31) | 0 (0) | 0 (0) |
| Murine adeno-associated virus (new) | 6 (19) | 0 (0) | 0 (0) |
| Murine adeno-associated virus 1 | 1 (3) | 0 (0) | 0 (0) |
| Murine adenovirus 1 | 2 (6) | 0 (0) | 0 (0) |
| Murine bocavirus | 1 (3) | 0 (0) | 0 (0) |
| Murine cytomegalovirus strain K181 | 10 (31) | 0 (0) | 0 (0) |
| Murine leukemia virus | 21 (66) | 0 (0) | 0 (0) |
| Mus musculus polyomavirus 2 | 5 (16) | 0 (0) | 0 (0) |
| Rat adeno-associated virus | 0 (0) | 2 (6) | 0 (0) |
| Rat bocavirus | 0 (0) | 1 (3) | 1 (2) |
| Rat cytomegalovirus | 0 (0) | 1 (3) | 2 (5) |
| Rat kobuvirus MM33 | 0 (0) | 1 (3) | 0 (0) |
| Rat minute virus | 0 (0) | 12 (34) | 2 (5) |
| Rat minute virus 1b | 0 (0) | 3 (9) | 0 (0) |
| Rat minute virus 1c | 0 (0) | 5 (14) | 0 (0) |
| Rat minute virus 2a | 0 (0) | 1 (3) | 0 (0) |
| Rat parvovirus 1a | 0 (0) | 2 (6) | 0 (0) |
| Rat parvovirus NTU1 | 0 (0) | 2 (6) | 3 (7) |
| Rattus norvegicus papillomavirus 1 | 0 (0) | 1 (3) | 0 (0) |
| Rodent bocavirus | 1 (3) | 0 (0) | 0 (0) |
| Rodent hepacivirus | 0 (0) | 15 (43) | 2 (5) |
| Rodent pegivirus | 0 (0) | 27 (77) | 0 (0) |
| Rodent torque teno virus 1 | 0 (0) | 21 (60) | 35 (81) |
| Rodent torque teno virus 2 | 0 (0) | 26 (74) | 10 (23) |
| Rodent torque teno virus 3 | 0 (0) | 10 (29) | 18 (42) |
| Torque teno mini virus 4 | 0 (0) | 0 (0) | 1 (2) |
| Torque teno mini virus 5 | 0 (0) | 0 (0) | 4 (9) |
| Torque teno virus | 0 (0) | 0 (0) | 4 (9) |
| Torque teno virus-like mini virus | 0 (0) | 3 (9) | 6 (14) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterson, A.C.; Sharma, H.; Kumar, A.; Ghersi, B.M.; Emrich, S.J.; Vandegrift, K.J.; Kapoor, A.; Blum, M.J. Rodent Virus Diversity and Differentiation across Post-Katrina New Orleans. Sustainability 2021, 13, 8034. https://doi.org/10.3390/su13148034
Peterson AC, Sharma H, Kumar A, Ghersi BM, Emrich SJ, Vandegrift KJ, Kapoor A, Blum MJ. Rodent Virus Diversity and Differentiation across Post-Katrina New Orleans. Sustainability. 2021; 13(14):8034. https://doi.org/10.3390/su13148034
Chicago/Turabian StylePeterson, Anna C., Himanshu Sharma, Arvind Kumar, Bruno M. Ghersi, Scott J. Emrich, Kurt J. Vandegrift, Amit Kapoor, and Michael J. Blum. 2021. "Rodent Virus Diversity and Differentiation across Post-Katrina New Orleans" Sustainability 13, no. 14: 8034. https://doi.org/10.3390/su13148034
APA StylePeterson, A. C., Sharma, H., Kumar, A., Ghersi, B. M., Emrich, S. J., Vandegrift, K. J., Kapoor, A., & Blum, M. J. (2021). Rodent Virus Diversity and Differentiation across Post-Katrina New Orleans. Sustainability, 13(14), 8034. https://doi.org/10.3390/su13148034






