Sustainability of Urban Soil Management: Analysis of Soil Physicochemical Properties and Bacterial Community Structure under Different Green Space Types
Abstract
1. Introduction
2. Materials and Methods
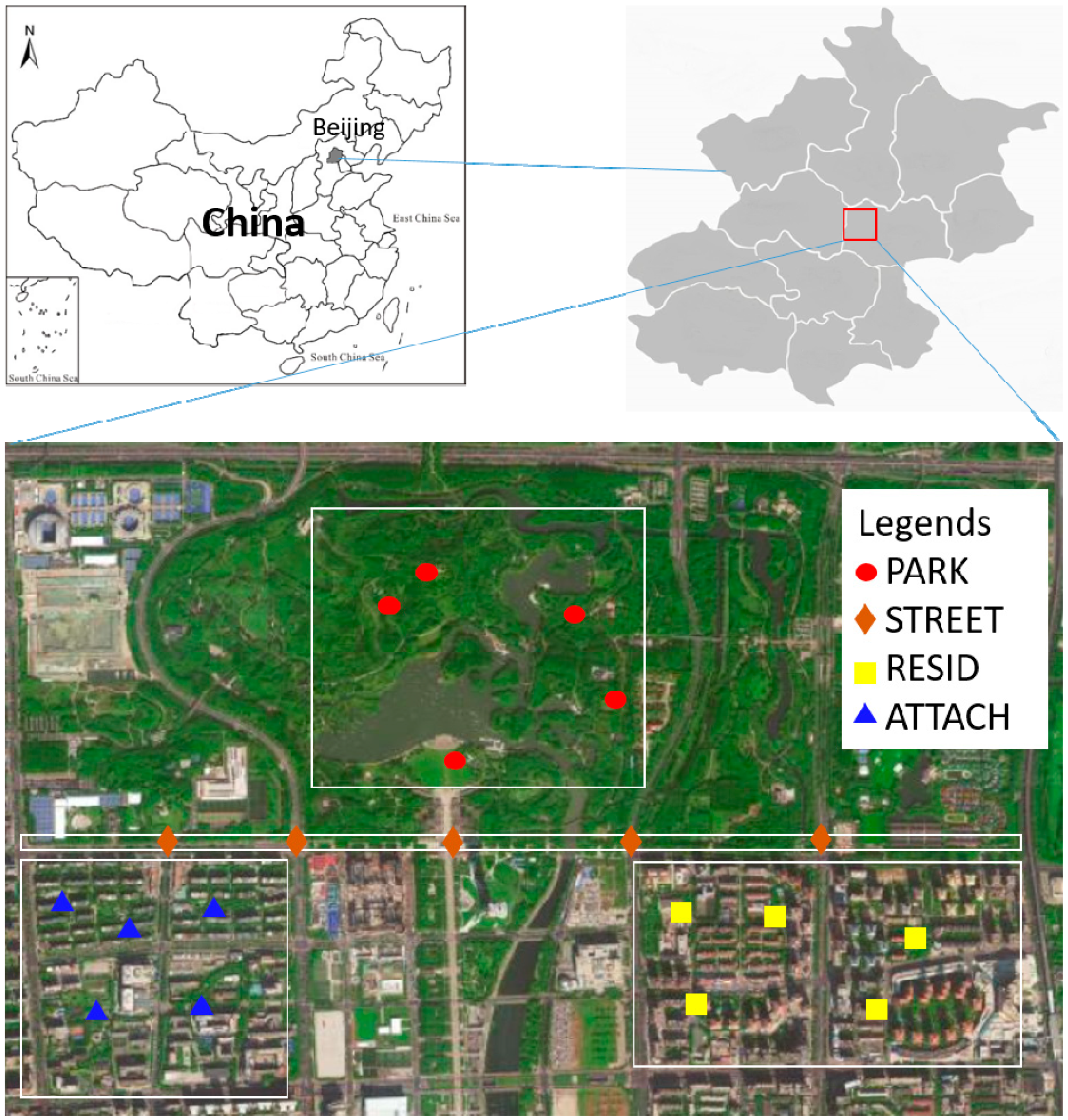
2.1. Study Area
2.2. Soil Sampling and Analysis
2.3. DNA Extraction and PCR Amplification
2.4. Illumina MiSeq Sequencing
2.5. Processing of Sequencing Data
2.6. Data Analysis
3. Results
3.1. Soil Physicochemical Characteristics
3.2. Analysis of 16S rRNA Sequencing Results
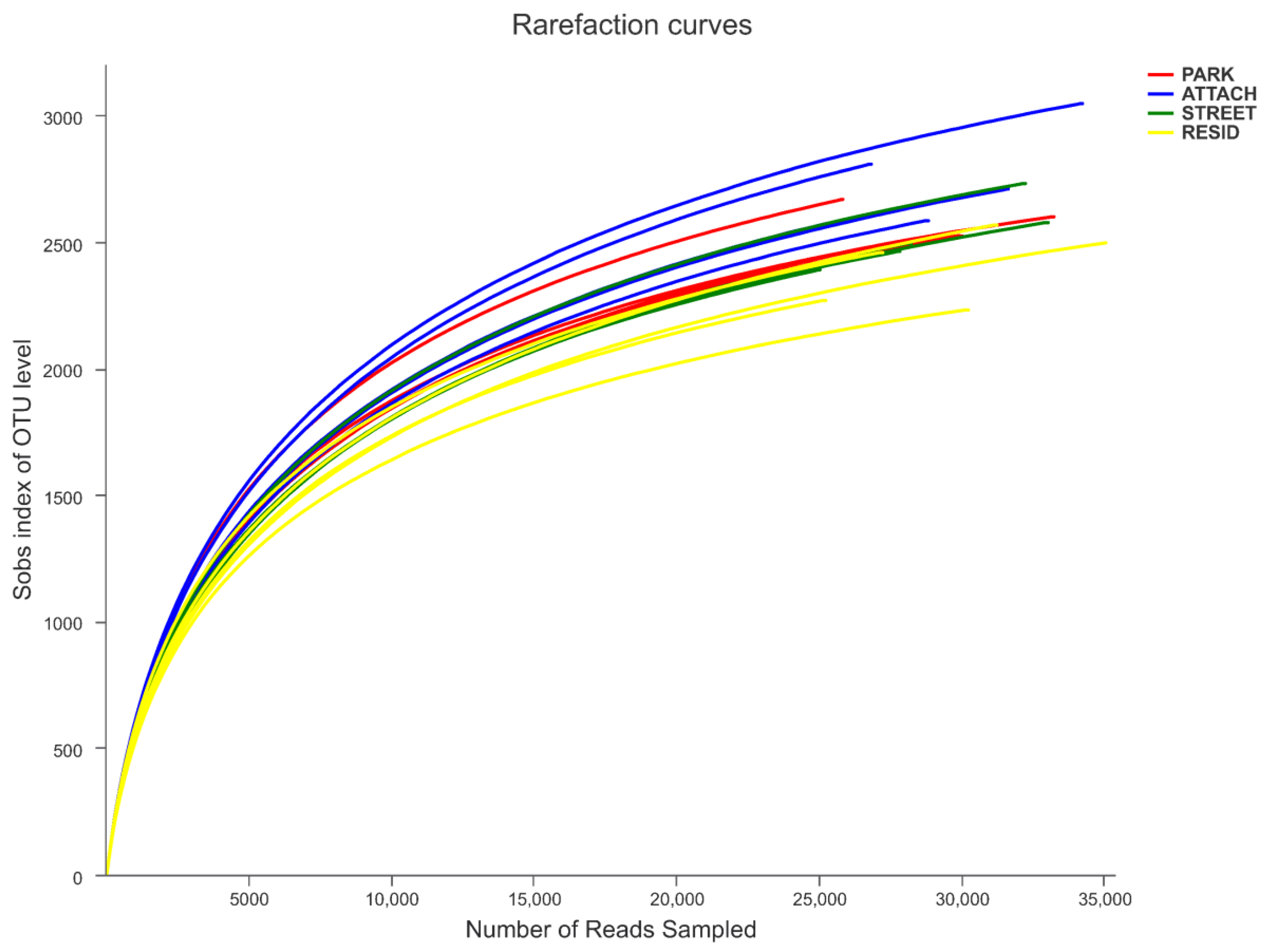
3.3. Richness and Diversity of Soil Bacterial Communities
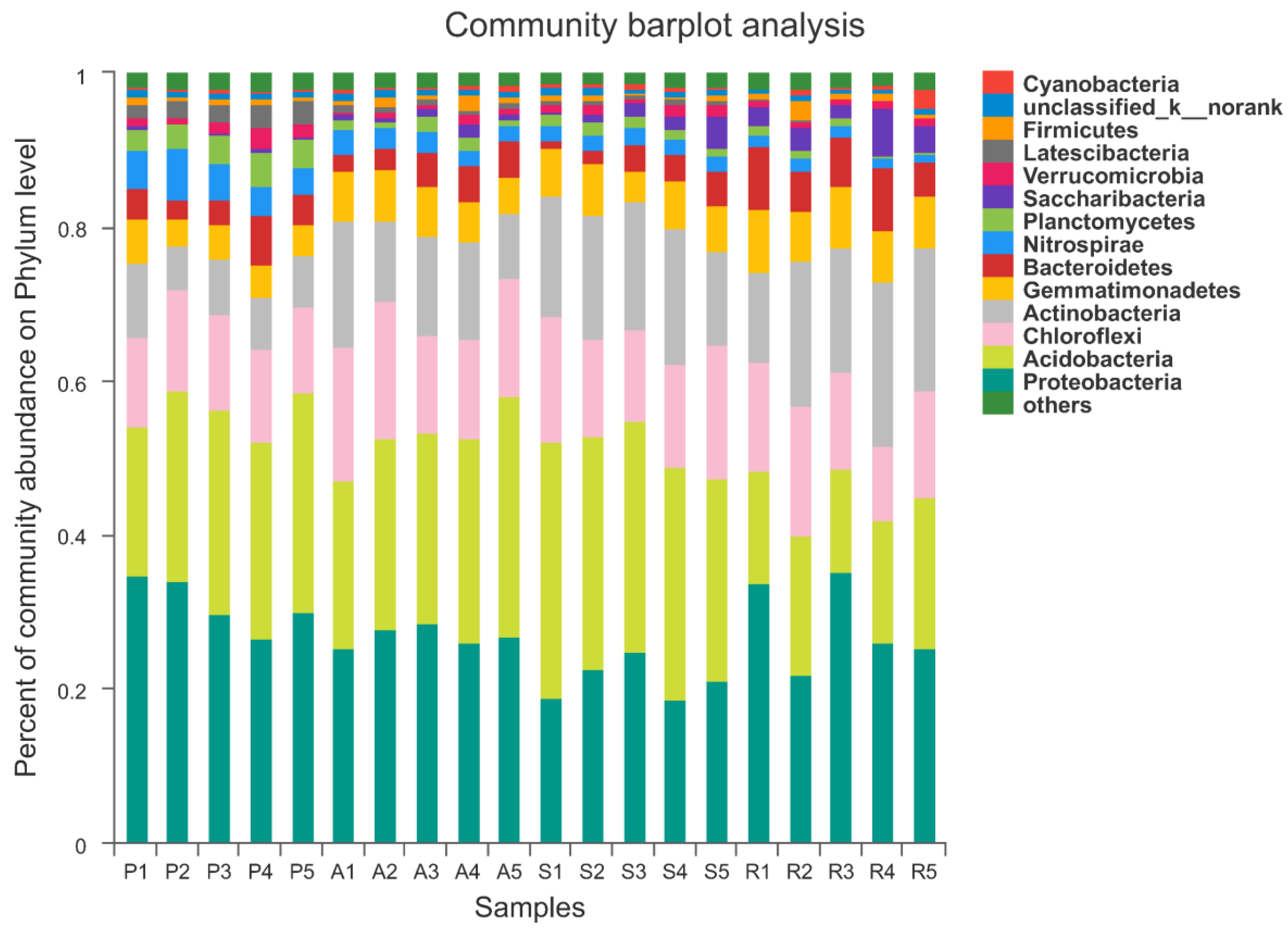
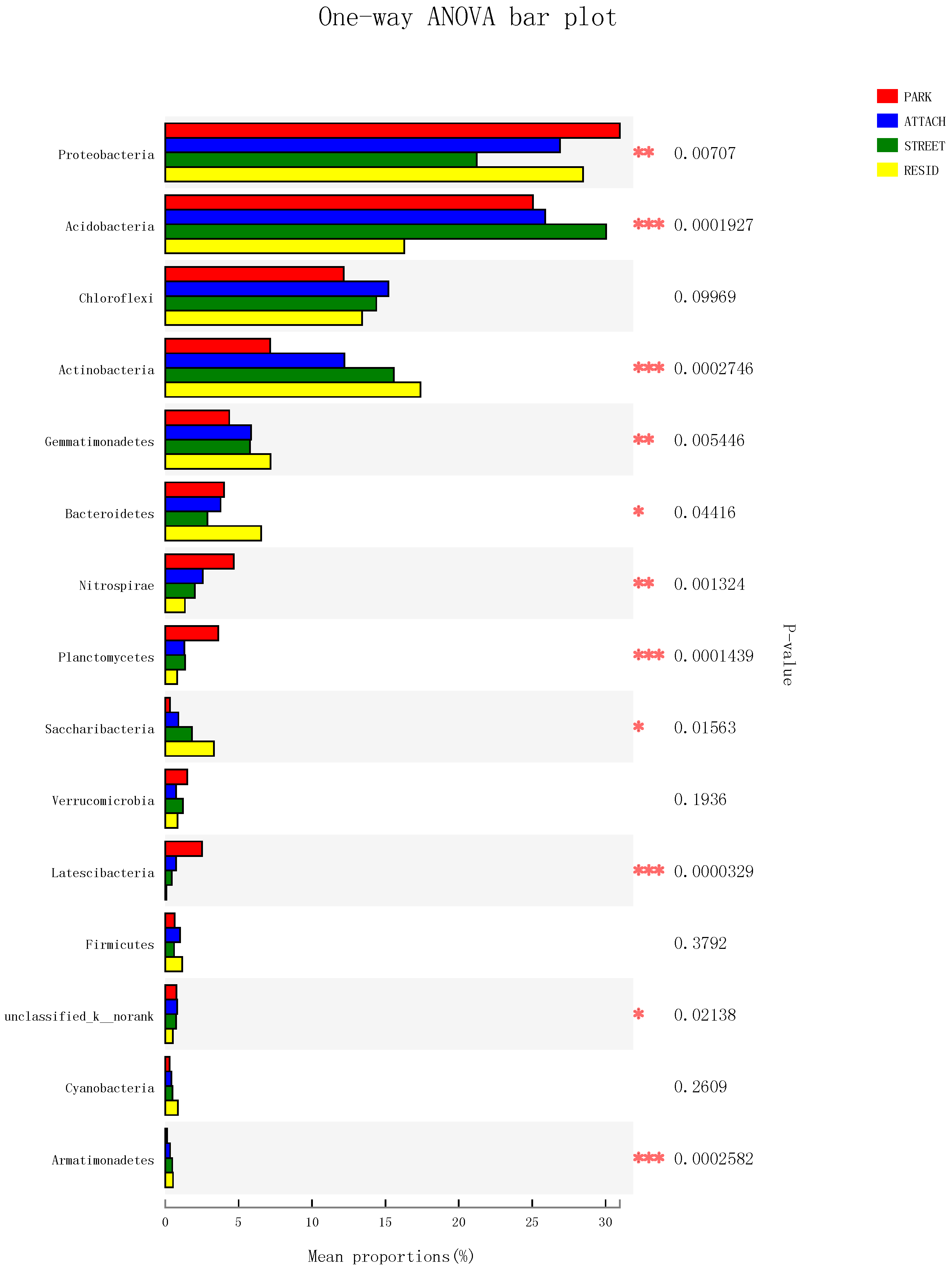
3.4. The Structure and Components of Soil Bacterial Community
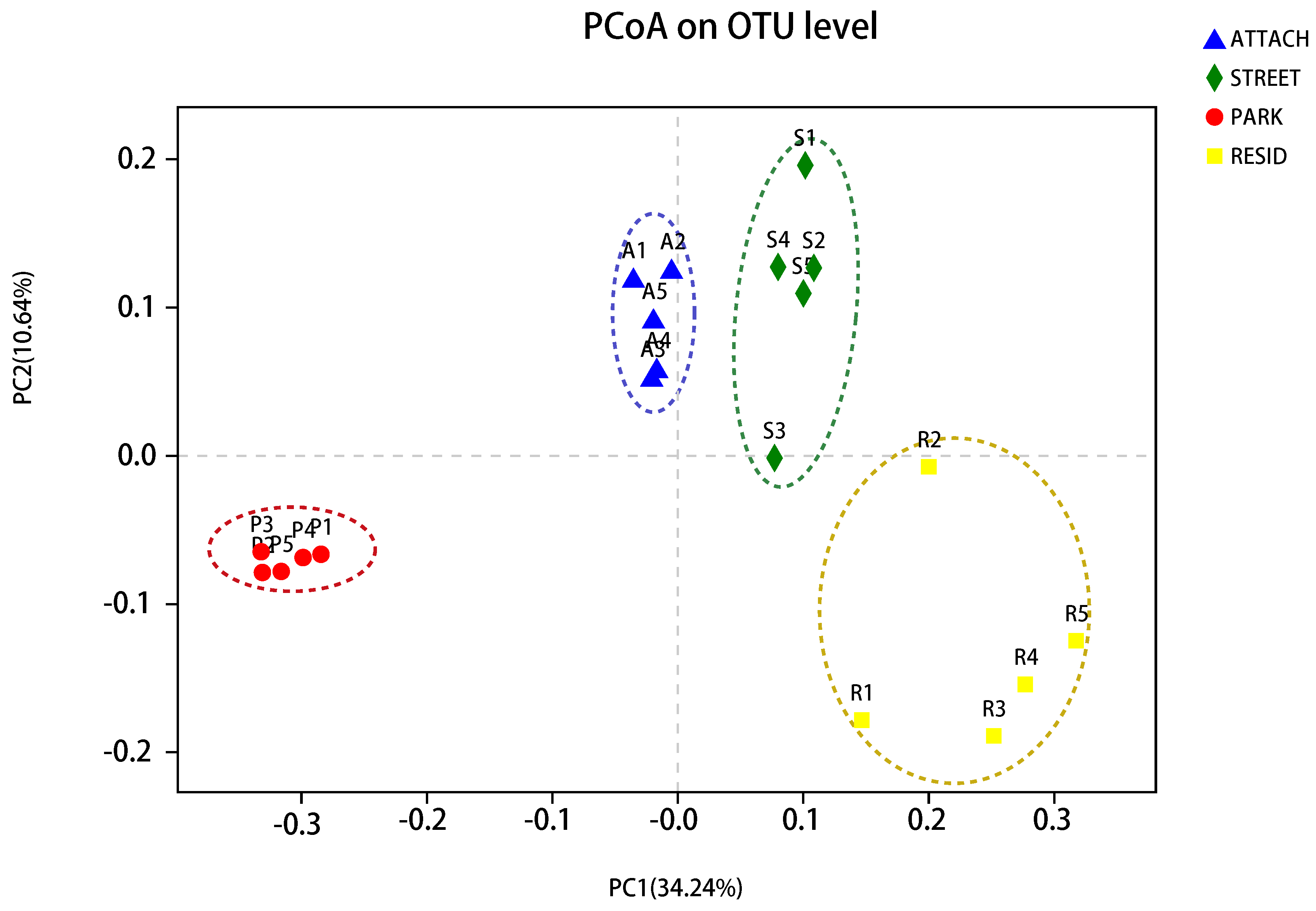
3.5. The Dissimilarity of Soil Bacterial Community
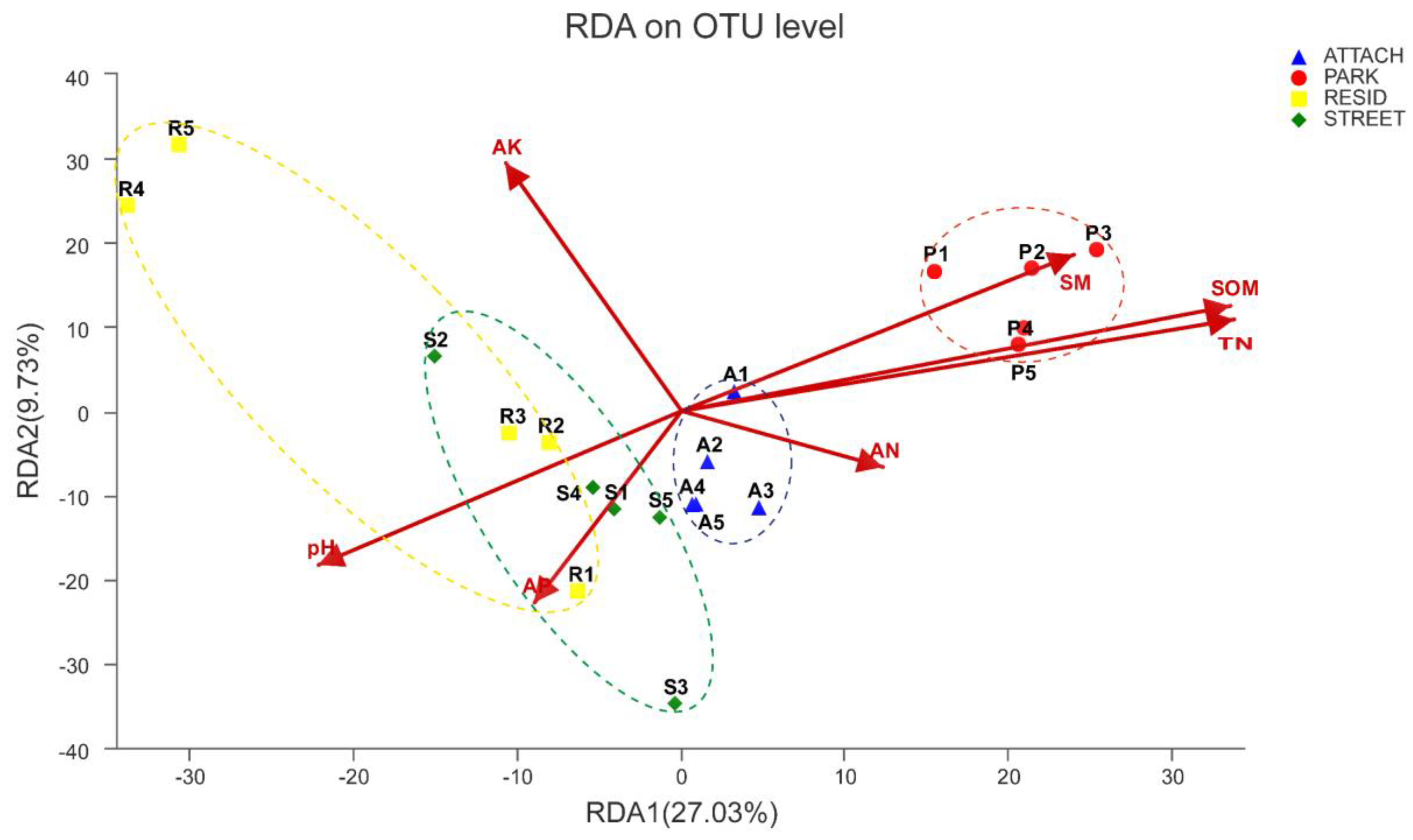
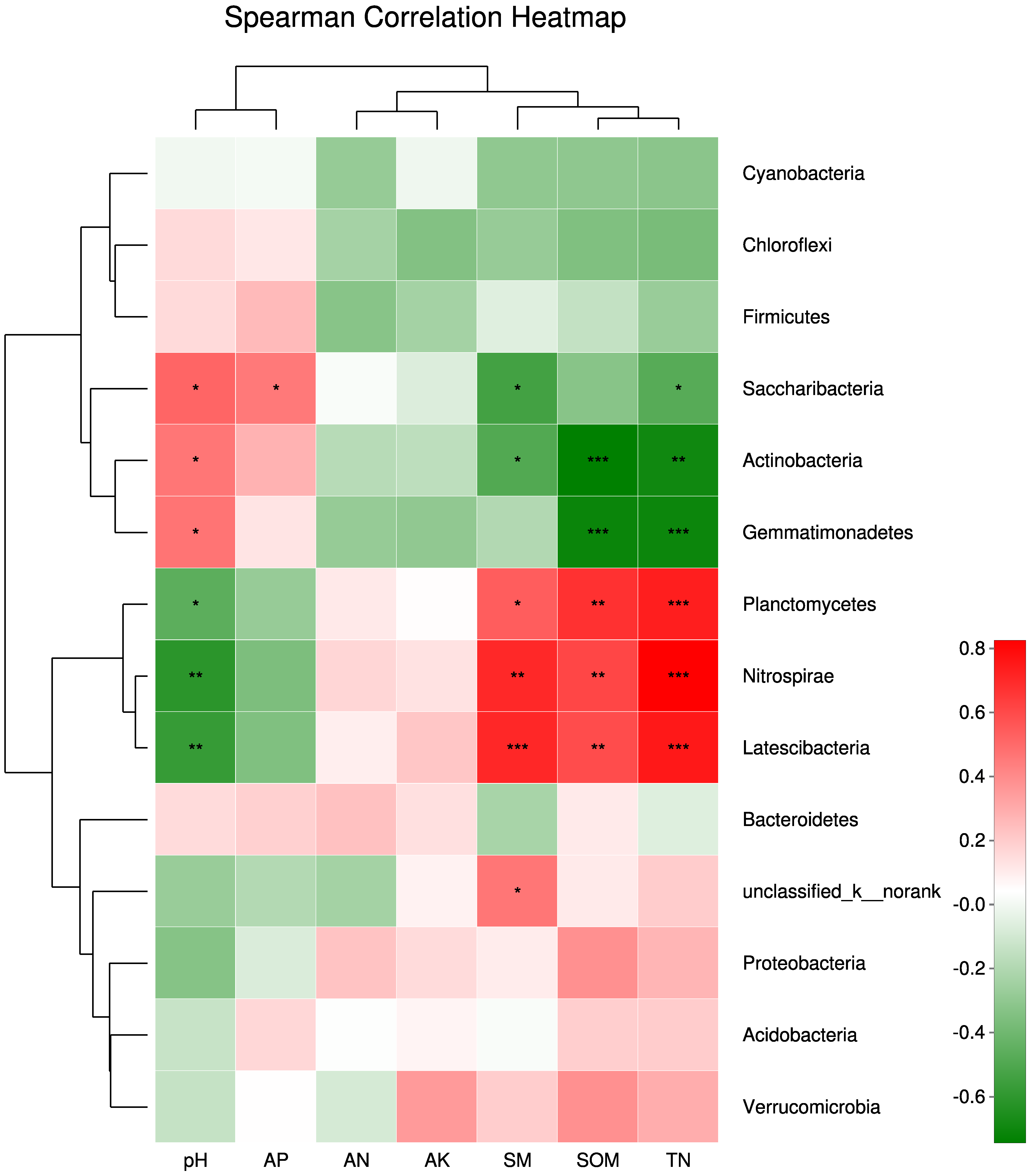
3.6. Relationships between Bacterial Community Structure and Soil Physicochemical Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ossola, A.; Aponte, C.; Hahs, A.K.; Livesley, S.J. Contrasting effects of urban habitat complexity on metabolic functional diversity and composition of litter and soil bacterial communities. Urban Ecosyst. 2016, 20, 595–607. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Peng, K.; Wu, S. Nutrients and heavy metals in urban soils under different green space types in Anji, China. Catena 2014, 115, 39–46. [Google Scholar] [CrossRef]
- De Kimpe, C.R.; Morel, J.-L. Urban soil management: A growing concern. Soil Sci. 2000, 165, 31–40. [Google Scholar] [CrossRef]
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Passey, T.; Xu, X. Amplicon-based metabarcoding reveals temporal response of soil microbial community to fumigation-derived products. Appl. Soil Ecol. 2016, 103, 83–92. [Google Scholar] [CrossRef]
- Zhao, D.; Li, F.; Wang, R. The effects of different urban land use patterns on soil microbial biomass nitrogen and enzyme activities in urban area of beijing, china. Acta Ecol. Sin. 2012, 32, 144–149. [Google Scholar] [CrossRef]
- Huang, X.; Liu, L.; Wen, T.; Zhang, J.; Wang, F.; Cai, Z. Changes in the soil microbial community after reductive soil disinfestation and cucumber seedling cultivation. Appl. Microbiol. Biotechnol. 2016, 100, 5581–5593. [Google Scholar] [CrossRef] [PubMed]
- Mhuireach, G.; Johnson, B.R.; Altrichter, A.E.; Ladau, J.; Meadow, J.F.; Pollard, K.S.; Green, J.L. Urban greenness influences airborne bacterial community composition. Sci. Total Environ. 2016, 571, 680–687. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, H.Y.H.; Meng, M.; Biswas, S.R.; Ye, L.; Zhang, J. Effects of land use change on the composition of soil microbial communities in a managed subtropical forest. For. Ecol. Manag. 2016, 373, 93–99. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in german forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, S. Polycentric development and the role of urban polycentric planning in china’s mega cities: An examination of beijing’s metropolitan area. Sustainability 2018, 10, 1588. [Google Scholar] [CrossRef]
- Pfeiffer, B.; Fender, A.-C.; Lasota, S.; Hertel, D.; Jungkunst, H.F.; Daniel, R. Leaf litter is the main driver for changes in bacterial community structures in the rhizosphere of ash and beech. Appl. Soil Ecol. 2013, 72, 150–160. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Xin, M.; Wang, L.; Jiang, H.; Li, N.; Wang, Z. The effects of erosion on the microbial populations and enzyme activity in black soil of northeastern china. Acta Ecol. Sin. 2014, 34, 295–301. [Google Scholar] [CrossRef]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer Science & Business Media: Berlin, Germany, 2007. [Google Scholar]
- Tan, K.H. Soil Sampling, Preparation, and Analysis; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Bremner, J.; Jenkinson, D. Determination of organic carbon in soil. Eur. J. Soil Sci. 1960, 11, 394–402. [Google Scholar] [CrossRef]
- Bremner, J. Determination of nitrogen in soil by the kjeldahl method. J. Agric. Sci. 1960, 55, 11–33. [Google Scholar] [CrossRef]
- Dewis, J.; Freitas, F. Physical and Chemical Methods of Soil and Water Analysis; Food and Agriculture Organization of the United Nations: Roam, Italy, 1970. [Google Scholar]
- Tsai, Y.-L.; Olson, B.H. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 1991, 57, 1070–1074. [Google Scholar] [PubMed]
- Claesson, M.J.; O’Sullivan, O.; Wang, Q.; Nikkilä, J.; Marchesi, J.R.; Smidt, H.; de Vos, W.M.; Ross, R.P.; O’Toole, P.W. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 2009, 4, e6669. [Google Scholar] [CrossRef] [PubMed]
- Neefs, J.-M.; Van de Peer, Y.; Hendriks, L.; De Wachter, R. Compilation of small ribosomal subunit rna sequences. Nucleic Acids Res. 1990, 18, 2237. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16s ribosomal rna gene pcr primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Lee, S.-M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Magurran, A.E. Ecological Diversity and Its Measurement; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Lynn, T.M.; Liu, Q.; Hu, Y.; Yuan, H.; Wu, X.; Khai, A.A.; Wu, J.; Ge, T. Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch. Microbiol. 2017, 199, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; Wang, Y.; Guan, J.; Liu, Q.; Lv, D.-A. Land use effects on soil quality along a native wetland to cropland chronosequence. Eur. J. Soil Biol. 2012, 53, 114–120. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, G.; Liang, J.; Zhang, J.; Cai, Q.; Huang, L.; Li, X.; Zhu, H.; Hu, C.; Shen, S. Changes of soil microbial biomass and bacterial community structure in dongting lake: Impacts of 50,000 dams of yangtze river. Ecol. Eng. 2013, 57, 72–78. [Google Scholar] [CrossRef]
- Zhang, C.; Nie, S.; Liang, J.; Zeng, G.; Wu, H.; Hua, S.; Liu, J.; Yuan, Y.; Xiao, H.; Deng, L.; et al. Effects of heavy metals and soil physicochemical properties on wetland soil microbial biomass and bacterial community structure. Sci. Total Environ. 2016, 557–558, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Meng, L.; Herman, U.; Lu, Z.; Crittenden, J. A survey of soil enzyme activities along major roads in beijing: The implications for traffic corridor green space management. Int. J. Environ. Res. Public Health 2015, 12, 12475–12488. [Google Scholar] [CrossRef] [PubMed]
- Seddaiu, G.; Porcu, G.; Ledda, L.; Roggero, P.P.; Agnelli, A.; Corti, G. Soil organic matter content and composition as influenced by soil management in a semi-arid mediterranean agro-silvo-pastoral system. Agric. Ecosyst. Environ. 2013, 167, 1–11. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, X.; Tu, S.; Lindström, K. Soil microbial biomass, crop yields, and bacterial community structure as affected by long-term fertilizer treatments under wheat-rice cropping. Eur. J. Soil Biol. 2009, 45, 239–246. [Google Scholar] [CrossRef]
- Shen, J.-P.; Cao, P.; Hu, H.-W.; He, J.-Z. Differential response of archaeal groups to land use change in an acidic red soil. Sci. Total Environ. 2013, 461, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.; Chen, W. Soil quality assessment of urban green space under long-term reclaimed water irrigation. Environ. Sci. Pollut. Res. Int. 2016, 23, 4639–4649. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.; Koranda, M.; Kitzler, B.; Fuchslueger, L.; Schnecker, J.; Schweiger, P.; Rasche, F.; Zechmeister-Boltenstern, S.; Sessitsch, A.; Richter, A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010, 187, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Q.; Li, Y.; Liu, J.; Pan, H.; Guan, X.; Xu, X.; Xu, J.; Di, H. Impact of mowing management on nitrogen mineralization rate and fungal and bacterial communities in a semiarid grassland ecosystem. J. Soils Sediments 2016, 17, 1715–1726. [Google Scholar] [CrossRef]
- Knops, J.M.; Tilman, D. Dynamics of soil nitrogen and carbon accumulation for 61 years after agricultural abandonment. Ecology 2000, 81, 88–98. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in tropical rain forests and coral reefs. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Hui, N.; Jumpponen, A.; Francini, G.; Kotze, D.J.; Liu, X.; Romantschuk, M.; Strommer, R.; Setala, H. Soil microbial communities are shaped by vegetation type and park age in cities under cold climate. Environ. Microbiol. 2017, 19, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xia, H.; Wu, J.; Yang, G.; Zhang, X.; Peng, H.; Yu, X.; Li, L.; Xiao, H.; Qi, H. Shifts in the relative abundance of bacteria after wine-lees-derived biochar intervention in multi metal-contaminated paddy soil. Sci. Total Environ. 2017, 599–600, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z. Assessing bacterial diversity in soil. J. Soils Sediments 2008, 8, 379–388. [Google Scholar] [CrossRef]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Qu, Y.; Su, X.; Zhou, J.; Li, D. Community structure and elevational diversity patterns of soil acidobacteria. J. Environ. Sci. 2014, 26, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Tan, G.; Wang, H.; Gai, X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- De Mandal, S.; Chatterjee, R.; Kumar, N.S. Dominant bacterial phyla in caves and their predicted functional roles in c and n cycle. BMC Microbiol. 2017, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Guo, L.; Niu, M.; Miao, F.; Jiao, S.; Hu, T.; Long, M. Ecological diversity and co-occurrence patterns of bacterial community through soil profile in response to long-term switchgrass cultivation. Sci. Rep. 2017, 7, 3608. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Diao, M.; Zhang, B.; Liu, H.; Wang, S.; Yang, M. Spatial distribution of vanadium and microbial community responses in surface soil of panzhihua mining and smelting area, china. Chemosphere 2017, 183, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Göransson, H.; Venterink, H.O.; Bååth, E. Soil bacterial growth and nutrient limitation along a chronosequence from a glacier forefield. Soil Biol. Biochem. 2011, 43, 1333–1340. [Google Scholar] [CrossRef]
- Shi, Y.-W.; Lou, K.; Li, C.; Wang, L.; Zhao, Z.-Y.; Zhao, S.; Tian, C.-Y. Illumina-based analysis of bacterial diversity related to halophytes salicornia europaea and sueada aralocaspica. J. Microbiol. 2015, 53, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Prendergast-Miller, M.T.; de Menezes, A.B.; Macdonald, L.M.; Toscas, P.; Bissett, A.; Baker, G.; Farrell, M.; Richardson, A.E.; Wark, T.; Thrall, P.H. Wildfire impact: Natural experiment reveals differential short-term changes in soil microbial communities. Soil Biol. Biochem. 2017, 109, 1–13. [Google Scholar] [CrossRef]
- Girvan, M.S.; Bullimore, J.; Pretty, J.N.; Osborn, A.M.; Ball, A.S. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 2003, 69, 1800–1809. [Google Scholar] [CrossRef] [PubMed]

| Types | SOM g/kg | TN g/kg | pH | SM % | AN mg/kg | AK mg/kg | AP mg/kg |
|---|---|---|---|---|---|---|---|
| PARK | 33.95a | 1.36a | 7.19b | 13.73a | 80.11a | 210.56a | 17.75a |
| ATTACH | 14.32b | 0.75b | 7.77a | 11.54a | 64.40b | 128.06a | 21.02a |
| STREET | 12.29b | 0.59bc | 7.83a | 6.66b | 57.85b | 128.85a | 22.14a |
| RESID | 10.09b | 0.43c | 7.85a | 6.31b | 54.56b | 215.85a | 22.36a |
| Sample | Shannon | Simpson | Ace | Chao1 | Coverage |
|---|---|---|---|---|---|
| PARK | 7.23ab | 0.0018b | 5434.58b | 5461.76b | 0.96ab |
| ATTACH | 7.30a | 0.0017b | 6093.74a | 6034.51a | 0.95b |
| STREET | 7.12b | 0.0024a | 5449.89b | 5460.28b | 0.96ab |
| RESID | 7.13b | 0.0023a | 5131.49b | 5183.30b | 0.96a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, S.; Sun, X.; Tong, J.; Fu, Z.; Li, J. Sustainability of Urban Soil Management: Analysis of Soil Physicochemical Properties and Bacterial Community Structure under Different Green Space Types. Sustainability 2019, 11, 1395. https://doi.org/10.3390/su11051395
Zhang J, Li S, Sun X, Tong J, Fu Z, Li J. Sustainability of Urban Soil Management: Analysis of Soil Physicochemical Properties and Bacterial Community Structure under Different Green Space Types. Sustainability. 2019; 11(5):1395. https://doi.org/10.3390/su11051395
Chicago/Turabian StyleZhang, Junda, Suyan Li, Xiangyang Sun, Jing Tong, Zhen Fu, and Jing Li. 2019. "Sustainability of Urban Soil Management: Analysis of Soil Physicochemical Properties and Bacterial Community Structure under Different Green Space Types" Sustainability 11, no. 5: 1395. https://doi.org/10.3390/su11051395
APA StyleZhang, J., Li, S., Sun, X., Tong, J., Fu, Z., & Li, J. (2019). Sustainability of Urban Soil Management: Analysis of Soil Physicochemical Properties and Bacterial Community Structure under Different Green Space Types. Sustainability, 11(5), 1395. https://doi.org/10.3390/su11051395




