Identification of Nitrogen, Phosphorus, and Potassium Deficiencies Based on Temporal Dynamics of Leaf Morphology and Color
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Materials
2.2. RGB Image Acquisition and Processing
2.3. Dynamic Analysis of Leaf Responses to NPK Deficiency in Different Leaf Positions
2.4. Quantification of Dynamic Characteristics
2.5. Identification Method
3. Results
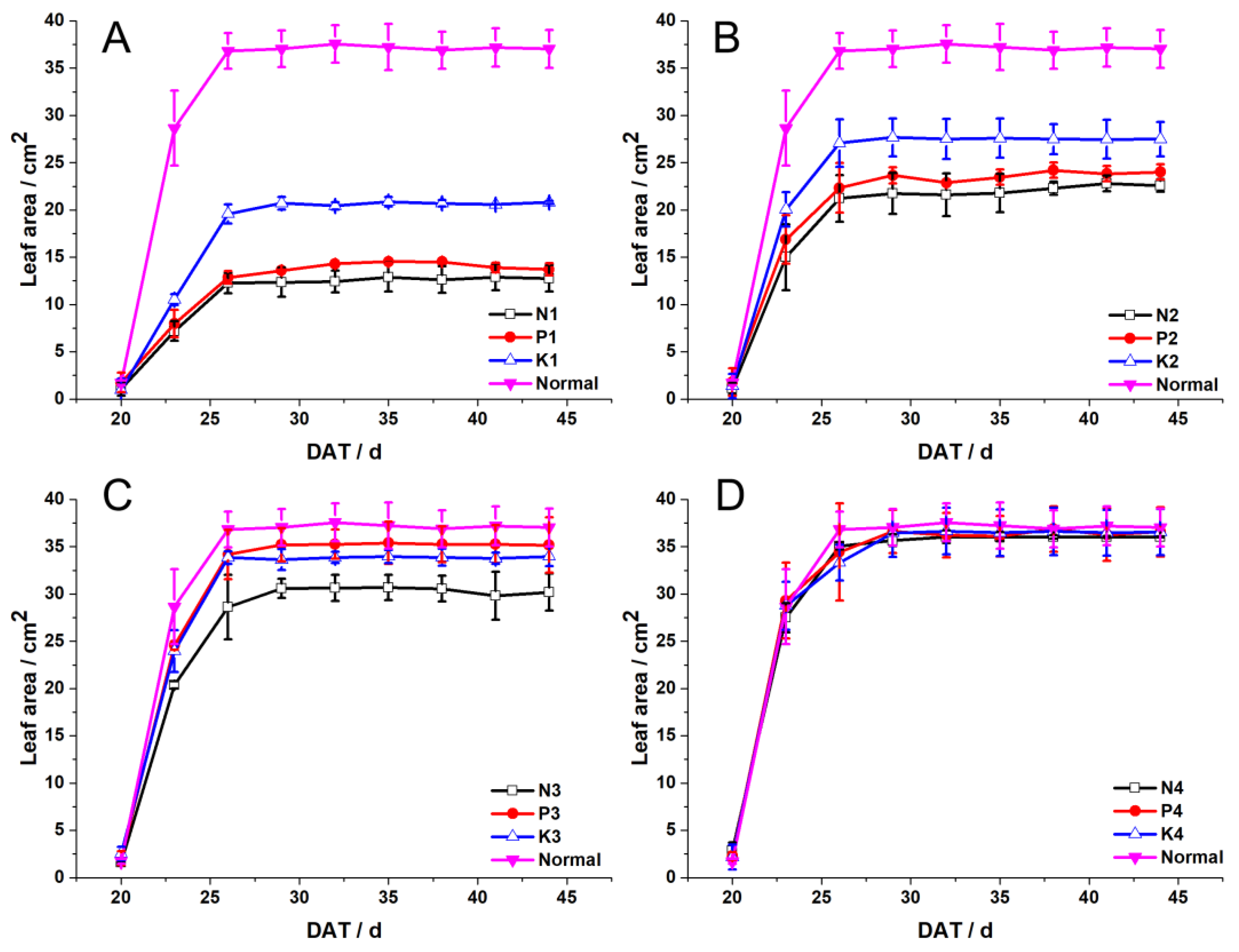
3.1. Leaf Responses of the First Incomplete Leaf and Fully Expanded Leaf to NPK Deficiencies
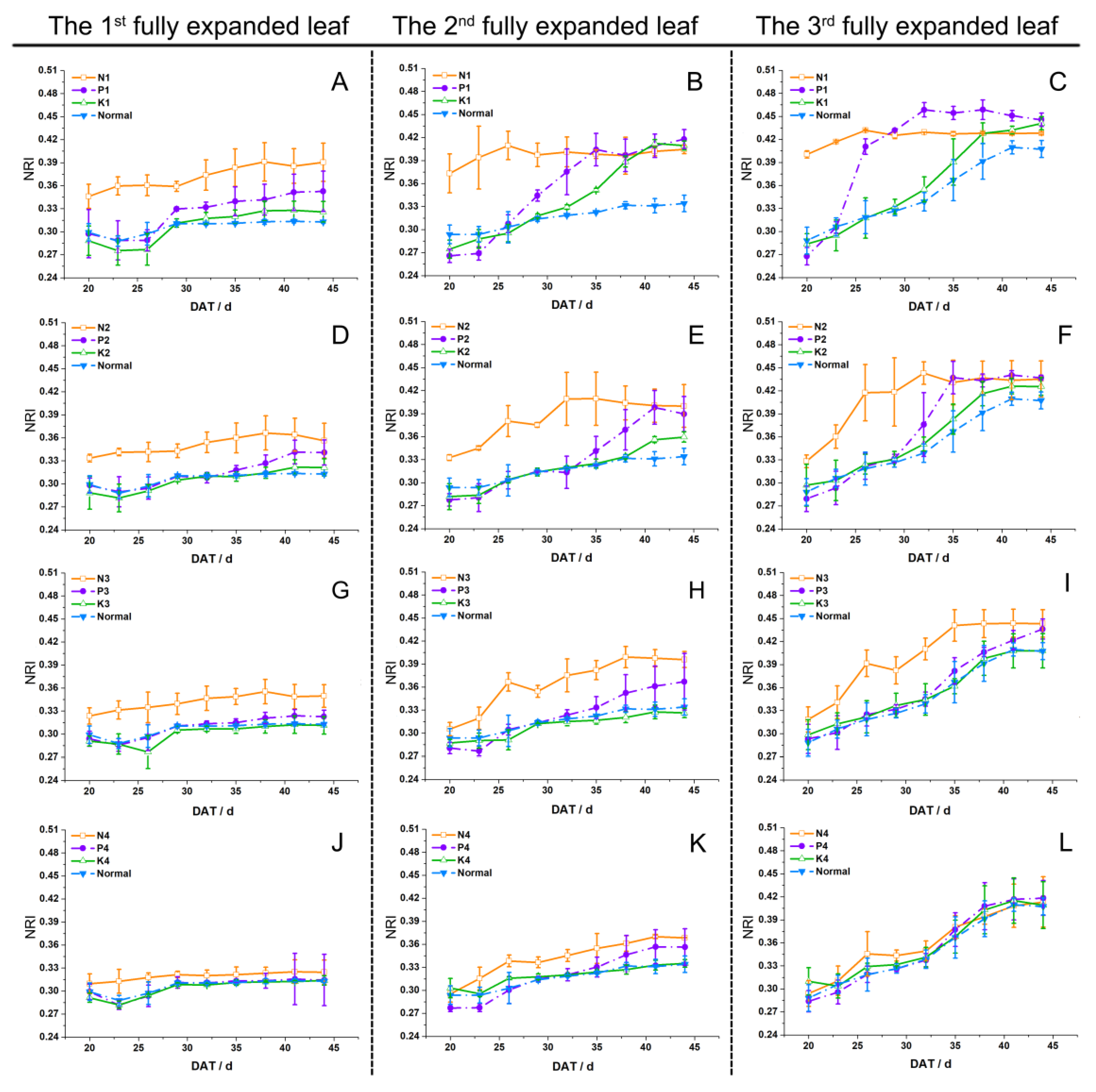
3.2. Selection of Optimal Indices
3.3. Identification of NPK Deficiency
4. Discussion
4.1. Different Influence of NPK Deficiencies on Leaf Extension and Senescence
4.2. Quantification Method of Dynamic Indices
4.3. Effectiveness of the Dynamic Nature of Different Leaf Positions in Identification
4.4. Future Work
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, Y.; Qiang, L.; He, S.; Yi, S.; Liu, X. Prediction of nitrogen and phosphorus contents incitrus leavesbased on hyperspectral imaging. Int. J. Agric. Biol. Eng. 2015, 8, 80–88. [Google Scholar]
- Rapaport, T.; Hochberg, U.; Shoshany, M.; Karnieli, A.; Rachmilevitch, S. Combining leaf physiology, hyperspectral imaging and partial least squares-regression (PLS-R) for grapevine water status assessment. ISPRS J. Photogramm. Remote Sens. 2015, 109, 88–97. [Google Scholar] [CrossRef]
- Xu, S.; He, J.; Ma, Y.; Liang, H.; Liu, G.; He, X. Research Progress of Hyperspectral Imaging Technology for Nondestructive Detection of Fruit Qualit. Food Res. Dev. 2013, 34, 4–8. [Google Scholar]
- Wang, Y.; Wang, D.; Shi, P.; Omasa, K. Estimating rice chlorophyll content and leaf nitrogen concentration with a digital still color camera under natural light. Plant Methods 2014, 10, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.; Zhang, G.; Wang, J. Estimating nitrogen status of rice using the image segmentation of G-R thresholding method. Field Crop. Res. 2013, 149, 33–39. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, G.; Wang, C. Digital camera-based image segmentation of rice canopy and diagnosis of nitrogen nutrition. Trans. Chin. Soc. Agric. Eng. 2012, 28, 131–136. (In Chinese) [Google Scholar]
- Sakamoto, T.; Shibayama, M.; Kimura, A.; Takada, E. Assessment of digital camera-derived vegetation indices in quantitative monitoring of seasonal rice growth. ISPRS J. Photogramm. Remote Sens. 2011, 66, 872–882. [Google Scholar] [CrossRef]
- Chen, L.; Lin, L.; Cai, G.; Sun, Y.; Huang, T.; Wang, K.; Deng, J. Identification of Nitrogen, Phosphorus, and Potassium Deficiencies in Rice Based on Static Scanning Technology and Hierarchical Identification Method. PLoS ONE 2014, 9, e11320011. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, K. Diagnosing of rice nitrogen stress based on static scanning technology and image information extraction. J. Soil Sci. Plant Nutr. 2014, 14, 382–393. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Walker, C.; Angus, J. Estimating the nitrogen status of crops using a digital camera. Field Crop. Res. 2010, 118, 221–227. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, G. Nitrogen Status Diagnosis of Rice Based on a Digital Camera. Chin. Agric. Sci. Bull. 2012, 28, 111–117. (In Chinese) [Google Scholar]
- Saberioon, M.; Amin, M.; Aimrun, W.; Gholizadeh, A.; Rahim Anuar, A. Assessment of colour indices derived from conventional digital camera for determining nitrogen status in rice plants. J. Food. Agric. Environ. 2013, 11, 655–662. [Google Scholar]
- Shi, Y. Rice Nutrition Diagnosis and Modeling Based on Digital Image. Doctoral Dissertation, Zhejiang University, Hangzhou, China, 2011. (In Chinese). [Google Scholar]
- Chen, L. Rice Nutrition Identification and Diagnosis Based on Machine Vision Technology. Doctoral Dissertation, Zhejiang University, Hangzhou, China, 2014. (In Chinese). [Google Scholar]
- Qi, W.; Wang, C.; Guo, X. Study on plant behavior perception based on computer vision: A review. Jiangsu Agric. Sci. 2017, 45, 20–26. (In Chinese) [Google Scholar]
- Neilson, E.; Edwards, A.; Blomstedt, C.; Berger, B.; Moller, B.; Gleadow, R. Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. J. Exp. Bot. 2015, 66, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Poiré, R.; Chochois, V.; Sirault, X.; Vogel, J.; Watt, M. Digital imaging approaches for phenotyping whole plant nitrogen and phosphorus response in Brachypodium distachyon. J. Integr. Plant. Biol. 2014, 56, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Tackenberg, O. A new method for non-destructive measurement of biomass, growth rates, vertical biomass distribution and dry matter content based on digital image analysis. Ann. Bot. 2007, 99, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Humplík, J.; Lazár, D.; Husičková, A.; Spíchal, L. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses—A review. Plant Methods 2015, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, L.; Liu, X.; Liu, L.; Cao, W.; Zhu, Y. Modeling Dynamics of Leaf Color Based on RGB Value in Rice. J. Intergr. Agric. 2014, 13, 749–759. [Google Scholar]
- Qin, G.; Jinson, D.; Cha, L.; Yuanyuan, S.; Ke, W.; Zhang, S. Diagnosis of Rice Nitrogen Nutrition Based on Spectral and Shape Characteristics of Scanning Leaves. Trans. Chin. Soc. Agric. Mach. 2012, 43, 170–174. (In Chinese) [Google Scholar]
- Saberioon, M.; Amin, M.; Anuar, A.; Gholizadeh, A.; Wayayok, A.; Khairunniza-Bejo, S. Assessment of rice leaf chlorophyll content using visible bands at different growth stages at both the leaf and canopy scale. Int. J. Appl. Earth Obs. Geoinf. 2014, 32, 35–45. [Google Scholar] [CrossRef]
- Li, L.; Zhang, M.; Ren, T.; Li, X.; Cong, R.; Wu, L.; Lu, J. Diagnosis of N nutrition of rice using digital image processing technique. J. Plant Nutr. Fertil. 2015, 21, 259–268. (In Chinese) [Google Scholar]
- Chen, J.; Yao, X.; Huang, F.; Liu, Y.; Yu, Q.; Wang, N.; Xu, H.; Zhu, Y. N status monitoring model in winter wheat based on image processing. Trans. Chin. Soc. Agric. Eng. 2016, 32, 163–170. (In Chinese) [Google Scholar]
- Gonzalez, R.; Woods, R. Digital Image Processing Using MATLAB, 2nd ed.; Pearson Education North Asia Limited: Hong Kong, China, 2002. [Google Scholar]
- Harbur, M.; Owen, M. Light and growth rate effects on crop and weed responses to nitrogen. Weed Sci. 2004, 52, 578–583. [Google Scholar] [CrossRef]
- Hunt, R. Plant Growth Curves: The Functional Approach to Plant Growth Analysis; Edward Arnold: London, UK, 1982. [Google Scholar]
- Xiong, X.; Yu, L.; Yang, W.; Liu, M.; Jiang, N.; Wu, D.; Chen, G.; Xiong, L.; Liu, K.; Liu, Q. A high-throughput stereo-imaging system for quantifying rape leaf traits during the seedling stage. Plant Methods 2017, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, J.; Lu, J.; Yu, D. An efficient renovation on kernel Fisher discriminant analysis and face recognition experiments. Pattern Recognit. 2004, 37, 2091–2094. [Google Scholar] [CrossRef]
- Na, W.; Ke, W.; Xie, R.; Lai, J.; Ming, B.; Li, S. Maize Leaf Disease Identification Based on Fisher Discrimination Analysis. Sci. Agric. Sin. 2009, 42, 3836–3842. (In Chinese) [Google Scholar]
- Ingestad, T. Nitrogen Stress in Birch Seedlings. Physiol. Plant 1979, 45, 149–157. [Google Scholar] [CrossRef]
- Mengel, K.; Kirkby, E. Principles of Plant Nutrition; The International Potash Institute (IPI): Bern, Switzerland, 1987. [Google Scholar]
- Du, L.; Gong, W.; Shi, S.; Yang, J.; Sun, J.; Zhu, B.; Song, S. Estimation of rice leaf nitrogen contents based on hyperspectral LIDAR. Int. J. Appl. Earth Obs. 2016, 44, 136–143. [Google Scholar] [CrossRef]
- Inoue, Y.; Sakaiya, E.; Zhu, Y.; Takahashi, W. Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sens. Environ. 2012, 126, 210–221. [Google Scholar] [CrossRef]
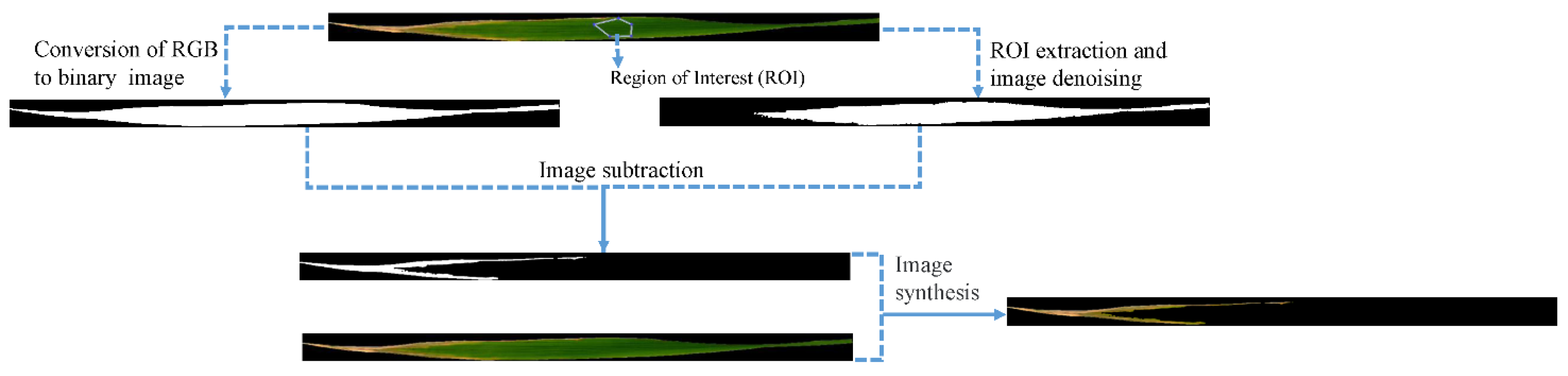
| Category | Index | Formula |
|---|---|---|
| Morphological indices | Leaf width (LW) | |
| Leaf length (LL) | ||
| Leaf area (LA or CLA) | ||
| Leaf perimeter (LP) | ||
| Eccentricity (EC) | Eccentricity = AxisLengthlong/AxisLengthshort | |
| Rectangularity (RE) | Rectangularity = Areaobject/Areabounding-box | |
| Area convexity (AC) | Area convexity = Area/Convex Area | |
| Circularity (CI) | Circularity = Rinscribedcircle/Rexcircle | |
| Form factor (FF) | Form fact = Perimeter/(4*√Area) | |
| Color indices | Red (R or CR) | |
| Green (G or CG) | ||
| Blue (B or CB) | ||
| Hue (H) | ||
| Saturability (S) | ||
| Brightness (BR) | Bightness = 0.3*R + 0.6*G + 0.1*B | |
| Normalized red index (NRI) | NRI = R/(R + G + B) | |
| Normalized green index (NGI) | NGI = G/(R + G + B) | |
| Normalized blue index (NBI) | NBI = B/(R + G + B) | |
| Dark green color index (DGCI) | DGCI = {(Hue − 60)/60 + (1−Saturation) + (1 − Brightness)}/3 | |
| Green-red vegetation index (GRVI) | GRVI = (G − R)/(G+R) | |
| Kawashima index (IKAW) | IKAW = (R − B)/(R + B) | |
| Principal component analysis index (IPCA) | IPCA = 0.994*|R − B|+0.961*|G − B| + 0.914*|G − R| |
| Category | DAT | 3 Days Interval Dataset | DAT | 6 Days Interval Dataset |
|---|---|---|---|---|
| Single dataset | 20–23 | P1 | 20–26 | P1′ |
| 23–26 | P2 | |||
| 26–29 | P3 | 26–32 | P2′ | |
| 29–32 | P4 | |||
| 32–35 | P5 | 32–38 | P3′ | |
| 35–38 | P6 | |||
| 38–41 | P7 | —— | —— | |
| Combined dataset | 20–26 | P1. P2 | 20–32 | P1′. P2′ |
| 20–29 | P1. P2. P3 (P1–P3) | |||
| 20–32 | P1. P2. P3. P4 (P1–P4) | |||
| 20–35 | P1. P2. P3. P4. P5 (P1–P5) | 20–38 | P1′. P2′. P3′ (P1′–P3′) | |
| 20–38 | P1. P2. P3. P4. P5. P6 (P1–P6) | |||
| 20–41 | P1. P2. P3. P4. P5. P6. P7 (P1–P7) | —— | —— |
| Leaf Position | Time Interval | Data Set | Selected Indices (RGR) |
|---|---|---|---|
| The first incomplete leaf | 3 days interval | P1 | LP, R, LL, AC, EC, CI, S, IKAW |
| P2 | NRI, NBI, LW, CI, IKAW | ||
| P3 | LA, R, NRI, H | ||
| P4 | LA, GRVI | ||
| P5 | IPCA, IKAW, G, S | ||
| P6 | GRVI, DGCI | ||
| P7 | NRI, NGI, NBI, IKAW | ||
| 6 days interval | P1′ | LA, G, B, NGI, DGCI, GRVI, IKAW, IPCA | |
| P2′ | LP, R, FF, IKAW, IPCA | ||
| P3′ | R, NGI, NBI, CI, BR, H, IKAW, IPCA | ||
| The 3rd fully expanded leaf | 3 days interval | P1 | CLA, LP |
| P2 | LA, R, B, NGI, NBI, AC, RE, S, GRVI, IPCA | ||
| P3 | B, NRI, NGI, NBI, BR, IKAW, CR, CB | ||
| P4 | LP, R, B, NRI, LL, BR, H, GRVI, IPCA, CR, CB | ||
| P5 | G, NGI, NBI, IKAW | ||
| P6 | R, G, NRI, NGI, NBI, H, DGCI, GRVI, IKAW, IPCA, CB | ||
| P7 | H, CLA | ||
| 6 days interval | P1′ | CLA, CG, NGI | |
| P2′ | NRI, NGI, NBI, LL, GRVI, IKAW | ||
| P3′ | NGI, NBI, CB, CR, IKAW, IPCA, H, S, AC |
| Category | Time Interval | Data Set | Optimal Leaf | Training (%) | Validation (%) |
|---|---|---|---|---|---|
| Single dataset | 3 days | P1 | the first incomplete leaf | 67.2 | 59.7 |
| P2 | the 3rd fully expanded leaf | 64.2 | 60.8 | ||
| P3 | the 3rd fully expanded leaf | 74.2 | 71.7 | ||
| P4 | the 3rd fully expanded leaf | 83.3 | 79.6 | ||
| P5 | the 3rd fully expanded leaf | 69.2 | 57.1 | ||
| P6 | the 3rd fully expanded leaf | 75.0 | 65.6 | ||
| P7 | the first incomplete leaf | 69.9 | 63.0 | ||
| 6 days | P1′ | the first incomplete leaf | 73.7 | 71.4 | |
| P2′ | the 3rd fully expanded leaf | 73.2 | 69.6 | ||
| P3′ | the 3rd fully expanded leaf | 80.2 | 69.1 | ||
| Combined dataset | 3 days | P1. P2 | the 3rd fully expanded leaf | 77.5 | 68.3 |
| P1–P3 | the 3rd fully expanded leaf | 81.7 | 77.5 | ||
| P1–P4 | the 3rd fully expanded leaf | 92.6 | 88.9 | ||
| P1–P5 | the 3rd fully expanded leaf | 95.1 | 92.6 | ||
| P1–P6 | the 3rd fully expanded leaf | 85.9 | 92.2 | ||
| P1–P7 | the 3rd fully expanded leaf | 96.8 | 96.8 | ||
| 6 days | P1′. P2′ | the 3rd fully expanded leaf | 83.2 | 76.8 | |
| P1′–P3′ | the first incomplete leaf | 88.5 | 83.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Tong, C.; He, S.; Wang, K.; Chen, L. Identification of Nitrogen, Phosphorus, and Potassium Deficiencies Based on Temporal Dynamics of Leaf Morphology and Color. Sustainability 2018, 10, 762. https://doi.org/10.3390/su10030762
Sun Y, Tong C, He S, Wang K, Chen L. Identification of Nitrogen, Phosphorus, and Potassium Deficiencies Based on Temporal Dynamics of Leaf Morphology and Color. Sustainability. 2018; 10(3):762. https://doi.org/10.3390/su10030762
Chicago/Turabian StyleSun, Yuanyuan, Cheng Tong, Shan He, Ke Wang, and Lisu Chen. 2018. "Identification of Nitrogen, Phosphorus, and Potassium Deficiencies Based on Temporal Dynamics of Leaf Morphology and Color" Sustainability 10, no. 3: 762. https://doi.org/10.3390/su10030762
APA StyleSun, Y., Tong, C., He, S., Wang, K., & Chen, L. (2018). Identification of Nitrogen, Phosphorus, and Potassium Deficiencies Based on Temporal Dynamics of Leaf Morphology and Color. Sustainability, 10(3), 762. https://doi.org/10.3390/su10030762





