Introduction
Aquatic invertebrates and algae are largely used in ecotoxicology studies. Laboratories spend large amounts of resources to maintain, grow and breed organisms. One of the most time-consuming tasks involves quantifying cultures and their feed (
e.g. algae and plankton) densities. New and emerging approaches for estimating plankton concentrations have been adopted to overcome the limitations of manual counting, such as coulter counter, optical particle counter, and digital zooplankton image analysis. However, automated systems have some limitations (
e.g. volume analysed) and can show higher variability than manual counts [
1]. In some situations estimating the number of organisms in a population still requires manually counting specimens. It has been demonstrated during the last decades, that the accuracy of the estimation density of algae and invertebrates is a function of the sample size, the volume of the concentrated sample, the fraction of the concentrated sample that is counted; it could also be related to samples counted by different people [
2,
3,
4,
5,
6]. Counting large volumes (0.5-1 L) should be ideal to get a good estimation of the plankton density but could require a team of analysts and becomes time-consuming and very expensive. Based on these issues, the field of ecotoxicology could benefit from a mechanized device that quickly and accurately estimates large volume samples. One such instrument is the XperCount (XPC), an optics-based device developed by XpertSea Solutions Inc. in collaboration with leaders in the aquaculture industry and
Aquarium du Québec. The XPC can count thousands to millions of small aquatic organisms within a few seconds and can analyze volumes up to 10 L in a single measurement. The goal of this paper is to present the potential of the XPC for reducing variability in population size estimation of live aquatic cultures.
Materials and Methods
The XPC technology consists of a sealed optoelectronic unit, installed on the lid of a plastic container, which is adapted and resistant to the marine environment. To perform the count, the lid is removed and a sample of the aquatic culture between 0.5 and 10 L is poured into the container. The lid is secured back into place and, upon activation an emitter positioned under the lid then sends an electromagnetic (EM) signal towards the bottom of the container. A receptor, located next to the emitter, measures the intensity of the signal that is returned by the organisms. The XPC uses preprogrammed algorithms to correlate the value recorded by the receptor to a concentration or total number of organisms in the container. The final output can be visualized on a simple LED screen and three buttons user interface located on top of the container lid.
Every time measurements are recorded with the XPC, they are stored into a dynamic data bank managed by custom software. Through this software, various statistical analyses and algorithm developments take place, yielding counting applications specific to each type of aquatic organism. This proprietary data bank is one of the key elements of the XPC technology and as it grows, so does the accuracy and overall performance of the system. To facilitate new development integration into the XPC, a data transfer interface serves as a vehicle between the software and hardware components. This interface generates electronic files that can be downloaded on an SD card and inserted into the XPC. This interface allows easy reconfiguration of the platform from a distance, allowing more interaction between the users and XpertSea.
To demonstrate how the XPC can reduce the variability in population size estimation of live aquatic cultures, the small crustacean Artemia salina nauplii (400 μm) and with Litopenaeus vannamei shrimps larvae (7 mm) serve as examples.
Using a micro mesh nylon net (100 m porosity), Artemia nauplii were harvested and concentrated into a 1 L sample, which was then poured into the container of the XPC. After uniformly mixing the organisms, three measurements were performed with the device; then three 10 mL sub-samples were taken with a LTS 20 mL micropipette (model L0935053A). Each sub-sample was identified and set aside for manual counting under a binocular using a Ward cell. This operation was repeated 20 times using different dilutions of the Artemia nauplii sample in the XPC container. For each of the 20 dilutions, a volume of Artemia mixture was removed from the container and replaced with clear seawater to keep 1 L volume constant. Again, three measurements were taken with the XPC and three 10 mL sub-samples were extracted with the micropipette. Statistical differences between distributions were realized with a Wilcoxon signed ranks test using for non-parametric and paired data. With the shrimp specimens, the impact of sample size on the estimation of a total population was investigated. The larvae were removed from a production tank and concentrated into a 100 liter holding tank. After uniform mixing of the animals, six 25 mL samples were taken with a spring-operated aliquot. Each 25 mL sample was manually counted and used to estimate the population in the 100 L holding tank. The XperCount was then used to count 5, 10 and 15 L samples from the holding tank. For each volume, six measurements involving new samples were taken (after each measurement the sample was poured back into the holding tank).
Results and Discussion
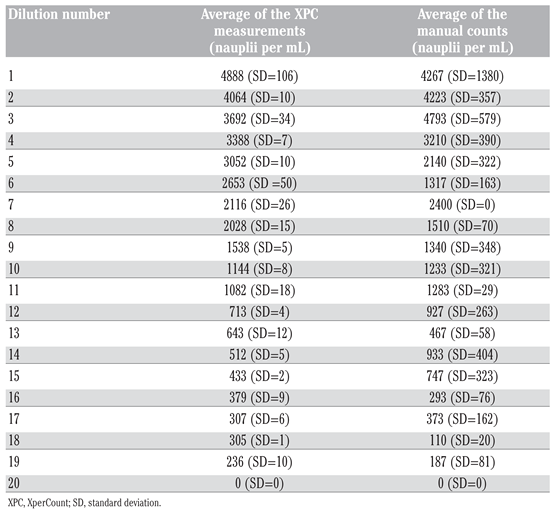
By physical principle, the number of
Artemia nauplii larvae per mL should gradually decrease with each dilution performed. This can be clearly observed with the XPC measurements but not with the manual counts (
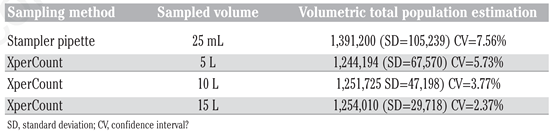
Table 1). At the highest densities, the manual counts did not properly reflect the dilutions of the nauplii samples. Although there is no significant difference between distributions (Wilcoxon signed ranks test, P=0.688), it can also be observed that the standard deviation is higher for the manual counts than for the XPC measurements at all dilution levels. The same trend can be observed with the shrimp larvae in
Table 2. The samples collected using a Stampler pipette yielded higher standard deviation and variation coefficient than the samples counted using the XPC method. These results are not surprising and have been clearly demonstrated by several studies; those indicated that smaller sub-sampled volumes analyzed lead to important errors in count, especially at high organism densities [
2,
3,
7].
Manual counts showed higher relative standard deviations at lower concentrations with smaller volumes of specimens (
Figure 1). For shrimp larvae, the concentration of specimens in the samples remained constant throughout the experiment but the sample sizes differed (5, 10 and 15 L;
Table 2). For
Artemia nauplii, the sampling volumes remained constant but the concentrations varied by successive dilutions. It is interesting to note that the performance of the XPC did not appear to be affected by the concentration of
Artemia specimens in a sample but appeared to be only impacted by the volume of the sample with shrimp larvae. In both cases, the relative standard deviations were higher for manual counts than for the XPC readings. This observation seems to suggest that an optimal ratio can be reached with the XPC based on the sample volume to the size and concentration of specimens. This outcome is especially important for researchers who need accurate results when working with large volumes of live organisms. To date, mathematical models are being used by researchers to determine appropriate rates of sampling for the populations of organisms they are working with. These models have shown benefits of limiting the time spent taking samples but can significantly compromise accuracy when used for larger populations [
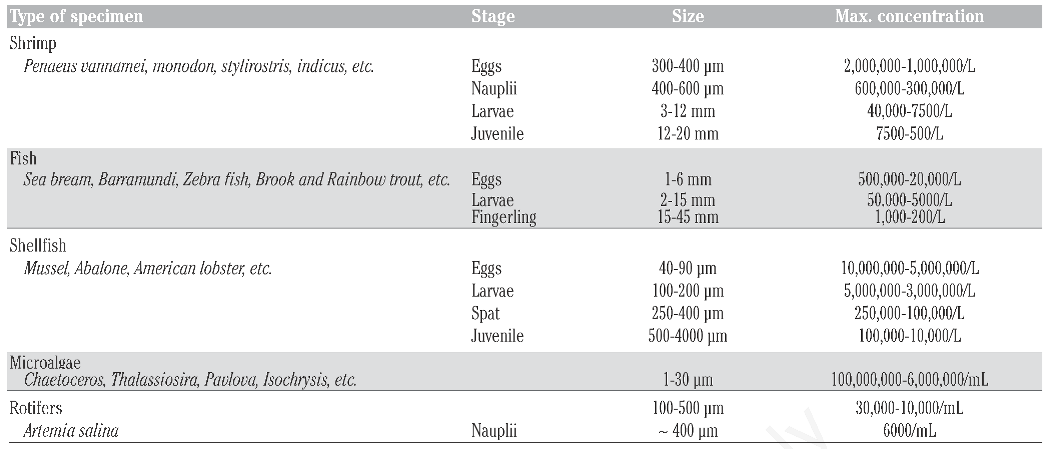
2]. The XPC alleviates this compromise between time and accuracy. Using the ability to count larger sample volumes with accuracy and speed, the XPC allows researchers to design and complete reliable experiments within a feasible time frame without sacrificing time or accuracy. The XPC saves time and money and gives users a high level of precision for several types of organisms. Counting applications for several different types of small aquatic organisms have already been developed for the XPC (
Table 3).
Conclusions
The coherence between the dilutions and XPC measurements, as well as the low coefficients of variation throughout the counts for both experiments, supports the hypothesis that performing counts on larger volumes can reduce the variability in population size estimation of live cultures, especially at lower concentrations. To confirm these results further, similar work is actually being conducted to validate the accuracy and the repeatability of the XPC system with Artemia and other organisms (e.g. algae, rotifers, mysis and fish larvae). Limitations of the XPC are also being investigated. Foreign particulates such as feed residuals, hydra and other contaminations and molts could potentially interfere with the EM signals emitted by the technology platform and have to be included into the product development strategy. Researchers can use the XPC to determine quickly and accurately the densities of algae, invertebrates and fish cultures at different growth stages. Also, the possibility of doing experiments directly in the container (such as toxicant additions) will enable unprecedented time series analysis and studies. So far in terms of better resource management, the XPC has demonstrated the potential to become an indispensable tool for aquatic ecotoxicology studies.