In Silico Analysis of Triamterene as a Potential Dual Inhibitor of VEGFR-2 and c-Met Receptors
Abstract
1. Introduction
2. Methodology
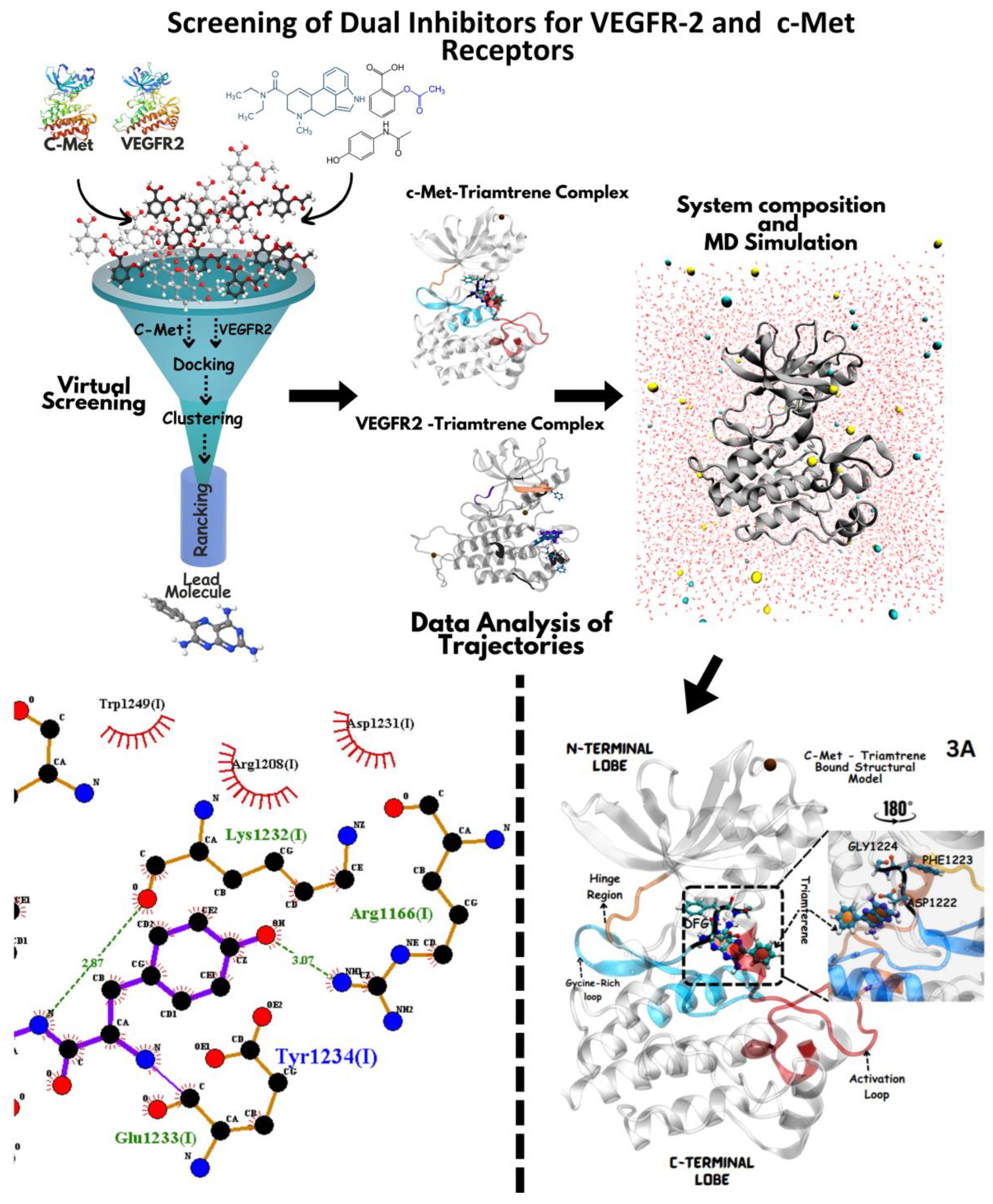
2.1. Software Tools
2.2. Drug Library Preparation for c-Met and VEGFR-2 Screening
2.3. VEGFR-2/c-Met Structural Refinement for Ligand Screening
2.4. Active Site Mapping via EPOS for Protein Inhibition
2.5. VEGFR-2/c-Met Docking with AutoDock
2.6. Protein/Ligand Preparation for AutoDock Docking
2.7. Grid Parameter Optimization for Precise Ligand Docking
2.8. AutoGrid/AutoDock execution and Binding Energy Evaluation
2.9. Ranking Docked Ligand Poses for VEGFR-2 and c-Met via Virtual Screening
2.10. LOD Scoring for Pose Ranking and Identification of Potential Ligands
2.11. MD Simulation of (VEGFR-2/c-Met) Ligand Complexes: Setup and Analysis
2.12. Visualization and Structural Analysis of Docking/MD Results
3. Results
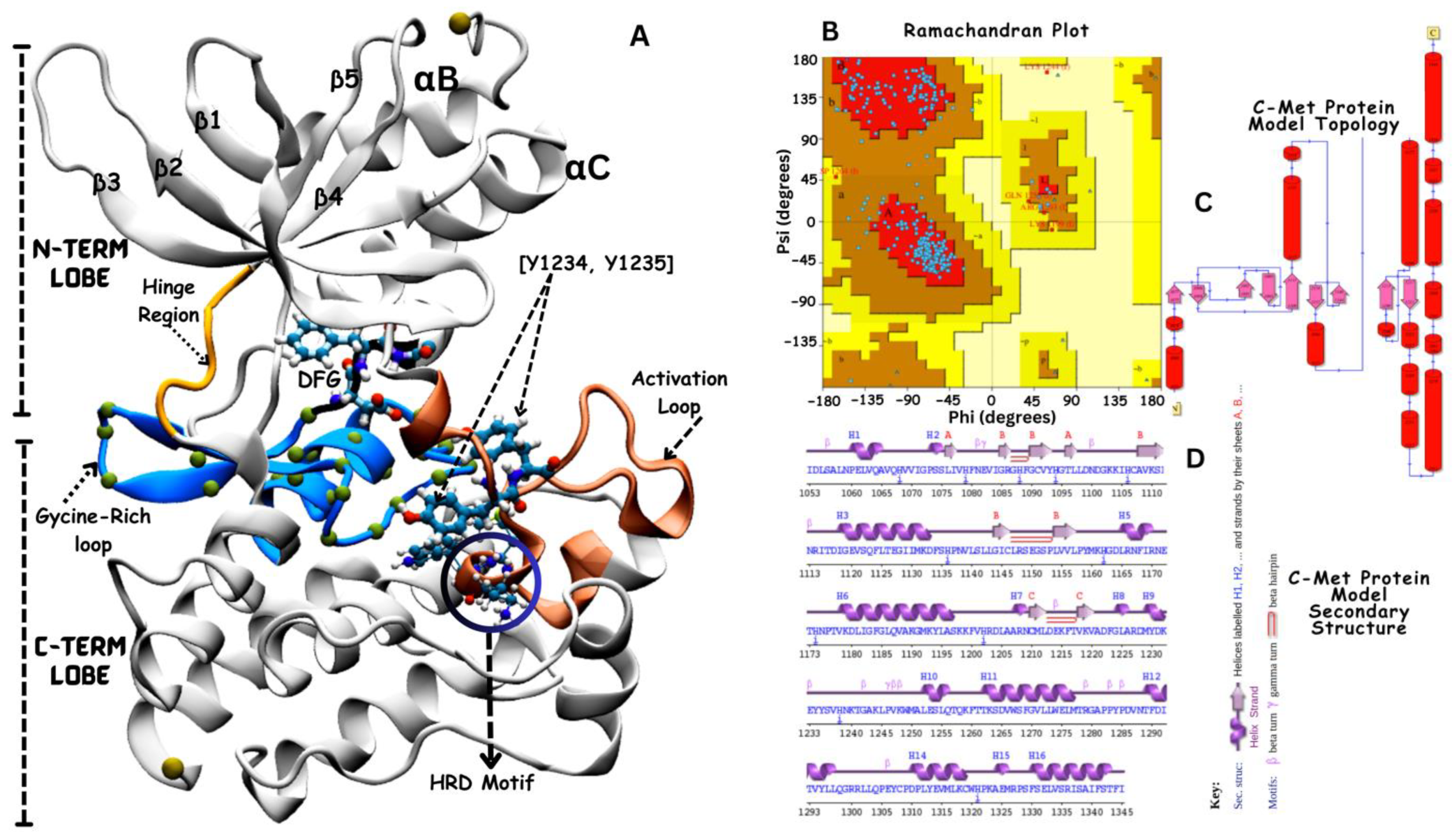
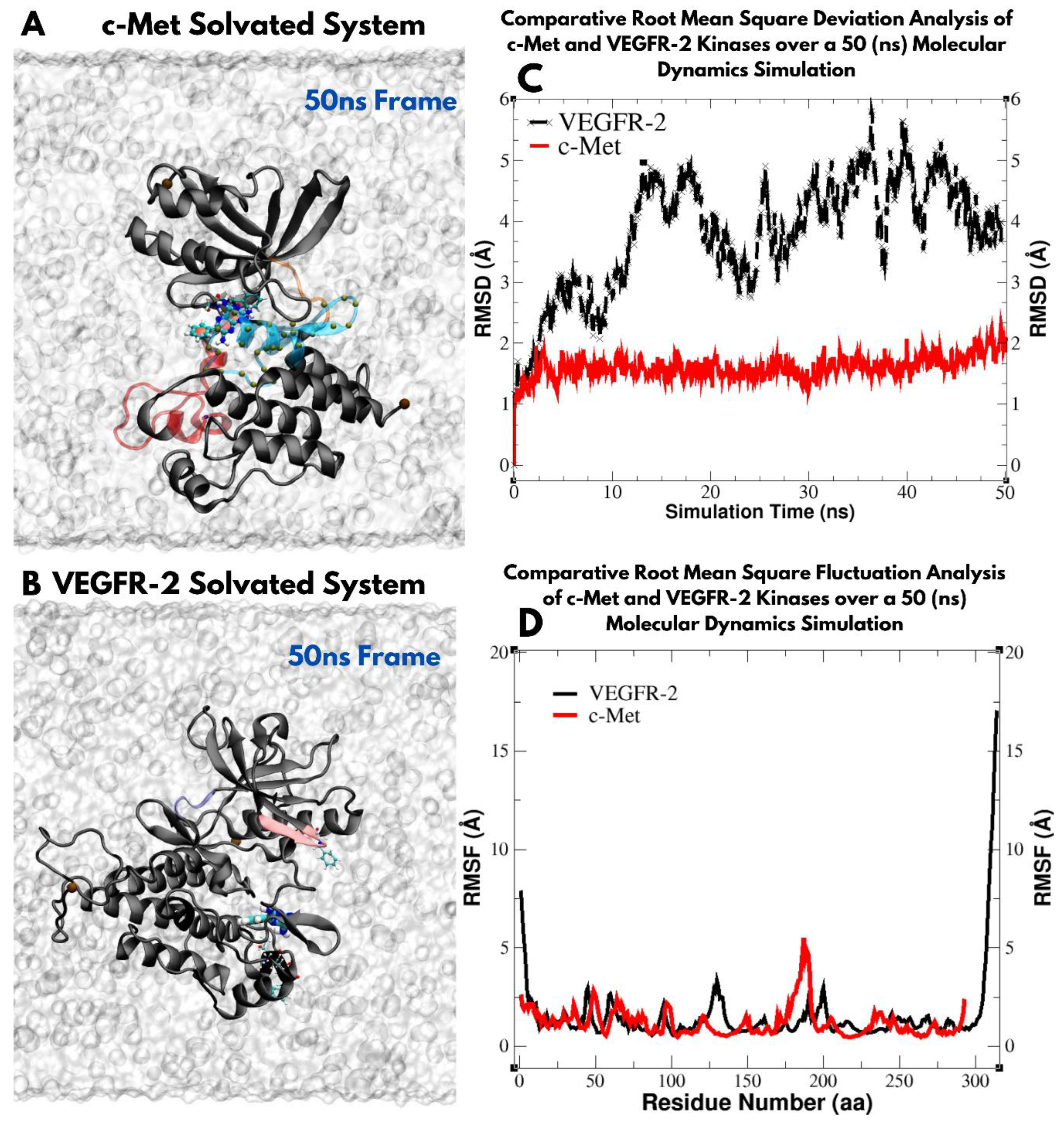
3.1. Structural Modeling of VEGFR-2 Kinase Domain
3.2. Structural Modeling of c-Met Kinase Domain
3.3. Hierarchical Ranking of Potential VEGFR-2 and c-Met Kinase Inhibitors
3.4. Virtual Screening: Top 11 Selected Ligands for c-Met and VEGFR-2
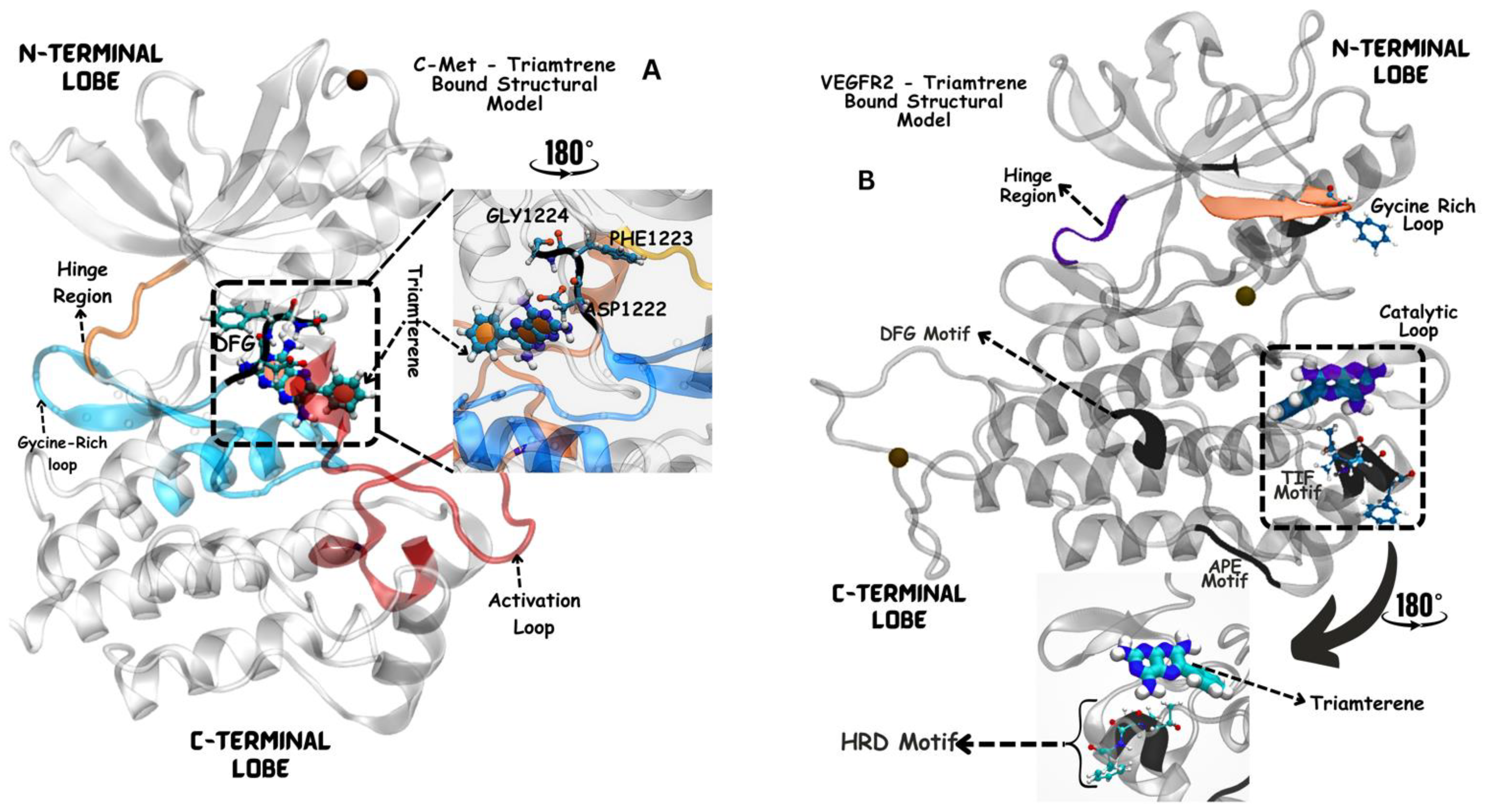
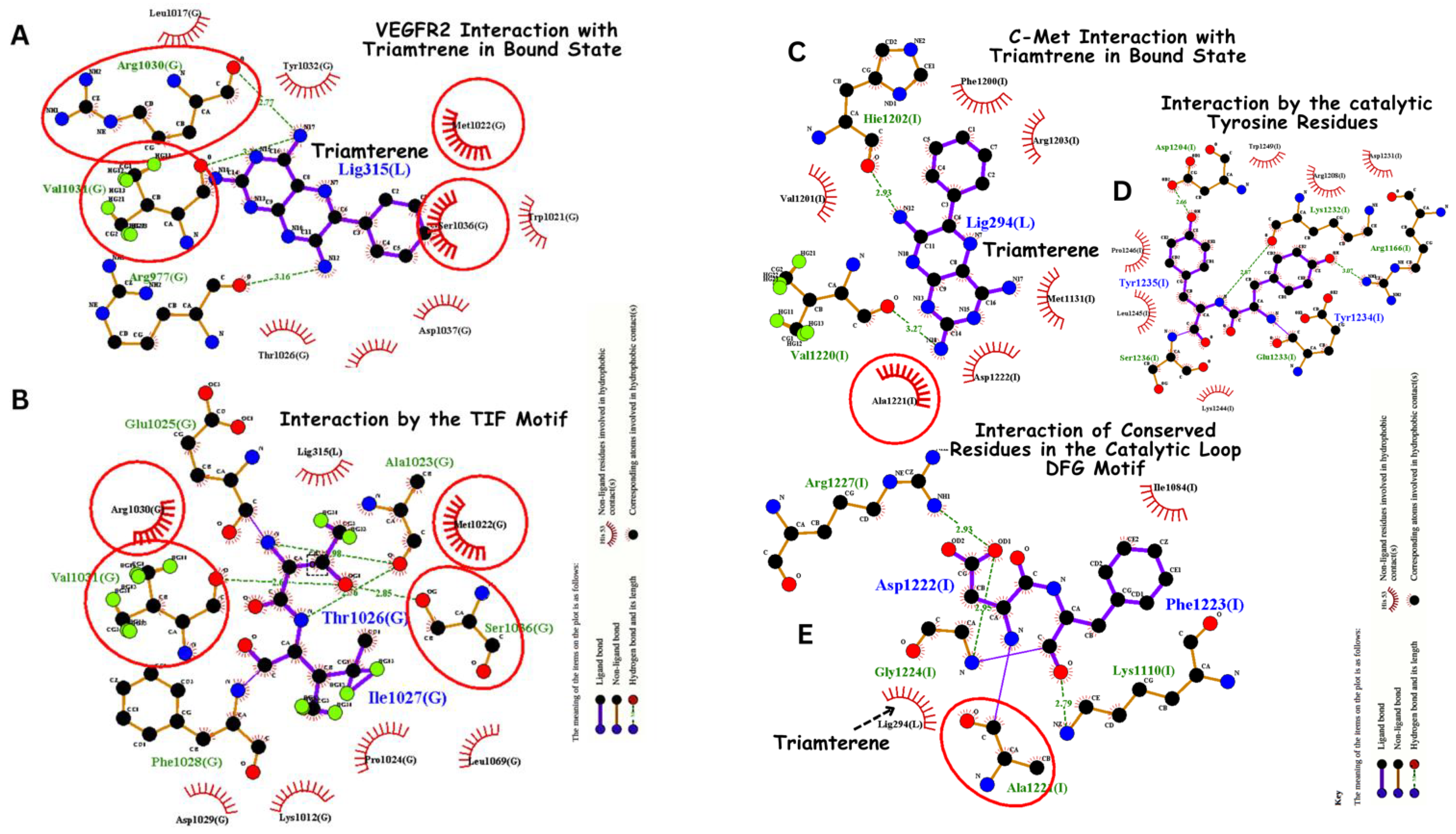
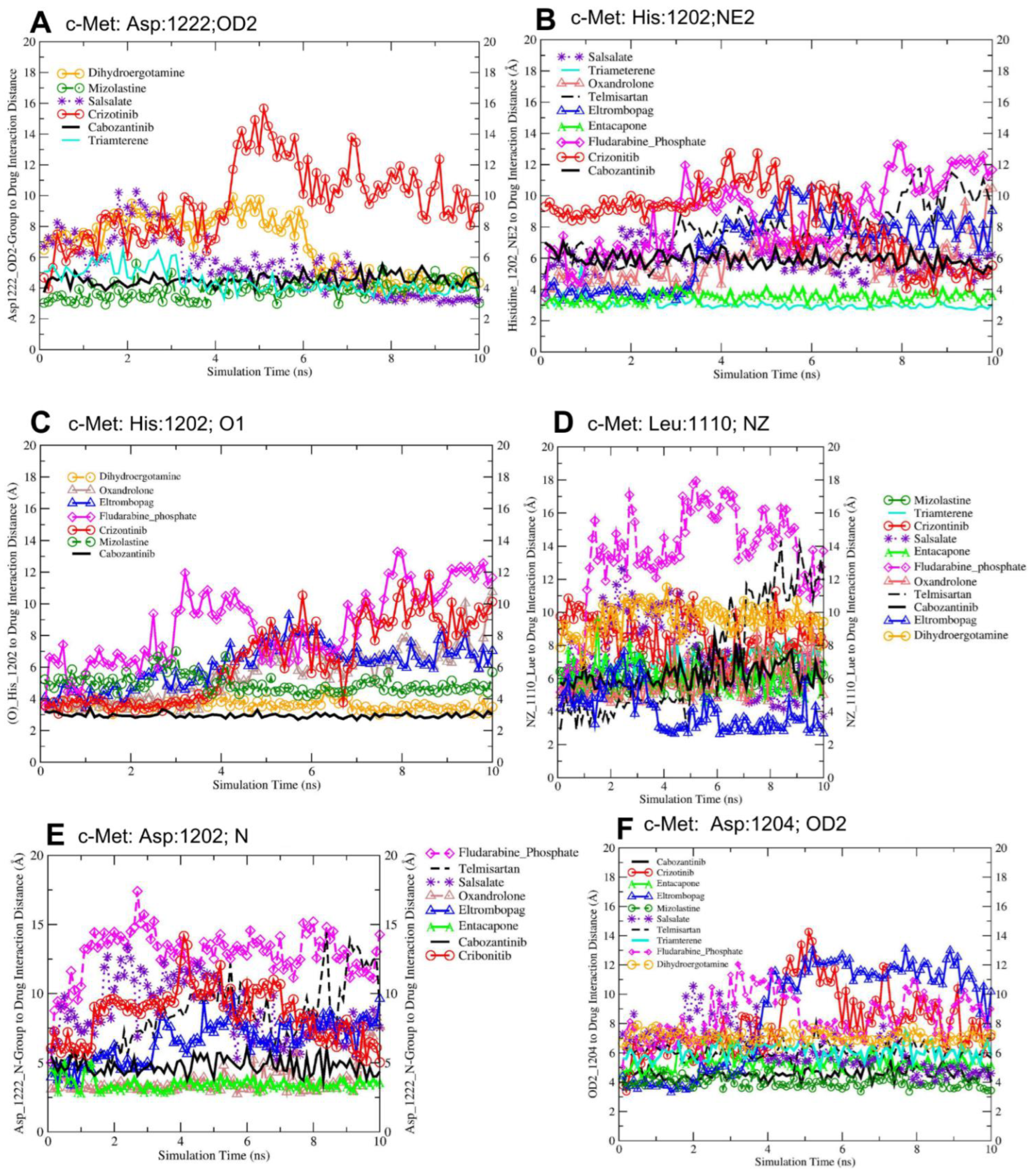
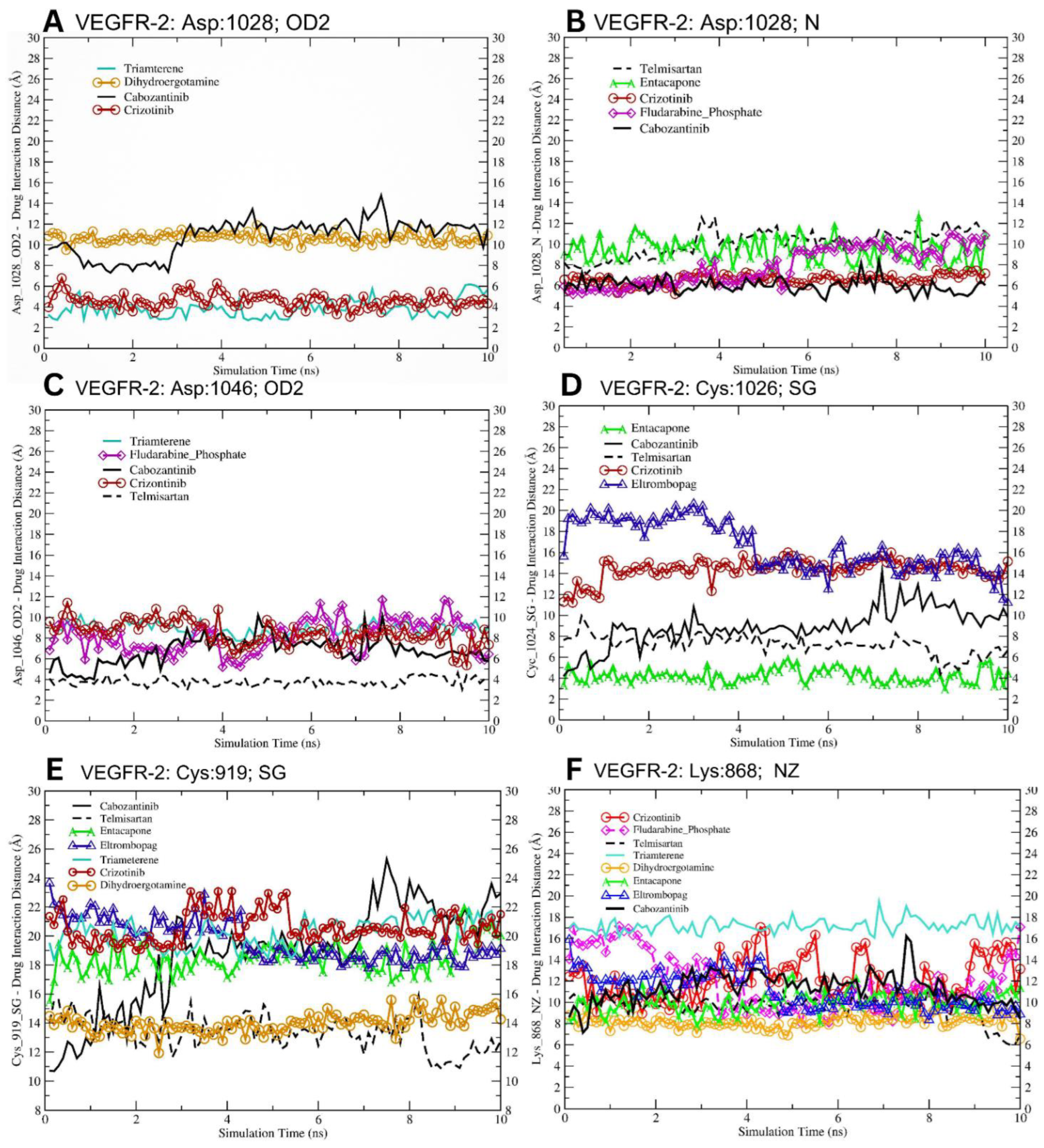
3.5. Molecular Dynamics-Driven Identification of lead Drug Interactions with c-Met and VEGFR-2 Catalytic Domains
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Oncogenic Signaling of Growth Factor Receptors in Cancer: Mechanisms and Therapeutic Opportunities; MDPI: Basel, Switzerland, 2023.
- Patel, S.A.; Nilsson, M.B.; Le, X.; Cascone, T.; Jain, R.K.; Heymach, J.V. Molecular Mechanisms and Future Implications of VEGF/VEGFR in Cancer Therapy. Clin. Cancer Res. 2023, 29, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, X.; Li, G.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.; Lopes, J.M.; Silva, R.; Peixoto, J.; Leitão, D.; Soares, P.; Fernandes, A.C.; Linhares, P.; Vaz, R.; Lima, J. The role of c-Met and VEGFR2 in glioblastoma resistance to bevacizumab. Sci. Rep. 2021, 11, 6067. [Google Scholar] [CrossRef]
- Sierra, J.R.; Tsao, M.-S. c-MET as a potential therapeutic target and biomarker in cancer. Ther. Adv. Med Oncol. 2011, 3, S21–S35. [Google Scholar] [CrossRef]
- Rizwani, W.; Allen, A.E.; Trevino, J.G. Hepatocyte growth factor from a clinical perspective: A pancreatic cancer challenge. Cancers 2015, 7, 1785–1805. [Google Scholar] [CrossRef]
- Mohamady, S.; Galal, M.; Eldehna, W.M.; Gutierrez, D.C.; Ibrahim, H.S.; Elmazar, M.M.; Ali, H.I. Dual Targeting of VEGFR2 and C-Met Kinases via the Design and Synthesis of Substituted 3-(Triazolo-thiadiazin-3-yl)indolin-2-one Derivatives as Angiogenesis Inhibitors. ACS Omega 2020, 5, 18872–18886. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, W.-Q.; Broussy, S.; Han, B.; Fang, H. Recent advances of anti-angiogenic inhibitors targeting VEGF/VEGFR axis. Front. Pharmacol. 2023, 14, 1307860. [Google Scholar] [CrossRef]
- Grojean, M.; Schwarz, M.A.; Schwarz, J.R.; Hassan, S.; von Holzen, U.; Zhang, C.; Schwarz, R.E.; Awasthi, N. Targeted dual inhibition of c-Met/VEGFR2 signalling by foretinib improves antitumour effects of nanoparticle paclitaxel in gastric cancer models. J. Cell. Mol. Med. 2021, 25, 4950–4961. [Google Scholar] [CrossRef]
- Cybele, C.L.; Razonable, R.R. Tyrosine-Kinase Inhibitors. In Infectious Complications in Biologic and Targeted Therapies; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Wang, B.; Wu, H.; Hu, C.; Wang, H.; Liu, J.; Wang, W.; Liu, Q. An overview of kinase downregulators and recent advances in discovery approaches. Signal Transduct. Target. Ther. 2021, 6, 423. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Yakes, F.M.; Chen, J.; Tan, J.; Yamaguchi, K.; Shi, Y.; Yu, P.; Qian, F.; Chu, F.; Bentzien, F.; Cancilla, B.; et al. Cabozantinib (XL184), a novel met and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 2011, 10, 2298–2308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Peng, C.; Wang, G.; Xu, Z.; Luo, Y.; Wang, J.; Zhu, W. Exploring binding mechanisms of VEGFR2 with three drugs lenvatinib, sorafenib, and sunitinib by molecular dynamics simulation and free energy calculation. Chem. Biol. Drug Des. 2019, 93, 934–948. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, S.; Wang, K.; Sun, S.-Y. MET inhibitors for targeted therapy of EGFR TKI-resistant lung cancer. J. Hematol. Oncol. 2019, 12, 63. [Google Scholar] [CrossRef]
- Kaboli, P.J.; Chen, H.-F.; Babaeizad, A.; Geraylow, K.R.; Yamaguchi, H.; Hung, M.-C. Unlocking c-MET: A comprehensive journey into targeted therapies for breast cancer. Cancer Lett. 2024, 588, 216780. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Park, S.A.; Jeong, M.S.; Ha, K.-T.; Jang, S.B. Structure and function of vascular endothelial growth factor and its receptor system. BMB Rep. 2018, 51, 73–78. [Google Scholar] [CrossRef]
- Uchikawa, E.; Chen, Z.; Xiao, G.-Y.; Zhang, X.; Bai, X.-C. Structural basis of the activation of c-MET receptor. Nat. Commun. 2021, 12, 4074. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.C.; Maulik, G.; Christensen, J.; Salgia, R. c-Met: Structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003, 22, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Abdel-Halim, H.; Traini, D.; Hibbs, D.; Gaisford, S.; Young, P. Modelling of molecular phase transitions in pharmaceutical inhalation compounds: An in silico approach. Eur. J. Pharm. Biopharm. 2011, 78, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System, Version 2.3; Schrödinger LLC: New York, NY, USA, 2020. [Google Scholar]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand in-teractions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to su-percomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.-P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., III; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Pro-grams for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Eyrisch, S.; Helms, V. Transient Pockets on Protein Surfaces Involved in Protein−Protein Interaction. J. Med. Chem. 2007, 50, 3457–3464. [Google Scholar] [CrossRef] [PubMed]
- Cousins, K.R. Computer review of ChemDraw ultra 12.0. J. Am. Chem. Soc. 2011, 133, 8388. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef]
- Southan, C.; Stracz, A. Extracting and connecting chemical structures from text sources using chemicalize.org. J. Chem. 2013, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.H.; Liu, L.; Lee, M.; Xi, N.; Fellows, I.; D’angelo, N.D.; Dominguez, C.; Rex, K.; Bellon, S.F.; Kim, T.-S.; et al. Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives. J. Med. Chem. 2012, 55, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Engst, S.; Yamaguchi, K.; Yu, P.; Won, K.-A.; Mock, L.; Lou, T.; Tan, J.; Li, C.; Tam, D.; et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009, 69, 8009–8016. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular Dynamics Simulations and Drug Discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Mehare, H.; Bin Anilkumar, J.P.; Usmani, N.A. The Python Programming Language. In A Guide to Applied Machine Learning for Biologists; Springer Nature: Berlin, Germany, 2023. [Google Scholar] [CrossRef]
- Sanner, F.M. Python: A programming Language for software integration and development. J. Mol. Graph. Model 1999, 17, 57–61. [Google Scholar] [PubMed]
- Alamri, A.S.; Alhomrani, M.; Alsanie, W.F.; Alyami, H.; Shakya, S.; Habeeballah, H.; Abdulaziz, O.; Alamri, A.; Alkhatabi, H.A.; Felimban, R.I.; et al. Spectroscopic and Molecular Docking Analysis of π-Acceptor Complexes with the Drug Barbital. Appl. Sci. 2022, 12, 10130. [Google Scholar] [CrossRef]
- Tsai, K.-L.; Chang, S.-Y.; Yang, L.-W. DRDOCK: A Drug Repurposing Platform Integrating Automated Docking, Simulations, and a Log-Odds-Based Drug Ranking Scheme. bioRxiv 2021. [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Studio, D. Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 4.5; Accelrys Software Inc.: San Diego, CA, USA, 2015. [Google Scholar]
- Miyata, T. Discovery studio modeling environment. Ensemble 2015, 17, 98–104. [Google Scholar]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Welsh, M. VEGFA and tumour angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Goel, H.L.; Pursell, B.; Shultz, L.D.; Greiner, D.L.; Brekken, R.A.; Kooi, C.W.V.; Mercurio, A.M. P-Rex1 Promotes Resistance to VEGF/VEGFR-Targeted Therapy in Prostate Cancer. Cell Rep. 2016, 14, 2193–2208. [Google Scholar] [CrossRef]
- Ferreira, J.J.; Lees, A.J.; Poewe, W.; Rascol, O.; Rocha, J.-F.; Keller, B.; Soares-Da-Silva, P. Effectiveness of opicapone and switching from entacapone in fluctuating Parkinson disease. Neurology 2018, 90, e1849–e1857. [Google Scholar] [CrossRef] [PubMed]
- Bladt, F.; Faden, B.; Friese-Hamim, M.; Knuehl, C.; Wilm, C.; Fittschen, C.; Grädler, U.; Meyring, M.; Dorsch, D.; Jaehrling, F.; et al. EMD 1214063 and EMD 1204831 constitute a new class of potent and highly selective c-Met inhibitors. Clin. Cancer Res. 2013, 19, 2941–2951. [Google Scholar] [CrossRef] [PubMed]
- Itatani, Y.; Kawada, K.; Yamamoto, T.; Sakai, Y. Resistance to anti-angiogenic therapy in cancer—Alterations to anti-VEGF pathway. Int. J. Mol. Sci. 2018, 19, 1232. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.R.; Rath, P.; Oyinlade, O.; Lopez, H.; Mughal, S.; Xia, S.; Li, Y.; Kaur, H.; Zhou, X.; Ahmed, A.K.; et al. Crizotinib and erlotinib inhibits growth of c-Met+/EGFRvIII+ primary human glioblastoma xenografts. Clin. Neurol. Neurosurg. 2018, 171, 26–33. [Google Scholar] [CrossRef]
- De Mello, R.A.; Neves, N.M.; Amaral, G.A.; Lippo, E.G.; Castelo-Branco, P.; Pozza, D.H.; Tajima, C.C.; Antoniou, G. The role of met inhibitor therapies in the treatment of advanced non-small cell lung cancer. J. Clin. Med. 2020, 9, 1918. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Aronov, A.M.; Deininger, D.D.; Giroux, S.; Lauffer, D.J.; Li, P.; Liang, J.; McGinty, K.; Ronkin, S.; Swett, R.; et al. Discovery of Potent, Selective Triazolothiadiazole-Containing c-Met Inhibitors. ACS Med. Chem. Lett. 2021, 12, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-X.; Liu, F.-R.; Zhang, Y.; Chen, Q.; Chen, Z.-Q.; Liu, Q.-W.; Huang, Y.; Yang, Y.-P.; Fang, W.-F.; Xi, N.; et al. Preclinical characterization and phase I clinical trial of CT053PTSA targets MET, AXL, and VEGFR2 in patients with advanced solid tumors. Front. Immunol. 2022, 13, 1024755. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Whang, Y.M.; Campbell, P.; Mulcrone, P.L.; Elefteriou, F.; Cho, S.W.; Park, S.I. Dual targeting c-met and VEGFR2 in osteoblasts suppresses growth and osteolysis of prostate cancer bone me-tastasis. Cancer Lett. 2018, 414, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Mosallam, A.M.; Ibrahim, A.O.A.; Badr, M.; Abdelmonsef, A.H. Novel 3-phenylquinazolin-2,4(1H,3H)-diones as dual VEGFR-2/c-Met-TK inhibitors: Design, synthesis, and biological evaluation. Sci. Rep. 2023, 13, 18567. [Google Scholar] [CrossRef]
- Navis, A.C.; Bourgonje, A.; Wesseling, P.; Wright, A.; Hendriks, W.; Verrijp, K.; van der Laak, J.A.W.M.; Heerschap, A.; Leenders, W.P.J. Effects of Dual Targeting of Tumor Cells and Stroma in Human Glioblastoma Xenografts with a Tyrosine Kinase Inhibitor against c-MET and VEGFR2. PLoS ONE 2013, 8, e58262. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Wang, Y.; Lin, C.; Zhang, D.; Chen, J.; Ouyang, L.; Wu, F.; Zhang, J.; Chen, L. Recent progress on vascular endothelial growth factor receptor inhibitors with dual targeting capabilities for tumor therapy. J. Hematol. Oncol. 2022, 15, 89. [Google Scholar] [CrossRef] [PubMed]

| Name and PubChem CID | Molecular Formular | Molecular Weight {g/mol} | Canonical SMILES | 2D Structure |
|---|---|---|---|---|
| Cabozantinib | C28H24FN3O5 | 501.5 | COC1=CC2=C(C=CN=C2C=C1OC)OC3=CC=C(C=C3)NC(=O)C4(CC4)C(=O)NC5=CC=C(C=C5)F | 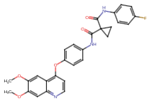 |
| Crizotinib | C21H22Cl2FN5O | 450.3 | CC(C1=C(C=CC(=C1Cl)F)Cl)OC2=C(N=CC(=C2)C3=CN(N=C3)C4CCNCC4)N |  |
| Entacapone | C14H15N3O5 | 305.29 | CCN(CC)C(=O)C(=CC1=CC(=C(C(=C1)O)O)[N+](=O)[O-])C#N | 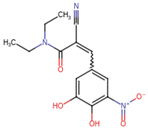 |
| Eltrombopag | C25H22N4O4 | 442.5 | CC1=NN(c2ccc(C)c(C)c2)C(=O)/C1=N\Nc1cccc(c2cccc(C(=O)O)c2)c1O |  |
| Telmisartan | C33H30N4O2 | 514.6 | CCCC1=NC2=C(N1CC3=CC=C(C=C3)C4=CC=CC=C4C(=O)O)C=C(C=C2C)C5=NC6=CC=CC=C6N5C |  |
| Triamterene | C12H11N7 | 253.26 | C1=CC=C(C=C1)C2=NC3=C(N=C(N=C3N=C2N)N)N | 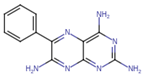 |
| Dihydroergotamine | C33H37N5O5 | 583.7 | CC1(C(=O)N2C(C(=O)N3CCCC3C2(O1)O)CC4=CC=CC=C4)NC(=O)C5CC6C(CC7=CNC8=CC=CC6=C78)N(C5)C | 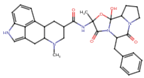 |
| Fludarabine Phosphate | C10H13FN5O7P | 365.21 | C1=NC2=C(N=C(N=C2N1C3C(C(C(O3)COP(=O(O)O)O)O)F)N | 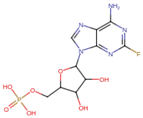 |
| Mizolastine | C24H25FN6O | 432.5 | CN(C1CCN(CC1)C2=NC3=CC=CC=C3N2CC4=CC=C(C=C4)F)C5=NC=CC(=O)N5 |  |
| Oxandrolone | C19H30O3 | 306.4 | CC12CCC3C(C1CCC2(C)O)CCC4C3(COC(=O)C4)C |  |
| Salsalate | C14H10O5 | 258.2 | C1=CC=C(C(=C1)C(=O)OC2=CC=CC=C2C(=O)O)O | 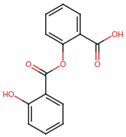 |
| No: | Candidate Drugs | Binding Affinity in c-Met Target (kcal/mol) | Binding Affinity in VEGFR-2 Target (kcal/mol) | Proximity to the Catalytic Site in c-Met (Å) | Proximity to the Catalytic Site in VEGFR-2 (Å) | Log Odds (LOD) Score in c-Met | Log Odds (LOD) Score in VEGFR-2 |
|---|---|---|---|---|---|---|---|
| 1. | Cabozantinib | −7.7 | −8.2 | 3.1 | 2.9 | −0.55 | −0.52 |
| 2. | Crizotinib | −7.9 | −7.5 | 2.4 | 3.1 | −0.64 | −0.60 |
| 3. | Telmisartan | −7.3 | −7.8 | 3.2 | 2.8 | −15.3 | −18.7 |
| 4. | Triamterene | −8.0 | −8.3 | 2.5 | 2.7 | −0.50 | −0.6 |
| 5. | Dihydroergotamine | −7.6 | −7.4 | 2.9 | 3.0 | −12.5 | −11.8 |
| 6. | Oxandrolone | −7.7 | −7.1 | 3.3 | 3.5 | −5.7 | −8.2 |
| 7. | Mizolastine | −7.0 | −7.3 | 3.1 | 2.7 | −8.2 | −10.4 |
| 8. | Fludarabine Phosphate | −7.4 | −7.0 | 2.3 | 2.6 | −6.64 | −9.62 |
| 9. | Eltrombopag | −7.8 | −8.0 | 2.7 | 2.5 | −22.3 | −25.1 |
| 10. | Entacapone | −7.5 | −7.2 | 2.2 | 2.4 | −19.0 | −17.5 |
| 11. | Salsalate | −6.8 | −6.6 | 3.4 | 3.6 | −37.18 | −40.5 |
| Lead Drug | Asp1222 (OD2) Å | His1202 (NE2) Å | His1202 (O1) Å | Leu1110(NZ) Å | Asp1202(N) Å | Asp1204 (OD2) Å |
|---|---|---|---|---|---|---|
| Cabozantinib | 4.54 ± 0.04 *** (−3.55) | 4.54 ± 0.05 *** (−5.15) | 5.95 ± 0.01 *** (−2.48) | 6.19 ± 0.07 *** (−2.41) | 4.77 ± 0.06 *** (−3.80) | 4.54 ± 0.14 *** (−5.15) |
| Crizotinib | 8.09 ± 0.19 | 9.69 ± 0.22 | 8.43 ± 0.25 | 8.60 ± 0.13 | 8.57 ± 0.19 | 9.69 ± 0.31 |
| Dihydroergotamine | 6.75 ± 0.21 *** (−1.34) | 6.75 ± 0.04 *** (−2.94) | - | - | - | 6.75 ± 0.31 *** (−2.94) |
| Eltrombopag | - | 8.75 ± 0.22 *** (−0.94) | 6.65 ± 0.13 *** (−1.78) | 3.86 ± 0.12 *** (−4.74) | 6.58 ± 0.15 *** (−1.99) | 8.75 ± 0.05 *** (−0.94) |
| Entacapone | - | 5.30 ± 0.03 *** (−4.39) | 3.51 ± 0.22 *** (−4.92) | 6.71 ± 0.08 *** (−1.89) | 3.47 ± 0.04 *** (−5.10) | 5.30 ± 0.62 *** (−4.39) |
| Fludarabine Phosphate | - | 8.49 ± 0.14 *** (−1.20) | 13.18 ± 0.23 *** (4.75) | 13.59 ± 0.30 *** (4.99) | 12.88 ± 0.17 *** (4.31) | 8.49 ± 0.29 *** (−1.20) |
| Mizolastine | 3.89 ± 0.06 *** (−4.20) | 4.02 ± 0.06 *** (−5.67) | - | 6.32 ± 0.08 *** (−2.28) | - | 4.02 ± 0.41 *** (−5.67) |
| Oxandrolone | - | - | 5.84 ± 0.16 *** (−2.59) | 5.83 ± 0.10 *** (−2.77) | 3.67 ± 0.11 *** (−4.90) | - |
| Salsalate | 6.32 ± 0.32 *** (−1.77) | 6.34 ± 0.21 *** (−3.35) | - | 7.33 ± 0.24 *** (−1.27) | 8.72 ± 0.18 (0.15) NS | 6.34 ± 0.43 *** (−3.35) |
| Telmisartan | - | 6.11 ± 0.23 *** (−3.58) | 7.85 ± 0.31 *** (−0.58) | 7.21 ± 0.22 *** (−1.39) | 8.39 ± 0.51 (−0.18) NS | 6.11 ± 0.33 *** (−3.58) |
| Triamterene | 4.54 ± 0.24 *** (−3.55) | 6.07 ± 0.17 *** (−3.62) | 3.05 ± 0.34 *** (−5.38) | 6.99 ± 0.12 *** (−1.61) | - | 6.07 ± 0.23 *** (−3.62) |
| Lead Dugs | Cys919 (SG) Å | Cys1024(SG) Å | Lys868 (NZ) Å | Asp1028 (OD2) Å | Asp1046 (OD2) Å |
|---|---|---|---|---|---|
| Cabozantinib | 18.76 ± 0.34 | 8.94 ± 0.18 | 11.32 ± 0.06 | 10.78 ± 0.14 | 6.81 ± 0.17 |
| Telmisartan | 13.35 ± 0.12 *** (−5.41) | 7.22 ± 0.10 *** (−1.72) | 9.64 ± 0.11 *** (−1.68) | - | 3.78 ± 0.04 *** (−3.03) |
| Entacapone | 18.50 ± 0.07 (−0.26) NS | 4.22 ± 0.08 *** (−4.72) | 10.02 ± 0.10 *** (−1.30) | - | - |
| Eltrombopag | 19.68 ± 0.137 *** (+0.92) | 16.54 ± 0.24 *** (+7.60) | 11.02 ± 0.16 *** (−0.30) | - | - |
| Triamterene | 20.27 ± 0.09 *** (+1.51) | - | 17.20 ± 0.06 *** (+5.88) | 3.84 ± 0.07 *** (−6.94) | 8.89 ± 0.06 *** (+2.08) |
| Crizotinib | 20.59 ± 0.11 *** (+1.83) | 14.39 ± 0.10 *** (+5.45) | 12.14 ± 0.20 *** (+0.82) | 4.59 ± 0.07 *** (−6.19) | 8.47 ± 0.12 *** (+1.66) |
| Dihydroergotamine | 13.99 ± 0.05 *** (−4.77) | - | 18.17 ± 0.05 *** (+6.85) | 10.66 ± 0.04 (−0.12) | - |
| Fludarabine Phosphate | - | - | 11.81 ± 0.26 *** (+0.49) | - | 8.15 ± 0.16 *** (+1.34) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lutimba, S.; Saleem, B.; Aleem, E.; Mansour, M.A. In Silico Analysis of Triamterene as a Potential Dual Inhibitor of VEGFR-2 and c-Met Receptors. J. Xenobiot. 2024, 14, 1962-1987. https://doi.org/10.3390/jox14040105
Lutimba S, Saleem B, Aleem E, Mansour MA. In Silico Analysis of Triamterene as a Potential Dual Inhibitor of VEGFR-2 and c-Met Receptors. Journal of Xenobiotics. 2024; 14(4):1962-1987. https://doi.org/10.3390/jox14040105
Chicago/Turabian StyleLutimba, Stuart, Baraya Saleem, Eiman Aleem, and Mohammed A. Mansour. 2024. "In Silico Analysis of Triamterene as a Potential Dual Inhibitor of VEGFR-2 and c-Met Receptors" Journal of Xenobiotics 14, no. 4: 1962-1987. https://doi.org/10.3390/jox14040105
APA StyleLutimba, S., Saleem, B., Aleem, E., & Mansour, M. A. (2024). In Silico Analysis of Triamterene as a Potential Dual Inhibitor of VEGFR-2 and c-Met Receptors. Journal of Xenobiotics, 14(4), 1962-1987. https://doi.org/10.3390/jox14040105








