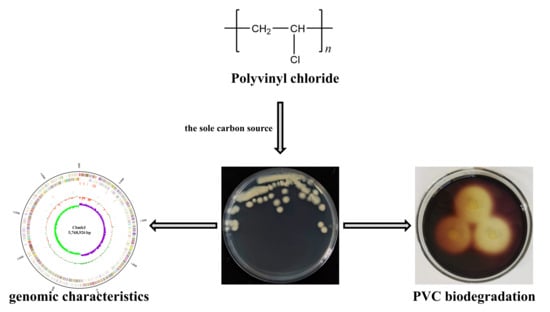
The Complete Genome Sequence of Bacillus toyonensis Cbmb3 with Polyvinyl Chloride-Degrading Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Medium
2.2. Genome Sequencing and Annotation
2.3. Phylogenetic Analysis
2.4. Genome Component and Gene Annotation
2.5. Biodegradation Characterization
2.6. Plant-Growth-Promoting Attributes
3. Results
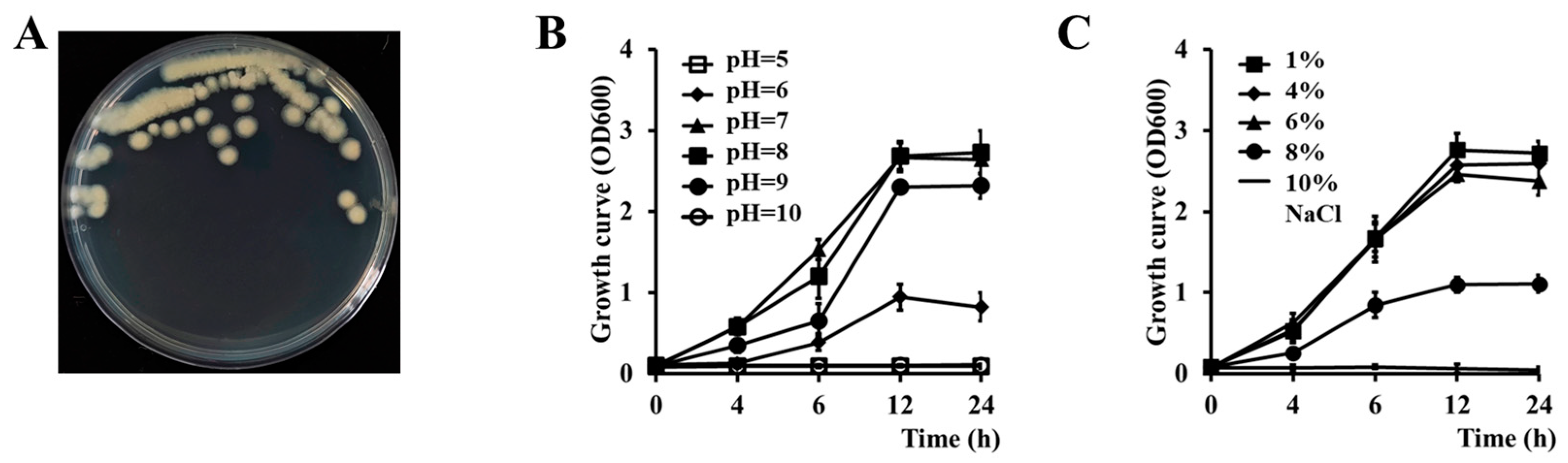
3.1. Isolation and Phenotypic Characteristics of Cbmb3
3.2. Genomic Characteristics of Cbmb3
3.3. Phylogenetic Analysis
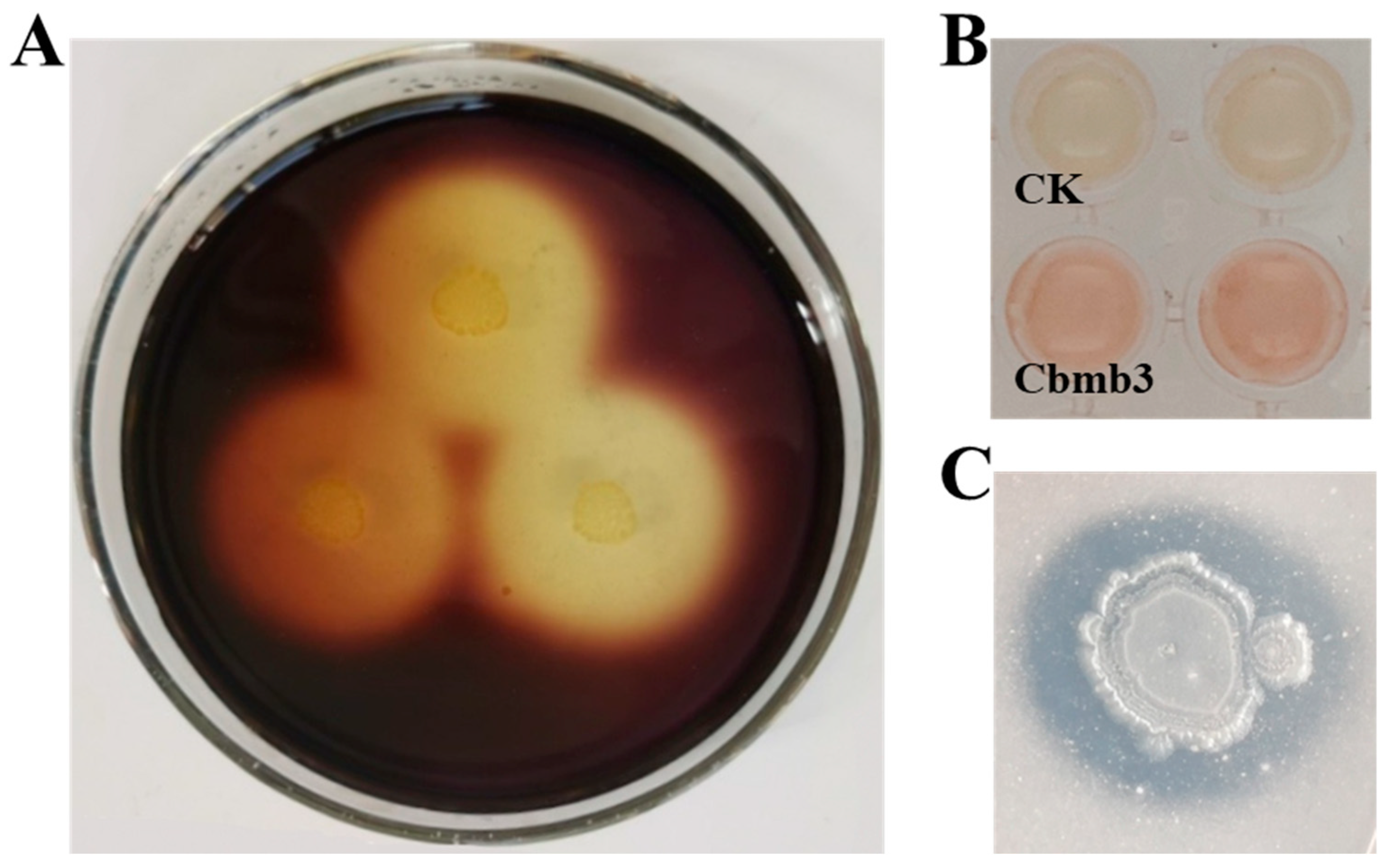
3.4. PVC-Degrading Properties of Cbmb3
3.5. Plant-Growth-Promoting Properties of Cbmb3
3.6. Antagonistic Activity of Cbmb3 against Plant Pathogens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Min, K.; Cuiffi, J.D.; Mathers, R.T. Ranking Environmental Degradation Trends of Plastic Marine Debris Based on Physical Properties and Molecular Structure. Nat. Commun. 2020, 11, 727. [Google Scholar] [CrossRef]
- Cai, L.; Zhao, X.; Liu, Z.; Han, J. The Abundance, Characteristics and Distribution of Microplastics (MPs) in Farmland Soil-Based on Research in China. Sci. Total Environ. 2023, 876, 162782. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; He, D.; Zhang, X.; Li, X.; Chen, Y.; Wei, G.; Zhang, Y.; Ok, Y.S.; Luo, Y. National-Scale Distribution of Micro(Meso)Plastics in Farmland Soils across China: Implications for Environmental Impacts. J. Hazard. Mater. 2022, 424, 127283. [Google Scholar] [CrossRef]
- Lu, L.; Li, W.; Cheng, Y.; Liu, M. Chemical Recycling Technologies for PVC Waste and PVC-Containing Plastic Waste: A Review. Waste Manag. 2023, 166, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Nizzetto, L.; Futter, M.; Langaas, S. Are Agricultural Soils Dumps for Microplastics of Urban Origin? Environ. Sci. Technol. 2016, 50, 10777–10779. [Google Scholar] [CrossRef]
- Tang, K.H.D. Microplastics in Agricultural Soils in China: Sources, Impacts and Solutions. Environ. Pollut. 2023, 322, 121235. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, Y.; Jiang, L.; Liu, C.; Zhu, H.; Qiu, D.; Wang, S. Quantification and Characterization of Microplastics in Farmland Soils of Jiangsu Province, East China. Environ. Sci. Pollut. Res. Int. 2023, 30, 120653–120662. [Google Scholar] [CrossRef] [PubMed]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic Biodegradation: Frontline Microbes and Their Enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Q.; Chen, Y.; Ren, X.-M.; Li, Y.-Y.; Zhang, Y.-J.; Zhang, H.; Han, H.; Chen, Z.-J. Plant Growth-Promoting Bacteria Modulate Gene Expression and Induce Antioxidant Tolerance to Alleviate Synergistic Toxicity from Combined Microplastic and Cd Pollution in Sorghum. Ecotoxicol. Environ. Saf. 2023, 264, 115439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl Chloride Degradation by a Bacterium Isolated from the Gut of Insect Larvae. Nat. Commun. 2022, 13, 5360. [Google Scholar] [CrossRef]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl Chloride Biodegradation by Pseudomonas Citronellolis and Bacillus Flexus. New Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef]
- El-Dash, H.A.; Yousef, N.E.; Aboelazm, A.A.; Awan, Z.A.; Yahya, G.; El-Ganiny, A.M. Optimizing Eco-Friendly Degradation of Polyvinyl Chloride (PVC) Plastic Using Environmental Strains of Malassezia Species and Aspergillus Fumigatus. Int. J. Mol. Sci. 2023, 24, 15452. [Google Scholar] [CrossRef]
- Rillig, M.C.; Kim, S.W.; Zhu, Y.-G. The Soil Plastisphere. Nat. Rev. Microbiol. 2024, 22, 64–74. [Google Scholar] [CrossRef]
- Olabemiwo, F.A.; Hagan, A.; Cham, M.; Cohan, F.M. Two Plant-Growth-Promoting Bacillus Species Can Utilize Nanoplastics. Sci. Total Environ. 2024, 907, 167972. [Google Scholar] [CrossRef]
- Liu, X.; Wang, D.; Yin, Z.; Sun, L.; Pang, S.; Liu, J.; Li, W.; Cui, S.; Huang, W.; Du, Y.; et al. Insights into Evolutionary, Genomic, and Biogeographic Characterizations of Chryseobacterium Nepalense Represented by a Polyvinyl Alcohol-Degrading Bacterium, AC3. Microbiol. Spectr. 2022, 10, e02179-22. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Stothard, P.; Wishart, D.S. Circular Genome Visualization and Exploration Using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded Coverage of Metagenomic, Viral and microRNA Families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene Ontology Consortium: Going Forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A Critical Examination of the Specificity of the Salkowski Reagent for Indolic Compounds Produced by Phytopathogenic Bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Rajawat, M.; Singh, S.; Tyagi, S.P.; Saxena, A.K. A Modified Plate Assay for Rapid Screening of Potassium-Solubilizing Bacteria. Pedosphere 2016, 26, 768–773. [Google Scholar] [CrossRef]
- Atashgahi, S.; Liebensteiner, M.G.; Janssen, D.B.; Smidt, H.; Stams, A.J.M.; Sipkema, D. Microbial Synthesis and Transformation of Inorganic and Organic Chlorine Compounds. Front. Microbiol. 2018, 9, 3079. [Google Scholar] [CrossRef]
- Novotný, Č.; Fojtík, J.; Mucha, M.; Malachová, K. Biodeterioration of Compost-Pretreated Polyvinyl Chloride Films by Microorganisms Isolated From Weathered Plastics. Front. Bioeng. Biotechnol. 2022, 10, 832413. [Google Scholar] [CrossRef]
- Shao, J.; Li, S.; Zhang, N.; Cui, X.; Zhou, X.; Zhang, G.; Shen, Q.; Zhang, R. Analysis and Cloning of the Synthetic Pathway of the Phytohormone Indole-3-Acetic Acid in the Plant-Beneficial Bacillus Amyloliquefaciens SQR9. Microb. Cell Fact. 2015, 14, 130. [Google Scholar] [CrossRef]
- Rasul, M.; Yasmin, S.; Suleman, M.; Zaheer, A.; Reitz, T.; Tarkka, M.T.; Islam, E.; Mirza, M.S. Glucose Dehydrogenase Gene Containing Phosphobacteria for Biofortification of Phosphorus with Growth Promotion of Rice. Microbiol. Res. 2019, 223–225, 1–12. [Google Scholar] [CrossRef]
- Stasi, R.; Neves, H.I.; Spira, B. Phosphate Uptake by the Phosphonate Transport System PhnCDE. BMC Microbiol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Furtwängler, K.; Tarasov, V.; Wende, A.; Schwarz, C.; Oesterhelt, D. Regulation of Phosphate Uptake via Pst Transporters in Halobacterium Salinarum R1. Mol. Microbiol. 2010, 76, 378–392. [Google Scholar] [CrossRef]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef]
- Mor, R.; Sivan, A. Biofilm Formation and Partial Biodegradation of Polystyrene by the Actinomycete Rhodococcus Ruber: Biodegradation of Polystyrene. Biodegradation 2008, 19, 851–858. [Google Scholar] [CrossRef]
- You, Y.; Ye, F.; Mao, W.; Yang, H.; Lai, J.; Deng, S. An Overview of the Structure and Function of the Flagellar Hook FlgE Protein. World J. Microbiol. Biotechnol. 2023, 39, 126. [Google Scholar] [CrossRef]
- Carpenter, P.B.; Zuberi, A.R.; Ordal, G.W. Bacillus Subtilis Flagellar Proteins FliP, FliQ, FliR and FlhB Are Related to Shigella Flexneri Virulence Factors. Gene 1993, 137, 243–245. [Google Scholar] [CrossRef]
- Inoue, Y.; Kinoshita, M.; Kida, M.; Takekawa, N.; Namba, K.; Imada, K.; Minamino, T. The FlhA Linker Mediates Flagellar Protein Export Switching during Flagellar Assembly. Commun. Biol. 2021, 4, 646. [Google Scholar] [CrossRef] [PubMed]
- Dergham, Y.; Sanchez-Vizuete, P.; Le Coq, D.; Deschamps, J.; Bridier, A.; Hamze, K.; Briandet, R. Comparison of the Genetic Features Involved in Bacillus Subtilis Biofilm Formation Using Multi-Culturing Approaches. Microorganisms 2021, 9, 633. [Google Scholar] [CrossRef]
- Terra, R.; Stanley-Wall, N.R.; Cao, G.; Lazazzera, B.A. Identification of Bacillus Subtilis SipW as a Bifunctional Signal Peptidase That Controls Surface-Adhered Biofilm Formation. J. Bacteriol. 2012, 194, 2781–2790. [Google Scholar] [CrossRef]
- Winkelman, J.T.; Bree, A.C.; Bate, A.R.; Eichenberger, P.; Gourse, R.L.; Kearns, D.B. RemA Is a DNA-Binding Protein That Activates Biofilm Matrix Gene Expression in Bacillus Subtilis. Mol. Microbiol. 2013, 88, 984–997. [Google Scholar] [CrossRef]
- Diehl, A.; Roske, Y.; Ball, L.; Chowdhury, A.; Hiller, M.; Molière, N.; Kramer, R.; Stöppler, D.; Worth, C.L.; Schlegel, B.; et al. Structural Changes of TasA in Biofilm Formation of Bacillus Subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, 3237–3242. [Google Scholar] [CrossRef]
- Roske, Y.; Lindemann, F.; Diehl, A.; Cremer, N.; Higman, V.A.; Schlegel, B.; Leidert, M.; Driller, K.; Turgay, K.; Schmieder, P.; et al. TapA Acts as Specific Chaperone in TasA Filament Formation by Strand Complementation. Proc. Natl. Acad. Sci. USA 2023, 120, e2217070120. [Google Scholar] [CrossRef]
- Alexander, R.P.; Lowenthal, A.C.; Harshey, R.M.; Ottemann, K.M. CheV: CheW-like Coupling Proteins at the Core of the Chemotaxis Signaling Network. Trends Microbiol. 2010, 18, 494–503. [Google Scholar] [CrossRef]
- Frutos-Grilo, E.; Marsal, M.; Irazoki, O.; Barbé, J.; Campoy, S. The Interaction of RecA with Both CheA and CheW Is Required for Chemotaxis. Front. Microbiol. 2020, 11, 583. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.-H. Bacillus Velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Chen, F.; Aqeel, M.; Khalid, N.; Nazir, A.; Irshad, M.K.; Akbar, M.U.; Alzuaibr, F.M.; Ma, J.; Noman, A. Interactive Effects of Polystyrene Microplastics and Pb on Growth and Phytochemicals in Mung Bean (Vigna radiata L.). J. Hazard. Mater. 2023, 449, 130966. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, M.; Shahbaz, M.; Hu, Z.; Zhu, Z.; Lu, S.; Yu, Y.; Yao, H.; Chen, J.; Ge, T. Microplastics in Soil Can Increase Nutrient Uptake by Wheat. J. Hazard. Mater. 2022, 438, 129547. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yang, X.; Riksen, M.; Xu, M.; Geissen, V. Response of Common Bean (Phaseolus vulgaris L.) Growth to Soil Contaminated with Microplastics. Sci. Total Environ. 2021, 755, 142516. [Google Scholar] [CrossRef]
- Song, R.; Sun, Y.; Li, X.; Ding, C.; Huang, Y.; Du, X.; Wang, J. Biodegradable Microplastics Induced the Dissemination of Antibiotic Resistance Genes and Virulence Factors in Soil: A Metagenomic Perspective. Sci. Total Environ. 2022, 828, 154596. [Google Scholar] [CrossRef]
- Lu, X.-M.; Chen, Y.-L. Varying Characteristics and Driving Mechanisms of Antibiotic Resistance Genes in Farmland Soil Amended with High-Density Polyethylene Microplastics. J. Hazard. Mater. 2022, 428, 128196. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The Role of the Copper-Binding Enzyme—Laccase—In the Biodegradation of Polyethylene by the Actinomycete Rhodococcus Ruber. Int. Biodeterior. Biodegradation 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Jeon, H.J.; Kim, M.N. Comparison of the Functional Characterization between Alkane Monooxygenases for Low-Molecular-Weight Polyethylene Biodegradation. Int. Biodeterior. Biodegrad. 2016, 114, 202–208. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Al-Harrasi, A. Plant Growth Promoting Bacteria as an Alternative Strategy for Salt Tolerance in Plants: A Review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Kulkova, I.; Dobrzyński, J.; Kowalczyk, P.; Bełżecki, G.; Kramkowski, K. Plant Growth Promotion Using Bacillus Cereus. Int. J. Mol. Sci. 2023, 24, 9759. [Google Scholar] [CrossRef]
- Dobrzyński, J.; Jakubowska, Z.; Dybek, B. Potential of Bacillus Pumilus to Directly Promote Plant Growth. Front. Microbiol. 2022, 13, 1069053. [Google Scholar] [CrossRef]
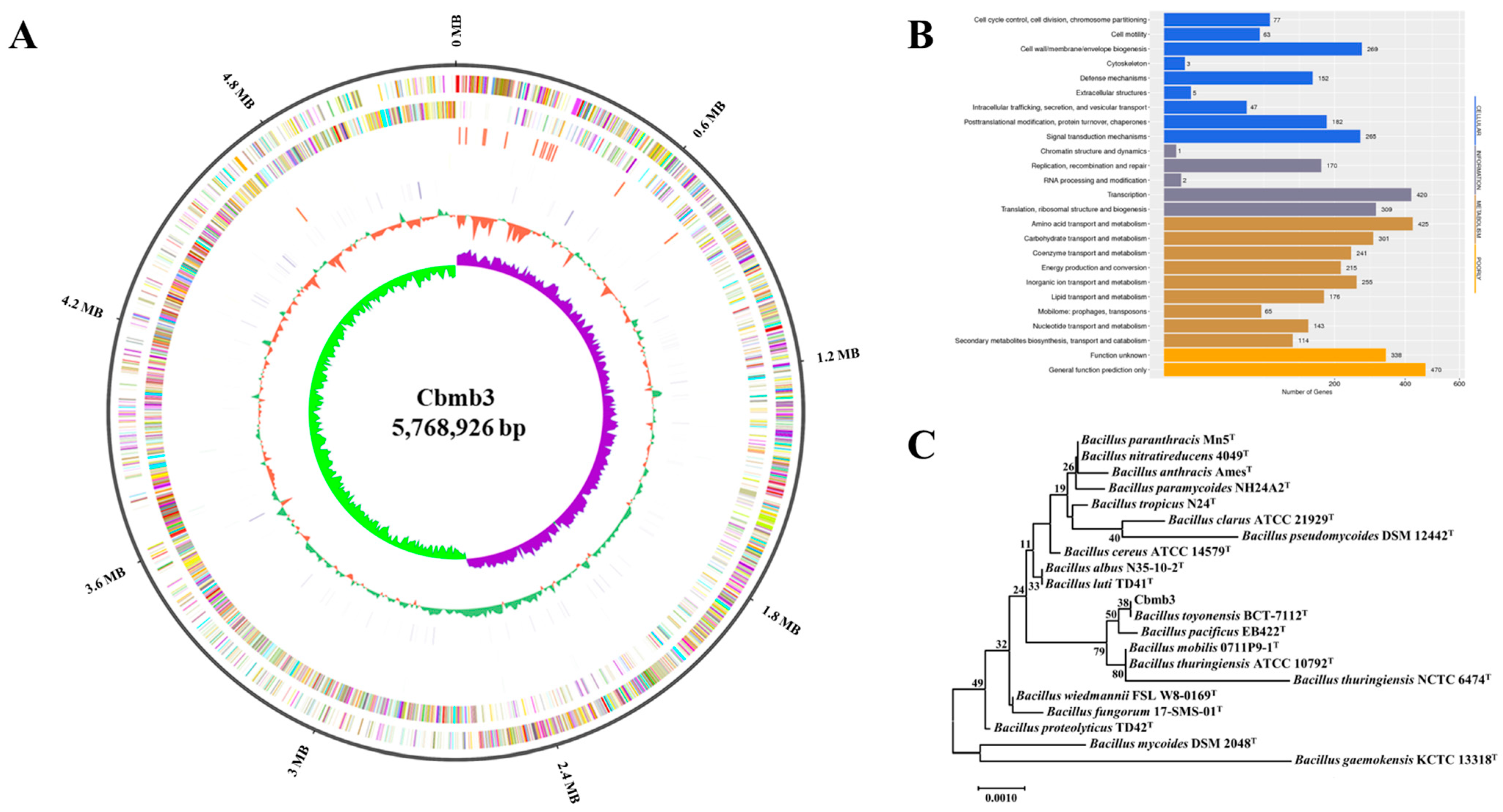

| Feature | Cbmb3 |
|---|---|
| Genome (bp) | 5,768,926 bp |
| C + G (%) | 35.95% |
| CDS number | 5835 |
| rRNA | 42 |
| tRNA | 105 |
| sRNA | 36 |
| Tandem Repeat | 405 |
| Prophage region | 15 |
| Gene island | 10 |
| CRISPR | 6 |
| Carbohydrate-Active enZYmes | 148 |
| Genes assigned to GO | 3163 |
| Genes assigned to COG | 4009 |
| Genes assigned to KEGG | 3017 |
| Strains | Accession No. | ANI (%) | dDDH (%) | Size (Mp) | G + C (%) |
|---|---|---|---|---|---|
| Cbmb3 | PRJNA992983 | 100.00 | 100.00 | 5.76 | 35.46 |
| Bacillus toyonensis BCT-7112 | PRJNA225857 | 98.47 | 99.52 | 4.94 | 33.78 |
| Bacillus toyonensis BPN45/4 | PRJNA521676 | 98.43 | 99.25 | 5.45 | 35.20 |
| Bacillus toyonensis P18 | PRJNA678769 | 98.53 | 98.92 | 5.25 | 35.19 |
| Bacillus wiedmannii SR52 | PRJNA490767 | 91.31 | 60.98 | 5.45 | 35.46 |
| Bacillus luti FJ | PRJNA515150 | 91.07 | 54.84 | 5.20 | 35.71 |
| Bacillus albus SXL388 | PRJNA960711 | 90.90 | 47.45 | 5.35 | 35.25 |
| Bacillus mobilis 16-00177 | PRJEB20065 | 90.79 | 34.26 | 5.69 | 35.28 |
| Bacillus thuringiensis ATCC 10792 | PRJNA29723 | 91.44 | 29.40 | 6.26 | 34.82 |
| Bacillus mobilis 0711P9-1 | PRJNA325888 | 90.75 | 27.99 | 5.66 | 35.30 |
| Bacillus proteolyticus TD42 | PRJNA325897 | 90.60 | 24.19 | 5.86 | 35.15 |
| Bacillus pacificus anQ-h4 | PRJNA762228 | 90.81 | 22.98 | 5.25 | 35.41 |
| Bacillus fungorum 17-SMS-01 | PRJNA408208 | 91.13 | 9.29 | 5.65 | 34.99 |
| Enzymes | Orthologous Genes | Locus Tags |
|---|---|---|
| Alcohol dehydrogenase [EC:1.1.1.1] | K00001, K13953 | 02186, 02529, 03409, 02226 |
| Glutathione dehydrogenase [EC:1.1.1.284] | K00121 | 03056 |
| Aldehyde dehydrogenase [EC:1.2.1.3] | K00128 | 01296, 02750, 03453 |
| Haloacid dehalogenase [EC:3.8.1.2] | K01560 | 03232, 05362 |
| Catalase-peroxidase [EC: 1.11.1.6] | K03781 | 01144, 04758 |
| Acetaldehyde dehydrogenase [EC:1.2.1.10] | K04072 | 04341 |
| Catechol 2,3-dioxygenase [EC:1.13.11.2] | K07104 | 03431, 04330 |
| Plant Growth Promotion Traits | Gene ID | Gene Name | Function |
|---|---|---|---|
| Indole-3-acetic acid (IAA) | 01174, 03242 | trpS | Tryptophan-tRNA ligase |
| 01229–01235 | trpABFCDE | Tryptophan biosynthesis operon | |
| Phosphonate solubilization | 03367, 04700 | gcd | Glucose dehydrogenase |
| 03582–03585 | phnCDEE1 | Phosphonate ABC transporter permease | |
| 04241 | phoU | Phosphate transport system regulatory protein | |
| 04242–04244 | pstCAB | Phosphate ABC transporter | |
| Biofilm | 01288–01290 | tapA-sipW-tasA | Amyloid fiber biosynthesis protein |
| 03614–03600 | epsA-O | Polysaccharide biosynthesis protein | |
| 03708–03710 | pgsABC | γ-poly-glutamate biosynthesis protein | |
| 04852–04856 | glgPADCB | Glycogen biosynthesis | |
| Chemotaxis and motility | 01638–01643 | che operon | Chemotaxis-associated protein |
| 01648–01679 | flg-fli-flh operon | Flagellar-associated operon | |
| 00419, 00573, 00591, 01956, 01956, 04999 | mcpAC | Methyl-accepting chemotaxis protein | |
| Quorum sensing | 04778 | luxS | Autoinducer 2 (AI-2) synthesis protein |
| 02889–02894 | lsrBDCA-RK | AI-2 uptake operon and transcriptional regulator |
| Region | Type | Cluster Position | Metabolites | MIBiG ID | Similarity |
|---|---|---|---|---|---|
| 1 | LAP | 1211148–1234654 | - | - | - |
| 2 | NI-siderophore | 1882901–1896614 | Petrobactin | BGC0000942 | 100% |
| 3 | NRP | 2237706–2289455 | Bacillibactin | BGC0000309 | 85% |
| 4 | NRP | 2433302–2458540 | Fengycin | BGC0001095 | 40% |
| 5 | Ripp | 2512857–2522101 | - | - | - |
| 6 | Ripp | 2574290–2584556 | - | - | - |
| 7 | terpene | 3377505–3399358 | Molybdenum cofactor | BGC0000916 | 17% |
| 8 | lassopeptide | 3509630–3533555 | Paeninodin | BGC0001356 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Yu, H.; Liu, X.; Sun, L.; Liu, X.; Hu, R.; Wang, C.; Zhuge, Y.; Xie, Z. The Complete Genome Sequence of Bacillus toyonensis Cbmb3 with Polyvinyl Chloride-Degrading Properties. J. Xenobiot. 2024, 14, 295-307. https://doi.org/10.3390/jox14010018
Wang D, Yu H, Liu X, Sun L, Liu X, Hu R, Wang C, Zhuge Y, Xie Z. The Complete Genome Sequence of Bacillus toyonensis Cbmb3 with Polyvinyl Chloride-Degrading Properties. Journal of Xenobiotics. 2024; 14(1):295-307. https://doi.org/10.3390/jox14010018
Chicago/Turabian StyleWang, Dandan, Hong Yu, Xinbei Liu, Li Sun, Xijian Liu, Ruilong Hu, Chao Wang, Yuping Zhuge, and Zhihong Xie. 2024. "The Complete Genome Sequence of Bacillus toyonensis Cbmb3 with Polyvinyl Chloride-Degrading Properties" Journal of Xenobiotics 14, no. 1: 295-307. https://doi.org/10.3390/jox14010018
APA StyleWang, D., Yu, H., Liu, X., Sun, L., Liu, X., Hu, R., Wang, C., Zhuge, Y., & Xie, Z. (2024). The Complete Genome Sequence of Bacillus toyonensis Cbmb3 with Polyvinyl Chloride-Degrading Properties. Journal of Xenobiotics, 14(1), 295-307. https://doi.org/10.3390/jox14010018