Abstract
Cytochrome P450s (CYPs) are the most prominent family of enzymes involved in NADPH- and O2-dependent hydroxylation processes throughout all spheres of life. CYPs are crucial for the detoxification of xenobiotics in plants, insects, and other organisms. In addition to performing this function, CYPs serve as flexible catalysts and are essential for producing secondary metabolites, antioxidants, and phytohormones in higher plants. Numerous biotic and abiotic stresses frequently affect the growth and development of plants. They cause a dramatic decrease in crop yield and a deterioration in crop quality. Plants protect themselves against these stresses through different mechanisms, which are accomplished by the active participation of CYPs in several biosynthetic and detoxifying pathways. There are immense potentialities for using CYPs as a candidate for developing agricultural crop species resistant to biotic and abiotic stressors. This review provides an overview of the plant CYP families and their functions to plant secondary metabolite production and defense against different biotic and abiotic stresses.
1. Introduction
Cytochrome P450s (CYPs) are one of the most prominent families of enzymes, which are the oxidoreductases class, which uses heme-thiolate as a cofactor. CYPs catalyze NADPH- and/or O2-mediated hydroxylation reactions in primary and secondary metabolism in various species. CYPs greatly influence the diversity of metabolites. The metabolites are produced via oxidation, reduction, hydroxylation, epoxidation, dealkylation, C-C cleavage, desaturation, decarboxylation, dimerization, isomerization, and ring extension processes [1]. In 1958, the pigment CYP was discovered in the microsomes of rat liver; today, it is known that CYP is also found in plants, microorganisms, insects, and mammals. They exhibit fantastic natural diversity, as evidenced by their presence in a wide range of organisms. The methods for identification and characterization of CYPs varied between the pre-and post-genomic era. In the pre-genomic era, biochemical methods, such as CYP separation from microsomal fractions and CYP activity inhibition, were utilized to study the function of CYPs. Characterizing CYPs using these methods could be difficult because the CYP superfamily comprises a large gene family and numerous isoforms, making it sometimes difficult to predict the substrate of the enzyme [2]. Later, novel approaches to understanding how CYPs are involved in multiple plant processes, including stress responses, were opened up by the development of next-generation sequencing technologies (NGS) and their advancement and affordability. For instance, there are several CYP genes in the genomes of plants. They represent about 1% of the total protein-encoding genes: 245 in Arabidopsis [3], 316 in grapevine [4], 332 in soybean [5], 313 in poplar [6], and 334 in rice [7].
There were 318 CYPs counted preliminary in maize, along with initial counts of 372 in sorghum, 142 in papaya, 172 in lotus, 174 in mulberry, 225 in Bracypodium distachyon, 270 in tomato, and 399 in potato [8]. Various phylogenetic clans, including single-family and multi-family lineages, have been used to categorize all CYP family proteins of plant origin. These large plant CYPs have developed due to significant evolutionary processes, including gene duplication and diversification. Cytochrome P450 monooxygenases are crucial in producing metabolites. These are necessary for healthy growth and development, and adaptive strategies that define biotic interactions, such as trophic interactions between plants, insects, mammals, fish, and their corresponding pathogens. Plants have accumulated many more CYPs throughout evolution. Throughout their life cycle, plants are subjected to a range of environmental stressors. Plants undergo multiple alterations in their transcriptional, translational, and biochemical levels due to biotic and abiotic stresses, including adjustment to their genetic and protein levels to facilitate adaptation to stressful conditions (Figure 1). It also adjusts enzymatic activities, regulates the concentration of phytohormones to achieve a balanced state, and employs mechanisms to scavenge reactive oxygen species (ROS) to maintain its equilibrium and counteract stress. These adaptations encompass a range of responses to enhance the plant’s resilience and survival in challenging environments. Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are three defense signaling chemicals and have complex cross-talk networks that control plant defense [9,10]. Additionally, specific CYP genes connected to the response of plants to stress have been found. These specific CYP genes hold significant promise as potential candidates for developing agricultural species resistant to abiotic and biotic stresses. This review presents an overview of plant CYPs and their functions in plant stress responses.
Figure 1.
Overview of the role of CYPs in different stress tolerance. CYPs are involved in a wide range of biochemical pathways in plants. Many secondary metabolites, such as alkaloids, flavonoids, and terpenoids, are synthesized by plant CYPs. Plant CYPs are also involved in the biosynthesis of several plant hormones, including auxins, gibberellins, and abscisic acid, and the detoxification of xenobiotics, such as herbicides, pesticides, and pollutants.
2. Cytochrome P450 Gene Families
Eukaryotes contain a variety of cytochrome P450 monooxygenases comprising a vast and diversified class of enzymes with more than 35,000 members [7]. CYPs participate in various biosynthetic and xenobiotic processes, including incorporating carbon sources, xenobiotic detoxification, and creating secondary metabolites [11,12]. The CYP superfamily is one of the most extensive gene families in plants; it has 245 genes in Arabidopsis, 332 full-length genes, and 378 pseudogenes in soybean [5,13,14]. Initially, the perception of CYP was that of a pigment rather than an enzyme. Later, it was discovered that the specific pigment had striking similarities to heme-protein properties. Because they contain a particular heme group, CYPs can participate in binding carbon monoxide. In plants, the catalytic process involves this specific heme group.
Regarding plants, CYPs were discovered for the first time in the cotton plant [15,16]. After being cloned and phylogenetically categorized, the structures and functions of CYPs from various plants have been revealed. Different CYP types were classified through phylogenetic analysis, utilizing a combination of family numbers and subfamily letters. Paralogs were assigned identical numbers within the same species, while orthologs across different species were assigned shared numbers. For instance, if the amino acid sequence is more than 40% identical to one or more known CYPs, the proteins are grouped into the same CYP family. Moreover, 2 CYPs are regarded as belonging to the same subfamily if they share more than 55% of their genetic makeup. Therefore, unless any particular descriptions are provided, CYPs with more than 97% similarity are assumed to be the same gene [2]. Based on prior findings, the plants’ CYP gene family was divided into eleven clans [7]. Our knowledge of CYP is continually expanding as more genetic data becomes accessible.
CYPs play a pivotal role in a wide range of cellular functions that profoundly impact plant growth and development. CYPs synthesize secondary metabolites, including terpenoids, flavonoids, steroids, alkaloids, phenylpropanoids, glucosinolate, and cyanogenic glycosides. Plant CYP is critical in numerous metabolic processes and can bind to various biological compounds. As a result of these reactions, different fatty acid conjugates, plant hormones, secondary metabolites, lignin, and many protective chemicals are produced [17,18]. Several physiologically active chemicals are attributed to the large variety of CYP genes and their functions [1]. CYP enzymes serve important roles in the evolution and metabolic diversity of plants. Plant CYPs were initially divided into two main clades, the A-type and the non-A-type [19]. Therefore, the A-type group appeared to contain the bulk of P450s producing secondary metabolites. The non-A-type, on the other hand, includes a significantly more divergent set of sequences made up of several distinct clades. They occasionally exhibit a more local similarity to non-plant CYPs than plant CYPs and are involved in the metabolism of lipids or hormones [20].
2.1. Non-A-Type CYPs
There are 93 non-A-type, and 7 potential non-A pseudogene has been identified in Arabidopsis. Four branch points were detected in the non-A-type CYP phylogenetic tree based on bootstrap values and intron conservation. However, the phylogenetic tree could not resolve all the deeper branches. The oldest plant CYPs found in Chlamydomonas are the CYP97s and they form a single-family clan [14]. All plants have copies of all three CYP97 subfamilies and are often connected to the synthesis of carotenoids. The CYP74 family also belongs to the non-A-type, which has its own clade. It is found in Rhizobacteria, Cnidaria, and Chordata (in Amphioxus), and is more ancient than plants [21]. However, the evolutionary development of this family is still unclear [14]. Since the CYP74’s main sequence is notably different from other plant CYPs, its position in the evolutionary tree is not firmly determined. The CYP711 is another single-family clade that does not substantially associate with other Arabidopsis clans. The CYP711 clan first appears in green algae, represented by two families, most likely its ancestors. The CYP51 and CYP710 belong to separate clades and are found in fungi, brown algae, and Chlamydomonas. The CYP85 clan is also an important member of non-A-type CYP. This clans consists of the CYP85, CYP87, CYP88, CYP90, CYP702, CYP707, CYP708, CYP716, CYP718, CYP720, CYP722, CYP724 and CYP728 families. Although ancestors of this clan first occur in green algae during the early stages of plant evolution, the families that currently exist in angiosperms first arise in liverworts or bryophytes. The CYP72 clan is made up of the members CYP72, CYP734, CYP714, CYP721, CYP709, CYP735, and CYP715. Liverworts contain the earliest members of this clan, although those families found in angiosperms (CYP72, CYP709, and CYP735) are only found in ferns [14].
2.2. A-Type CYPs
In Arabidopsis, there are 152 A-type CYPs and 20 potential A-type pseudogenes. These A-type CYPs are typically distinguished by a single highly conserved intron, M, that is absent from all the other CYP subtypes. Additional conserved introns are ob-served within the various A-type families and subfamilies. However, compared to non-A CYPs, fewer introns are typically present in the A-type. With 52 members in the Arabidopsis genome, the CYP71 family is the most expanded group in the A-type clade. It is the best example of a blooming pattern of gene duplication. The CYP79 and CYP703 are related to the CYP71 because they have the Phe to His substitution in the PERF domain. On the other hand, locating the four families CYP73, CYP77, CYP89, and CYP701 in the CYP71 clan evolutionary trees is challenging. They exhibit distinct intron-exon boundaries compared to other genes within the CYP71 clan. The PERF and heme sequences found in these families exhibit a closer resemblance to the core A-type CYP members, such as the CYP705 and CYP71 families. The conserved M intron is absent from the CYP73, CYP77, CYP89, and CYP701. Although they have different intron arrangements, these four families are more closely linked to the A-type CYPs than to the non-A-type CYPs and non-plant CYPs [14]. With the rapid advancement of DNA sequencing technology, around 200,000 plant CYP gene sequences have been found and compiled in a database [22]. Nevertheless, only a small percentage of plant CYPs have been functionally characterized because screening CYP enzymes for a given catalytic function is expensive. Specifically, for plant CYP enzymes, Wang et al. 2021, developed the PCPCM (plant cytochrome P450 comparative modeling) and PCPLD (plant cytochrome P450 ligand docking) processes, where they used template-based structure prediction and ligand docking, respectively. They conducted cross-docking experiments that effectively demonstrated the validation of PCPLD for virtual screening of CYPs [23].
3. Chemical Diversity and Evolution
Plants have substantially greater CYPomes (the cumulative quantity of CYPs present in a specific species) than mammals or microbes. Early in the history of plants, the CYPs began to diversify. In the green algae Chlamydomonas reinhardtii, 39 CYP genes were identified, while in Physcomitrium patens, a moss species, the number of identified CYP was 71 [24,25]. Green algae, land plants, and a newly identified phylum of sea algae, the Prasinodermophyta, all belong to the plant lineage Viridiplantae. The Prasinodermophyta has been considered to be the oldest diverging phylum within Viridiplantae [26]. Green algae are comprised of charophyte and chlorophyte algae. Twelve recognized algal classes are represented by chlorophytes, whereas charophytes represent five known algal classes. Successful development and evolution of terrestrial plants require changes in biochemistry along with plant architecture [25]. Some CYP families’ emergence during plant evolution is correlated to the appearance of particular metabolite classes, such as carotenoids, phytohormones, oxygenated lipids, or phenylpropanoids. The CYPs are very diverse in green algae. There is no indication of saturation in the count of algal CYP families, suggesting that numerous families are still yet to be discovered. The stark difference between the charophyte and chlorophyte algae can be seen in the fact that only 41 CYP families (around 12%) are shared by these two groups.
Along with high sequence identity, the presence of fungal CYP55 (nitric oxide synthase) in chlorophytes but not charophytes may indicate lateral transmission of CYP55 after the two algal clades split [27]. Families shared by two or more taxonomic groups and families unique to a particular taxon comprise most of the known CYP families. The estimated numbers of species in each of the nine categories are as follows: gymnosperms: 1000; ferns: 10,500; mosses: 12,000; lycophytes: 1300; hornworts: 200–250; liverworts: 7500; ferns: 10,500; ferns: 12,000; and angiosperms: 300,000. It was believed that CYP clans 51, 74, 97, 710, 711, and 746 comprise of single-family clans. However, this perception has been revised with the annotation of additional algal sequences. It has been discovered that within the 51 clan, there are 3 CYP families unique to green algae, 4 families within the 710 clan, a minimum of 46 families within the 711 clan, and possibly 2 families within the 746 clan. The CYP51, CYP710, CYP97, CYP74, and CYP86 families carry out crucial catalytic processes in metabolism and are conserved in green algae and land plants (Figure 2). Five families (CYP51, CYP710, CYP97, CYP74, and CYP704) are found across several kingdoms. However, green algae do not have CYP704. Despite the recruitment to a specialized pathway and the conservation of duplicates, the families are inclined to be retained as a single copy or at low copy numbers. Gene duplication events have been linked to the growth of the plant CYP superfamily, which was then followed by functional divergence and adaptation to particular metabolic pathways. Whole-genome duplication (WGD) events, which have occurred several times in plant genomes, have helped to enlarge gene families, including the CYP superfamily. One of the largest CYP superfamilies in plants, for instance, the Arabidopsis thaliana genome, comprises of more than 250 CYP genes. It has been established that the diversification of plant secondary metabolites and the development of plant defense systems against herbivores and pathogens are connected with expanding CYP superfamily in plants. Utilizing comparative genomics and transcriptomics, a number of research has been conducted on the evolution of plant CYPs [28]. As an illustration, the model tree species Populus trichocarpa genome reveals the expansion of CYP families involved in synthesizing lignin and secondary metabolites. Similar growth of CYP families involved in the synthesis of carotenoids and flavonoids was seen in the genome of Solanum lycopersicum. These investigations have shed light on the functions and adaptive evolution of plant CYPs. Gene duplication and WGD incidents have aided in the growth of the plant CYP superfamily. Tandem duplication events, which cause the duplication of closely related genes like CYPs, have been demonstrated to be a key mechanism of gene duplication in plants. For instance, simultaneous duplication events within the CYP93 family of A. thaliana led to the emergence of two unique subfamilies, CYP93A and CYP93B. While the CYP93B enzymes are engaged in the biosynthesis of flavonoids and lignin, the CYP93A enzymes are involved in synthesizing plant hormones [29].
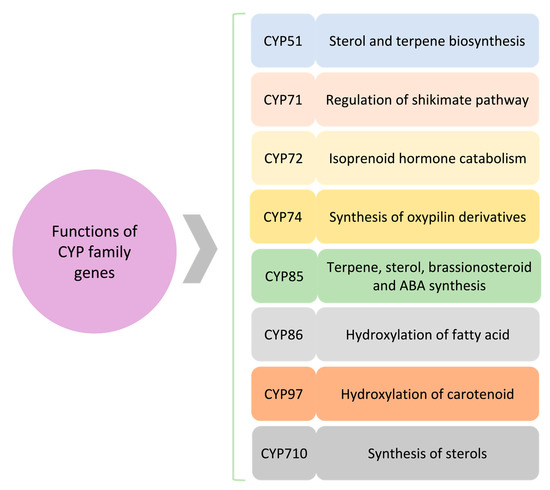
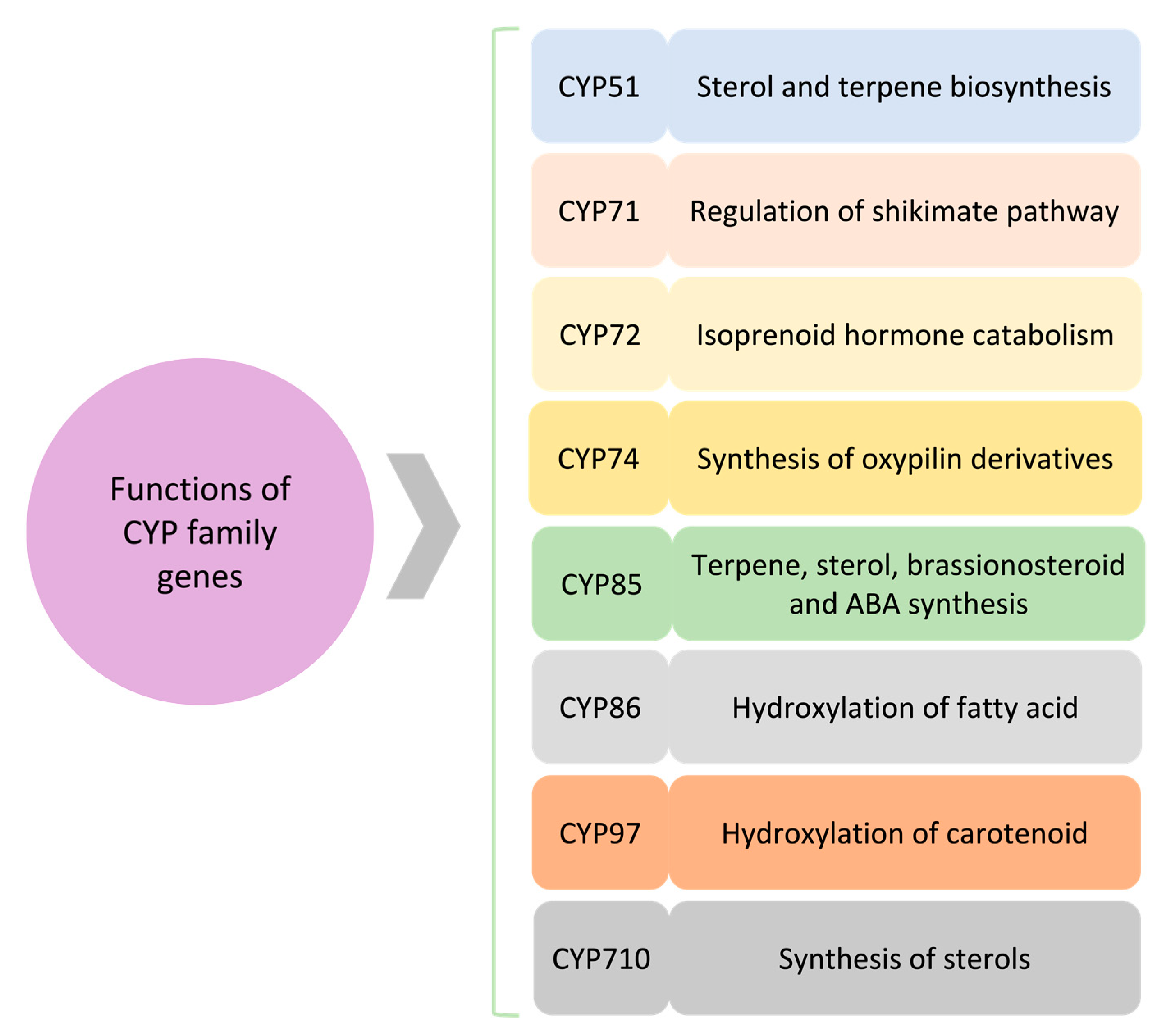
Figure 2.
A schematic of the role of major CYP clans. Several CYP clans are involved in plant secondary metabolites and hormone biosynthesis. Synthesis of terpene and sterols is being regulated through CYP51, CYP85, and CYP710. CYP85 also affects hormone regulation, i.e., brassinosteroid and abscisic acid (ABA). CYP86 and CYP97 are involved in fatty acid and carotenoid hydroxylation, respectively. CYP71 is associated with the shikimate pathway, whereas CYP72 and CYP74 impact hormone signaling.
4. Metabolite Biosynthesis
Secondary metabolites are widely present across various plants and play essential roles as compounds for defense and signaling purposes. To date, more than 200,000 secondary metabolites (SMs) have been discovered and characterized. These SMs belong to diverse compound classes, including alkaloids, flavonoids, terpenoids, and glucosinolates [30]. Plants produce different primary or secondary metabolites as a defensive strategy under stress. The CYP gene families have diverse roles in the biosynthesis of metabolic compounds and promote the plant’s development and defense (Table 1). The CYP gene contributes significantly to photosynthesis and photo-protection, which are closely linked to plant growth [31]. Xanthophylls, the yellow pigments, play a vital role in light harvesting during photosynthesis. CYP97A3 and CYP97C1 play an active role in the biosynthesis of xanthophylls by catalyzing the hydroxylation of the beta and gamma rings of carotenoids. In the Sorghum plant, specific CYP genes such as CYP97C1 and CYP97A3 determine the pathway of lutein. This pathway is crucial for protecting chlorophyll as it facilitates energy absorption and transfer [32]. Moreover, the CYP gene family plays a significant role in auxin biosynthesis in specific plant species. This involvement in auxin biosynthesis is crucial for various tropic behaviors, such as growth towards the light (phototropism) or gravity (gravitropism), as well as root initiation and apical dominance regulation [33]. Furthermore, CYP734, CYP724, and CYP90 participate in glycoalkaloids biosynthesis in the Solanaceae family, whereas CYP74D genes are essential in the biosynthesis of antimicrobial substances such as oxylipins [34]. These oxylipins are fatty acid derivatives that protect plants against particular stressors.
The biosynthesis of cutin and suberin in plants, like Symphytum tuberosum and A. thaliana, heavily relies on the active involvement of CYP genes, particularly CYP86A1, CYP86A2, CYP86A8, and CYP86A33 [35,36,37]. Cutin and suberin are complex biopolymers that protect plants against infections and UV by regulating water evaporation and transpiration. The expression of CYP72A59v1 and CYP72A66 is responsible for saponin biosynthesis in response to drought/salt stress in M. truncatula [38]. In Arabidopsis, 13-hydroperoxy linolenic acid (13-HPOT) is converted to the plant defense hormone, jasmonic acid, by the allene oxide synthase (AOS) encoded by the CYP74A1 gene [39]. Meanwhile, CYP74B4 and CYP94A52 may be involved in the oxylipin biosynthesis pathway, and CYP83D3 in the glucosinolates biosynthesis pathway, which may enhance drought/NaCl stress tolerance by minimizing water loss due to stomatal closure. Additionally, it has been observed that the CYP704G6 gene has a potential role in the biosynthesis of fatty acids and can impact flower development when plants are subjected to drought stress. Meanwhile, the CYP71A31 and CYP71AU56 genes are likely responsible for the biosynthesis of indole-3-acetic acid (IAA), a plant hormone associated with various growth processes. These genes may significantly improve plant drought tolerance by enhancing IAA production [38]. The ABA-independent SnRKs (Sucrose non-fermenting 1-related protein kinases) impact the mRNA levels of aquaporins and the enzyme CYP97B2. Aquaporins regulate water transport across cell membranes, while CYP97B2 plays a role in auxin biosynthesis. The activity of ABA-independent SnRKs influences the expression of the gene, thereby affecting water transport and auxin biosynthesis in plants [40]. Under salt stress, CYP79B2 and CYP79B3 are induced in Arabidopsis shoots and roots, suggesting a role for the indole-3-acetaldoxime (IAOx) pathway in regulating salt stress responses [40]. Additionally, CYPs facilitate the production of benzoxazinoids, such as 2,4-dihydroxy1,4-benzoxazin-3-one (DIBOA) and 2,4-dihydroxy7-methoxy-1,4-benzoxazin-3-one (DIMBOA), which are essential for enhancing plant defense against diseases, herbivores, and weeds [41,42].

Table 1.
Potential CYP genes responsible for secondary metabolites biosynthesis and biotic and abiotic stress tolerance.
Table 1.
Potential CYP genes responsible for secondary metabolites biosynthesis and biotic and abiotic stress tolerance.
| Plant Species | CYP Genes | Function | Useful Trait | References |
|---|---|---|---|---|
| Arabidopsis thaliana | CYP97C1 | Lutein biosynthesis | Abiotic stress tolerance | [43] |
| CYP83B1 | Biosynthesis of indole glucosinolates | Abiotic stress tolerance | [44] | |
| CYP86A1 | Suberin biosynthesis | Insect resistance | [35] | |
| CYP82G1 | Biosynthesis of homoterpene volatiles | Herbivory resistance | [45] | |
| CYP86A2 | Biosynthesis of fatty acids | Biotic stress tolerance | [37] | |
| CYP83A1 and CYP83B1 | Glucosinolates biosynthesis | Insect tolerance | [46] | |
| CYP86A2, A8 | Cutin biosynthesis | Insect tolerance | [37] | |
| CYP51H, CYP71A,D, CYP72A, CYP81Q, CYP87D, CYP88D,L, CYP93E, CYP705A, CYP708A, and CYP716A,C,E,S,U,Y | Specialized triterpenes metabolism | [47] | ||
| Solanum tuberosum | CYP72A188, CYP72A208 | Biosynthesis of steroidal glycoalkaloids | Abiotic stress tolerance | [48] |
| Oryza sativa | CYP714A3 | Gibberellin biosynthesis | Heavy metal stress resistant | [49] |
| Triticum aestivum | CYP88A | Biosynthesis of gibberellin | Heavy metal stress resistance | [49] |
| CYP71 | Biosynthesis of phytotoxin e.g., 2,4-dihydroxy1,4-benzoxazin-3-one (DIBOA) and 2,4-dihydroxy7-methoxy-1,4-benzoxazin-3-one (DIMBOA) | Biotic stress tolerance | [50] | |
| Pisum sativum | CYP88A | Biosynthesis of gibberellin | Heavy metal stress tolerance | [44] |
| CYP96A15 | Biosynthesis of epicuticular wax | Biotic stress tolerance | [51] | |
| Panax ginseng | CYP71 | Biosynthesis of secondary metabolites, alkaloids, and flavonoids | Heavy metal stress resistance | [52] |
| Zea mays | CYP71 | Biosynthesis of phytotoxin, e.g., DIBOA and DIMBOA | Biotic stress tolerance | [50] |
| CYP96A15 | Biosynthesis of epicuticular wax | Biotic stress tolerance | [51] | |
| CYP1Capsicum annuum | Synthesis of dehydrin | Biotic stress tolerance | [53] | |
| Nicotiana benthamiana | CYP51H10 | Biosynthesis of triterpenes | Biotic stress tolerance | [54] |
| Nicotiana rustica | CYP51 | Biosynthesis of triterpene | [54] | |
| Helianthus tuberosum | CYP96A15 | Biosynthesis of epicuticular wax | Biotic stress tolerance | [51] |
| Sorghum bicolor | CYP79A1 and CYP71E1 | Biosynthesis of the cyanogenic glucoside | Insect tolerance | [55] |
| CYP79 | Cyanogenic glucosides biosynthesis | [56] | ||
| Catharanthus roseus | CYP72A1 | Alkaloid biosynthesis | Disease resistance | [57] |
| Ocimum basilicum | CYP82D | Biosynthesis of different secondary metabolites | [58] | |
| Vitis vinifera | CYP75 | Flavonoid biosynthesis | [59] | |
| Gossypium arboreum | CYP706B1 | Alteration of the isoprenoid pathway | [60] | |
| Symphytum tuberosum | CYP86A33 | Biosynthesis of suberin | [36] | |
| Pogostemon cablin | CYP71, CYP77, CYP81, CYP82 | Biosynthesis of sesquiterpenes | [61] | |
| Citrus sp. | UV-B-induced CYP gene | Biosynthesis of 3′-hydroxylated flavonoids | [62] | |
| Cajanus cajan | CYP75B165 | Biosynthesis of flavonoids | [63] | |
| Glycine max | CYP82D26 | Isoflavonoid pathway biosynthesis | [64] | |
| Legumes | CYP93C | Synthesis of legume specific isoflavonoid | [65] | |
| Land plants | CYP93B, CYP93E, CYP93G | Saponin biosynthesis | [66] | |
| Higher plants | CYP74A | Synthesis of allene oxide | Biotic stress resistance | [67] |
5. Xenobiotic Metabolism and Hormone Regulations
CYPs are one of the main enzymes in charge of detoxifying exogenous molecules in plants and other organisms. Toxins, including pesticides, heavy metals, allelochemicals, organic pollutants, and other pollutants, are frequently exposed to plants. Plants build specialized and coordinated defense mechanisms to survive in unfavorable growing conditions. CYP’s involvement in the biosynthesis of plants’ hormones, lipids, and secondary compounds is better understood than its function in detoxifying exogenous chemicals [68]. On the other hand, they have the potential to serve as a crucial metabolic reservoir for environmental contaminants [69], presenting a promising approach to regulate and control herbicide selectivity and tolerance [70], and could represent a good potential for bioremediation.
The ability of some monocotyledonous plants’ herbicide detoxification system to be induced by a class of synthetic chemicals known as herbicide safeners. Safeners can shield grass fields from herbicide damage without lowering herbicide activity in target weed species. They protect the target plant from herbicide injury while inducing the expression of genes involved in plant defense and detoxification, such as glutathione S-transferases (GSTs) and CYPs. This indicates that safeners use a previously unrecognized signaling pathway to detoxify xenobiotics or endogenous toxins [71]. CYPs mediate the primary metabolism of several important chemical classes of herbicides, frequently resulting in crop selectivity [72]. The connections between the physiological and detoxification roles of various CYP isoforms and herbicide selectivity have been well studied in rice and maize [73,74,75]. The safener responsiveness was primarily attributed to clan 71 and clan 72 members within the CYP family, with fewer outliers in clan 86. Most of the upregulated CYP71 clans in Arabidopsis belong to the family CYP81. In maize, CYP81A9 and CYP81A6 have been linked to herbicide sensitivity in sweetcorn cultivars with five different mechanisms of action [76]. Further evidence demonstrated that Echinochloa phyllopogon and Lolium rigidum are resistant to graminicides and had an increased expression of clan 71 and 72 CYPs [77,78].
Herbicide metabolism is linked to several members of the CYP family. The involvement of CYPs is crucial in the metabolic transformation of target sites, leading to structural changes. Herbicide metabolism is separated into three phases, i.e., conversion (phase 1), conjugation (phase 2), and compartmentation (phase 3). The Phase 1 metabolism of herbicides is mostly mediated by CYPs [79]. In general, there are different criteria which are considered to demonstrate that CYP was involved in the development of resistance. For instance, the position in the microsomal palate, NADPH and oxygen requirements, CO-induced light-reversible inhibition, antibody-mediated inhibition of CYP reductase and CYP inhibitor-induced suppression. It is established that a positive correlation exists between elevated CYP enzyme activity and herbicide resistance in weed plants [79]. Recent research has shown that overexpression of the ginseng-derived CYP736A12 in Arabidopsis results in resistance to chlortoluron and isoproturon. The functional study of PgCYP76B93 provides information on using this gene as a marker for genetically modifying multipurpose crop species that can detoxify agrochemicals and environmental toxins [80]. The broader exploration of gene activity and regulation in various xenobiotic metabolizing enzymes has provided a comprehensive understanding of the functioning of plant detoxification systems across multiple species. This information will be highly advantageous in unrevealing the genetics and molecular underpinnings of plant herbicide selection.
Plant CYP enzymes tightly regulate the biosynthesis and catabolism of plant hormones like auxins, gibberellins, cytokinins, and abscisic acid (Table 2). The hydroxylation, epoxidation, and oxidative cleavage of hormones by these enzymes can alter their bioactivity or functions. Recent developments in molecular genetic analysis led to identifying and characterizing genes involved in producing plant hormones. In Arabidopsis, CYP79B2/B3 and CYP83 control the production of auxin. Gibberellin metabolism involves several CYPs in different plants, including CYP88A and CYP714A in Arabidopsis and CYP701A8 and CYP714B in rice. Moreover, the Arabidopsis CYP735A1 and CYP735A2 genes code for cytokinin hydroxylases that catalyze trans-zeatins, and the CYP707A gene code for an enzyme called ABA 8′-hydroxylase that regulates the amount of abscisic acid in Arabidopsis and barley (Table 2). The polyhydroxysteroid class, known as brassinosteroids (BRs), has been recognized as the sixth class of plant hormones. Numerous CYPs control BR biosynthesis, for instance, AtCYP72B1 and AtCYP72C1, control photomorphogenesis and plant steroid signal transduction by deactivating BR, and AtCYP85A controls the production of BR C-6 oxidase. AtCYP90A1 and AtCYP90B1 enhance vegetative growth and seed yield by regulating the biosynthesis of BRs. On the other hand, CYP90B3 and CYP724B2 catalyze the initial C-22 hydroxylation reactions in the BR biosynthesis pathway of Tomatoes [81].

Table 2.
List of several CYP genes in hormone biosynthesis.
6. Plant Defense against Herbivory and Pathogenicity
Biotic stress from living organisms—such as insects, pathogens, weeds, etc.—results in plant damage by disrupting the general dynamics of plants. These stresses deprive the host plant of the nutrients and minerals it needs, which can, in some severe circumstances, result in plant death. In addition to interfering with plant growth, biotic stressors also impact seed quality, root rot, and crop yield. Terpenoids, phytoalexins, alkaloids, cyanogenic glucosides, etc., are vital for a plant’s ability to respond to biotic stresses—they are synthesized proportionally by CYPs.
6.1. Cytochrome P450 in Plant-Pathogen Interaction
Among the various biotic stresses, the disease is regarded as the most predominant. Fungi, viruses, and bacteria play a substantial role in causing disease in plants.
6.1.1. Cytochrome P450 in Plant-Bacteria Interaction
In Arabidopsis, the AtCYP76C2 gene causes faster cell death to cope with stress caused by Pseudomonas syringae because it prevents the spread and proliferation of the pathogens [91]. The hypersensitive response (HR), driven by a P. syringae infection in A. thaliana, increases the expression of the CYP76C2 gene. The CYP76C2 gene expression is linked to injury, aging cell culture, and leaf senescence [92]. Phytoalexins are low molecular weight antibacterial chemicals with a wide range of structural variations produced rapidly by many plants in response to bacterial and fungal attacks. In Arabidopsis, CYP71 and CYP79 are reported to be associated with producing the phytoalexin ‘camalexin’ [93]. In Capsicum annuum, CaCYP1 is crucial for recognizing HR for safeguarding the plant against infection by Xanthomonas axonopodis. The CYP93C gene facilitates plant tolerance to disease stress by increasing the biosynthesis of isoflavonoids for plants, including Medicago truncatula, Glycyrrhiza echinata, Glycine max, and Cicer arietnum. Other CYP genes, such as CYP81E1, CYP81E3, CYP81E7, and CYP93A1, are also reported as significant players in the production of isoflavonoids [94]. In potatoes, the pathogen-inducible divinyl ether synthase from the CYP74 family produces the antibacterial substance oxylipin [34]. Oxylipins are derived from polyunsaturated fatty acids, play a crucial role in plant physiology. Among the major oxylipins produced by the plant, methyl jasmonate and jasmonic acid are particularly prominent. These compounds are primarily synthesized in response to physical injury and serve as key components in the plant’s defense against diseases. Withania somnifera has been studied to understand the role of CYPs in plant defense. The stalk and root have been found to express large amounts of the elicitor-responsive genes, including WsCYP98A and WsCYP76A. The role of WsCYPs in metabolite biosynthesis and defense against bacteria has also been reported in a prior study [95].
6.1.2. Cytochrome P450 in Plant-Fungi Interaction
The evolution of fungal pathogenicity has been linked to the diversification and extension of the CYP family. When pathogens attack host plants, CYP enzymes respond by inhibiting the formation of antibiotic phytoalexins [96]. Pisatin is a kind of phytoalexin produced by Pea, has been inactivated by CYPs in response to fungal attack [97]. Numerous crop plants are affected by the vascular wilt disease caused by the fungus Fusarium oxysporum. Antagonistic F. oxysporum strains defend plants from fungal attacks. In Lettuce, the pathogenic formae specialis of F. oxysporum expresses CYP505 in the host plant, which hydroxylates saturated fatty acid and helps plant tolerance. Lauric, palmitic, and stearic acids in the cortical cell membranes of the plant can be mono-hydroxylated by CYP505A1, and these hydroxylated substances may trigger the plant’s defensive mechanism [98]. The pepCYP gene participates in the defense mechanism against the pathogenic fungus Colletotrichum gloeosporioides which attack in the pepper. A CYP found in pepper called CYP89, is essential for the plant’s defense against pathogen infestations [53]. CYPs are involved in the signaling pathways that are triggered by jasmonic acid and methyl jasmonate. For example, methyl jasmonate activates the CYP82A3 gene in soybeans, which linked to fungal infections. N. benthamiana transgenic line overexpressing the CYP82A3 gene is highly resistant to gray mold and black shank disease [99]. Growing evidence shows that CYP enzymes are essential for plant defense against pathogenic fungi. Biochemical roles of CYPs associated with the metabolism of plant triterpenes have also been identified. CYP51H10 was required to synthesize anti-microbial oleanane-triterpene saponins (avenacins), which gave oats disease resistance against the fungi that infected the roots [100].
6.2. Cytochrome P450 in Plant-Insect Interaction
The number of CYP genes in plant genomes has expanded more significantly than in insect and vertebrate genomes. Numerous plant pathways evolved to produce ecologically significant secondary metabolites as defenses against insects. Maize has been developed into an important model species in the past two decades for research on physiological and molecular aspects of plant–insect interactions. The comprehensive defense mechanisms in maize have been studied in response to caterpillar feeding. The results revealed that the maize plant accumulates secondary metabolites for defense [101]. The simulated herbivory increases the expression of CYP79A61, and produces aldoxime, and phenyl acetaldoxime, which helps maize to defend against insect infestation [102]. In the arms race against herbivores, plants constantly evolve new defense chemicals to survive. One such class of specialized chemicals is saponins. The structure of saponins comprises a hydrophobic triterpenoid backbone with one or more hydrophilic saccharide groups. Saponins are poisonous or repellent for some insects, mollusks, fungi, and other microbes. In Barbarea vulgaris, a tandem repeat of eight CYP72A CYP colocalizes with a quantitative trait locus (QTL) for saponin accumulation. The function of CYP72A552 has evolved to catalyze the production of saponins based on hederagenins, which mediate plant defense against herbivores [103].
A multimeric complex containing a 13-lipoxygenase, allene oxide synthase (AOS), and allene oxide cyclase was found in A. thaliana. AOS is a member of the CYP74A family of CYP enzymes. AOS faces competition from hydroperoxide lyase (HPL, CYP74B), another CYP found in chloroplasts, for the same substrate. The ability of plants to respond to various biotic stresses depends on their AOS and HPL activities. The accumulation of JA is observed in plants as a reaction to various abiotic and biotic stresses. Additionally, JA is actively involved in wound responses and is a critical component in plant defense mechanisms. The AOS and HPL complexes regulate the production of JA during plant growth and stress response [104].
Insect pests cause physical damage to plants and act as vectors for viruses and bacteria. CYP genes have been involved in wound-induced defense. In sorghum, CYP71E1 and CYP79A1 enzymes catalyze the conversion of tyrosine, improving pest resistance and food safety [12]. Few plants, including Arabidopsis, have CYP86A1, which is involved in the manufacture of suberin that play a vital role against insect stress [35]. In Arabidopsis, the CYP family PAD3 gene is associated with the production of camalexin, conferring resistance against the green peach Aphid [105]. In P. trichocarpa, the synthesis of aldomixes is triggered by the CYP79D gene family under herbivore infection [102]. An unidentified CYP hydroxylase was found to convert cembratriene-ol (CBT-ol) into cembratriene-diol (CBT-diol) in the trichome glands of N. tabacum. By suppressing this, CYP increased the concentration of CBT-ol and displayed Aphid resistance [106]. In Coptis japonica, an alkaloid called berberine is produced by the CYP719, which is harmful to insects, including the Diamondback Moth.
7. Plant Abiotic Stress Tolerance
Plants can function normally in optimal conditions but often experience different biotic and abiotic stresses [107]. Exposure to different stresses interferes with their natural growth, leading to a significant loss in agricultural production [108]. It has been discovered that CYPs are involved in hormone signaling, hence governing plant response to different stresses [109,110]. CYPs defend plants from various stresses caused by environmental variables such as drought, heat, salt, and heavy metals [111,112,113]. Furthermore, CYPs play a direct role in plant secondary metabolism, actively by detoxifying external pollutants or those generated as metabolic byproducts in response to stress conditions [107].
7.1. Temperature Stress
Temperature stress, either heat or cold stress in plants, results in several undesirable occurrences, such as disruptions in respiration and photosynthesis, decreased cell membrane stability, and protein denaturation, ultimately leading to oxidative damage mediated by ROS [114]. In plants, heat stress leads to the denaturation of proteins by promoting the degradation of cellular proteins. It also affects the membrane’s permeability as plants’ cell walls disintegrate under high heat stress conditions [115]. Many findings indicated that heat or cold stress impacted the expression level of most flavonoid-related transcripts. Expression of CYP73A, CYP75A, and CYP75B genes involved in the production of flavonoid antioxidants such as trans-cinnamate 4-monooxygenase, flavonoid 3′,5′-hydroxylase, and flavonoid 3′-monooxygenase, respectively, was considerably increased in Lolium perenne and Festuca arundinacea in extreme low and extremely high temperatures [116]. The upregulation of CYP99A1 and CYP709C1 in cold-tolerant Sorghum indicates their involvement in low-temperature stress tolerance [117].
It has been reported that during prolonged cold stress in Arabidopsis, the CYP83A1 gene was found to be more expressed, playing a vital role in flavonoid (phenylpropanoids) metabolism [118]. It was reported in other studies that the CYP707A genes in Arabidopsis play a role in increasing the levels of ABA, specifically through the action of ABA 8′-hydroxylases. This increase in ABA levels facilitated the response of Arabidopsis to temperature stress [114,119]. In Panicum virgatum, 11 CYPs were differentially expressed under prolonged heat (38/30 °C, day/night, for 50 days), where, for the production of indole alkaloid secologanin, 2 CYP71A1 genes need to be present [120]. In contrast, the expression of CYP71 was significantly reduced in the leaves of Rhazya stricta when exposed to elevated temperatures (40–42.4 °C) [121]. Under high-temperature stress, CYP71A23 is responsible for pollen sterility in Brassica napus [122] and the expression of this gene increased in Panicum maximum [123].
7.2. Salinity Stress
Salinity stress refers to the inability of plants to uptake water due to the presence of excessive salts, particularly soluble salts, such as sodium chloride (NaCl), in the soil. The negative impacts of NaCl on plants are primarily due to decreased water availability due to Na+ accumulation in soil, increased oxidative stress, and the toxic effects of Na+ and Cl− ions [124]. Salt stress affects germination, growth and development, photosynthesis, and yield, eventually leading to death [125]. Plants have developed numerous methods to survive in highly salinized soil. It has also been demonstrated that CYP proteins are crucial in plant responses to salt stress by regulating hormone signaling and maintaining ROS homeostasis [126,127]. For example, expression of the CYP81D5 in maize and wheat at vegetative and reproductive stages enhanced salinity tolerance through ROS scavenging activity. The salinity stress response is closely associated with the induction of ABA. ABA may reduce transpiration, wilting, water uptake, and stomatal conductance. In contrast, ABA is favorable to maintaining low Na+/K+ and enhancing the activity of protecting enzymes, which ultimately keeps an intact cell membrane structure and reduces the damage caused by salt [128,129]. It has been reported that CYP707A protects plants from salt stress and increases salt tolerance by regulating ABA levels. The over-expression of the CYP709B3 gene improved salt-stress tolerance in transgenic Arabidopsis [130].
In a separate study, CYP736B, the homolog of CYP736A12, conferred enhanced salt tolerance by reducing H2O2 accumulation, increased carotenoid levels, and ABA biosynthesis in Arabidopsis [131]. Similarly, upregulated expression of CYP709 was observed in salt stressed Robinia pseudoacacia [132]. In a salt-tolerant strain of Physcomitrella patens, 50 CYPs modified their protein level under salinity stress to lessen cell wall deterioration and scavenge ROS [133]. The expression of CYP75A3 and CYP73A11 genes in cotton indicated salt tolerance with a reduced level of hydrogen peroxide and a significant increase in glutathione, ascorbate peroxidase, and proline. It also reported that knocking down these two genes downregulates several stress-responsive genes, making them more susceptible to salt stress [134]. Furthermore, it was revealed that salt stress significantly induced PtCYP714A3 from P. trichocarpa, and transgenic rice demonstrated enhanced salt tolerance [135]. In the mangrove tree Avicennia officinalis, AoCYP94B3 and AoCYP86B1 were found to reduce the suberin of the roots, thus leading to salt tolerance [136]. Another study revealed that heterologous expression of CYP94B1 in rice, Avicennia officinalis, and Arabidopsis enhances seedling salt tolerance by improving root suberin deposition [137].
7.3. Drought Stress
Water or drought stress refers to the insufficiency of moisture, recognized as a critical prerequisite for the optimal growth of a plant. The survival of plants during drought stress relies on the maintenance of cell homeostasis in a water-deficit environment. In response to such stress, plants activate specific hormones as a defense mechanism. ABA is an important plant hormone linked to drought stress and fluctuates dramatically during dehydration and rehydration. Nevertheless, in such scenarios, the catabolism of ABA plays a crucial role in maintaining the balance of ABA levels [138]. During drought stress, the presence of enzyme ABA 8′-hydroxylase (ABA8Ox) of the CYP707 family catalyzes the hydroxylation of the ABA, resulting in the formation of phaseic acid (PA) [139]. When maize was exposed to water deficit conditions, the CYP707A (ABA8Ox) gene was upregulated [140]. Similarly, CYP707A1 and CYP707A2 were significantly upregulated in Arachis hypogaea and Populus simonii under osmotic stress [141,142]. Moreover, it has been demonstrated that Arabidopsis exhibits upregulation of the same genes in response to drought stress [138].
During drought, the CYP96A8 gene is involved in the lignin biosynthesis of maize leaf, forming the necessary structural materials supporting plant tissues and other functions related to drought tolerance [143]. The plant cuticle is an important structure required for controlling non-stomatal water loss. In Arabidopsis, CYP86A8 is associated with the omega-hydroxylation of fatty acids and produces the monomer of cutin [144]. In contrast, CYP86A2 plays a significant role in epicuticular lipids synthesis, including cutin, reducing the cuticle membrane thickness. This reduction enhances water permeability, thereby aiding in drought tolerance mechanisms [37]. The specific role of CYP86A2 in drought tolerance mechanisms may vary among plant species. However, further research is needed to fully understand the specific mechanisms and the extent of CYP86A2’s impact on drought tolerance. During flower development in M. truncatula, CYP704G6 was highly expressed, suggesting it plays a similar role to CYP704B2 in fatty acid biosynthesis under drought conditions. In water-deficit states, CYPs participate directly or indirectly in the biosynthesis of antioxidants that can reduce oxidative damage [38]. In Citrus sinensis, CsCYT75B1 is upregulated in drought conditions, and overexpression of this gene is responsible for increased total flavonoid content and antioxidant activity [145]. In Arabidopsis, isoflavone and flavonoid 30-monooxygenase biosynthesis catalyzed by CYP81E9 and CYP78A126 may improve plant resistance to drought by lowering scavenge ROS [146]. Some other CYPs are involved in flavonoid biosynthesis, for instance, the CYP75 family regulates flavonoids biosynthesis in grapevines and ferns [6], heterologous expression of CYP82G24 in Carthamus tinctorius induces flavonoid biosynthesis genes [147].
It has been revealed that loss of CYP85A2 lowered the brassinolide (BL) accumulation and enhanced drought tolerance in Arabidopsis [148]. In another research, overexpression of SoCYP85A1 indicated drought stress tolerance due to higher castasterone (CS) (the precursor of the BL biosynthesis) accumulations. An increased abundance of CS results in a sharp decrease of BL, which enhances root growth, regulates ROS and controls the expression of genes that respond to stress [149].
7.4. Heavy Metal Toxicity
Crop production and quality are significantly hampered by heavy metal contamination of the soil and water, which causes toxicity and stress. Among all the heavy metals, lead, mercury, chromium, arsenic, and cadmium are the most toxic [150]. CYPs play an important role in detoxification when exposed to heavy metal stress [151]. The genes CYP71 in P. ginseng, CYP2E1 in Medicago sativa, CYP81D8 in A. thaliana, CYP88A in T. aestivum, and P. sativa have been associated with heavy metal stress tolerance. In Hg-contaminated soils, M. sativa plants that have been genetically altered with the human CYP2E1 and GST genes showed the potential for phytoremediation. Additionally, transgenic M. sativa expressing both of these genes showed a synergistic impact that improved metabolism in plants, enabling them to survive mercury exposure [152]. In P. ginseng, heavy metal interactions with the cell membrane produced several defense-related genes through the jasmonic acid pathway. The plant also has cross-talk networks with other defense-related pathways [153]. P. ginseng exhibited upregulation of CYP71 when exposed to heavy metals, particularly nickel and cadmium [52]. In maize, the CYP88A facilitates the synthesis of gibberellins, which integrates multiple hormone-signaling pathways in response to heavy metal toxicity stress [49].
7.5. Herbicide Stress
Herbicide resistance refers to a plant’s innate capacity to withstand while applying herbicides for weed control. The capacity of a species to live and reproduce following exposure to an herbicide at a regular application rate is known as herbicide tolerance. Herbicide exposure may cause a variety of physiological and biochemical reactions in plants. CYPs are involved in the physiological mechanism of hormones and secondary metabolites biosynthesis that confers herbicide resistance [79,154,155]. For instance, the overexpression of CYP736A12 in Arabidopsis has been found to slightly reduce plant height and contribute to the metabolism of phenylurea herbicides such as chlorotoluron and isoproturon, thus conferring herbicide resistance [156]. Both chlorotoluron and isoproturon, and phenylurea herbicides, actively metabolize the functionally defined CYPs CYP73A1 and CYP81B1 from Jerusalem artichoke. Following its identification in Jerusalem artichoke, CYP76B1 was subsequently discovered to possess the ability to metabolize herbicides belonging to the phenylurea family. CYP71A10, an enzyme derived from soybeans, may metabolize the phenylurea herbicide, chlortoluron, when expressed in yeast. However, it does so with ten times less effectiveness than CYP76B1 [122,157,158,159]. In addition, transgenic Arabidopsis and Nicotiana tabacum having rice CYP81A6 functionally characterized that CYP81A6 is involved in the betazon metabolism [74]. The CYP76C1, CYP76C2, and CYP76C4 genes also demonstrated herbicide tolerance in Arabidopsis. The heterologous expression of Wheat CYP71C6v1 was responsible for the metabolism of herbicides, including chlorsulfuron, triasulfuron, metsulfuron-methyl, bensulfuron-methyl, and tribenuron-methyl [160]. The gene CYP81A9 in maize has been identified as responsible for the metabolism of the acetolactate synthase (ALS) inhibitor herbicide nicosulfuron [161]. Maize has also demonstrated CYP-mediated metabolism of herbicides such as tembotrione, mesotrione, dicamba (synthetic auxin), diflufenzopyr (auxin transport inhibitor), and carfentrazone (protoporphyrinogen oxidase inhibitor) through the 4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitors [75,162,163,164].
8. Future Research
CYPs are multifunctional enzymes that contribute significantly to plant stress response, growth, and development processes. Through biosynthesis and regulation, CYPs enable plants to produce and control the levels of hormones, fatty acids, sterols, cell wall components, biopolymers, and other defense chemicals. Many CYPs have been identified, but their specific functions and contribution to the physiological process are still unknown. Understanding the roles of individual CYP is necessary to elucidate their particular biological processes. Future studies can use cutting-edge genomics, transcriptomics, proteomics, metabolomics, and bioinformatics technologies to study CYP family genes. Through genomics and transcriptomics, researchers can identify stress-responsive genes involved in stress response mechanisms and analyze the expression patterns of target CYP genes under diverse environmental conditions and developmental stages. Omics data can provide resources of molecular markers associated with desirable traits. These markers can be used for marker-assisted selection, allowing breeders to identify and select plants with specific traits more efficiently. GWAS has been extensively used to study CYPs in many plant species. GWAS at the epigenome, transcriptome, protein, and metabolic levels for crop breeding might help distinguish the beneficial and detrimental alleles. Metabolomics can provide insights into the metabolic pathways and biochemical processes associated with stress tolerance and productivity. Research on the protein–protein interactions of CYPs with additional enzymes, regulatory elements, and protein complexes would reveal important information about their molecular mechanisms and possible interacting partners in metabolic pathways. To find interacting partners and investigate the dynamics of CYP-mediated protein complexes, methods including yeast two-hybrid experiments, co-immunoprecipitation, and protein–protein interaction networks can be used. Alphafold is a powerful computational tool that uses deep learning algorithms to predict protein structures with remarkable accuracy. Alphafold multimer could be used to predict protein–protein interaction, which is efficient, cost-effective, and less time-consuming [165]. Despite getting the interaction through the alphafold, the result needs functional validation. Investigating the regulatory processes that control the CYP genes’ expression and function is another important area of research. A comprehensive understanding of the regulatory networks governing the expression of CYP would result from identifying the transcription factors, cis-regulatory elements, microRNAs, and signaling pathways involved. The integrated omics approach has tremendous potential in crop breeding to bridge the gap between mitigating plant stresses and achieving food security through increased production. Integrating data from different omics layers allows for a systems biology approach, enabling researchers to build comprehensive models of crop systems. These models can simulate plant responses to different stresses, predict crop performance under various conditions, and optimize breeding strategies accordingly. In recent years, several CYPs have been investigated as potential candidate genes for biotechnological uses. Despite plant CYP characterization and genetic engineering improvements, there is still a significant gap between current research and the technology’s commercialization. By incorporating the resistant trait in the cultivar, crop productivity could be increased and used for commercial purposes. We can improve our knowledge of CYPs in plants, elucidate their functional roles in various biological processes, and open the door for developing crop varieties with enhanced growth, productivity, and resistance to biotic and abiotic stresses.
Author Contributions
Conceptualization, S.C. and P.C.; writing—original draft preparation, S.C., P.C., A.B., S.D. and T.B.; writing—review and editing, S.C., P.C., A.B., S.D. and T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mizutani, M.; Sato, F. Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 2011, 507, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Chapple, C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Biol. 1998, 49, 311–343. [Google Scholar] [CrossRef]
- Schuler, M.A.; Duan, H.; Bilgin, M.; Ali, S. Arabidopsis cytochrome P450s through the looking glass: A window on plant biochemistry. Phytochem. Rev. 2006, 5, 205–237. [Google Scholar] [CrossRef]
- Jiu, S.; Xu, Y.; Wang, J.; Wang, L.; Liu, X.; Sun, W.; Sabir, I.A.; Ma, C.; Xu, W.; Wang, S.; et al. The cytochrome P450 monooxygenase inventory of grapevine (Vitis vinifera L.): Genome-wide identification, evolutionary characterization and expression analysis. Front. Genet. 2020, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Guttikonda, S.K.; Trupti, J.; Bisht, N.C.; Chen, H.; An, Y.Q.C.; Pandey, S.; Xu, D.; Yu, O. Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 2010, 10, 243. [Google Scholar] [CrossRef]
- Nelson, D.R.; Ming, R.; Alam, M.; Schuler, M.A. Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 2008, 1, 216–235. [Google Scholar] [CrossRef]
- Nelson, D.R.; Schuler, M.A.; Paquette, S.M.; Werck-Reichhart, D.; Bak, S. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 2004, 135, 756–772. [Google Scholar] [CrossRef]
- Schuler, M.A. P450s in Plants, Insects, and Their Fungal Pathogens. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano, P.R., Ed.; Springer: New York, NY, USA, 2015; Volume 1, pp. 409–449. [Google Scholar]
- Rojo, E.; Solano, R.; Sánchez-Serrano, J.J. Interactions between signaling compounds involved in plant defense. J. Plant Growth Regul. 2003, 22, 82–98. [Google Scholar] [CrossRef]
- Walling, L.L. The myriad plant responses to herbivores. J. Plant Growth Regul. 2000, 19, 195–216. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59. [Google Scholar] [CrossRef]
- Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S.; Werck-Reichhart, D. Cytochromes P450. TAB 2011, 9, e0144. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Sauveplane, V.; Olry, A.; Ginglinger, J.F.; Provart, N.J.; Werck-Reichhart, D. An extensive (co-) expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 47. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Frear, D.; Swanson, H.; Tanaka, F. N-demethylation of substituted 3-(phenyl)-1-methylureas: Isolation and characterization of a microsomal mixed function oxidase from cotton. Phytochemistry 1969, 8, 2157–2169. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes: II. Solubilization, purification, and properties. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Durst, F.; Nelson, D.R. Diversity and evolution of plant P450 and P450-reductases. Drug Metabol. Drug Interact. 1995, 12, 189–206. [Google Scholar] [CrossRef]
- Paquette, S.M.; Bak, S.; Feyereisen, R. Intron–exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000, 19, 307–317. [Google Scholar] [CrossRef]
- Lee, D.S.; Nioche, P.; Hamberg, M.; Raman, C. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 2008, 455, 363–368. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Q.; Liu, Y.; Liao, X.; Chu, H.; Chang, H.; Cao, Y.; Li, Z.; Zhang, T.; Cheng, J.; et al. PCPD: Plant cytochrome P450 database and web-based tools for structural construction and ligand docking. Synth. Syst. Biotechnol. 2021, 6, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 2006, 5, 193–204. [Google Scholar] [CrossRef]
- Rensing, S.A. How plants conquered land. Cell 2020, 181, 964–966. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Wang, H.; Sahu, S.K.; Marin, B.; Li, H.; Xu, Y.; Liang, H.; Li, Z.; Cheng, S.; et al. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat. Ecol. Evol. 2020, 4, 1220–1231. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- Wang, S.; Li, L.; Li, H.; Sahu, S.K.; Wang, H.; Xu, Y.; Xian, W.; Song, B.; Liang, H.; Cheng, S.; et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2020, 6, 95–106. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Mizutani, M. Impacts of diversification of cytochrome P450 on plant metabolism. Biol. Pharm. Bull. 2012, 35, 824–832. [Google Scholar] [CrossRef]
- Kim, J.E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry 2010, 71, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Feyereisen, R. The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001, 127, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Stumpe, M.; Kandzia, R.; Göbel, C.; Rosahl, S.; Feussner, I. A pathogen-inducible divinyl ether synthase (CYP74D) from elicitor-treated potato suspension cells. FEBS Lett. 2001, 507, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef]
- Serra, O.; Soler, M.; Hohn, C.; Sauveplane, V.; Pinot, F.; Franke, R.; Schreiber, L.; Prat, S.; Molinas, M.; Figueras, M. CYP86A33-targeted gene silencing in potato tuber alters suberin composition, distorts suberin lamellae, and impairs the periderm’s water barrier function. Plant Physiol. 2009, 149, 1050–1060. [Google Scholar] [CrossRef]
- Xiao, F.; Mark Goodwin, S.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef]
- Xia, Y.; Yang, J.; Ma, L.; Yan, S.; Pang, Y. Genome-wide identification and analyses of drought/salt-responsive cytochrome P450 genes in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 9957. [Google Scholar] [CrossRef]
- Bate, N.J.; Sivasankar, S.; Moxon, C.; Riley, J.M.; Thompson, J.E.; Rothstein, S.J. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol. 1998, 117, 1393–1400. [Google Scholar] [CrossRef]
- Kawa, D.; Meyer, A.J.; Dekker, H.L.; Abd-El-Haliem, A.M.; Gevaert, K.; Van De Slijke, E.; Maszkowska, J.; Bucholc, M.; Dobrowolska, G.; De Jaeger, G.; et al. SnRK2 Protein Kinases and mRNA Decapping Machinery Control Root Development and Response to Salt. Plant Physiol. 2020, 182, 361–377. [Google Scholar] [CrossRef]
- Butrón, A.; Chen, Y.; Rottinghaus, G.; McMullen, M. Genetic variation at bx1 controls DIMBOA content in maize. Theor. Appl. Genet. 2010, 120, 721–734. [Google Scholar] [CrossRef]
- Dick, R.; Rattei, T.; Haslbeck, M.; Schwab, W.; Gierl, A.; Frey, M. Comparative analysis of benzoxazinoid biosynthesis in monocots and dicots: Independent recruitment of stabilization and activation functions. Plant Cell 2012, 24, 915–928. [Google Scholar] [CrossRef]
- Tian, L.; Musetti, V.; Kim, J.; Magallanes-Lundback, M.; DellaPenna, D. The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 2004, 101, 402–407. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Chandler, P.M.; Poole, A.; Dennis, E.S.; Peacock, W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 2065–2070. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Badieyan, S.; Bevan, D.R.; Herde, M.; Gatz, C.; Tholl, D. Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 21205–21210. [Google Scholar] [CrossRef] [PubMed]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S. Triterpene structural diversification by plant cytochrome P450 enzymes. Front. Plant Sci. 2017, 8, 1886. [Google Scholar] [CrossRef]
- Umemoto, N.; Nakayasu, M.; Ohyama, K.; Yotsu-Yamashita, M.; Mizutani, M.; Seki, H.; Saito, K.; Muranaka, T. Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol. 2016, 171, 2458–2467. [Google Scholar] [CrossRef]
- Guo, P.; Bai, G.; Carver, B.; Li, R.; Bernardo, A.; Baum, M. Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum tolerance under aluminum stress. Mol. Genet. Genom. 2007, 277, 1–12. [Google Scholar] [CrossRef]
- Nomura, T.; Ishihara, A.; Imaishi, H.; Endo, T.; Ohkawa, H.; Iwamura, H. 2002. Molecular characterization and chromosomal localization of cytochrome P450 genes involved in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Mol. Genet. Genom. 2002, 267, 210–217. [Google Scholar] [CrossRef]
- Frank, M.R.; Deyneka, J.M.; Schuler, M.A. 1996. Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol. 1996, 110, 1035–1046. [Google Scholar] [CrossRef]
- Seitz, C.; Eder, C.; Deiml, B.; Kellner, S.; Martens, S.; Forkmann, G. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol. 2006, 61, 365–381. [Google Scholar] [CrossRef]
- Kim, Y.C.; Kim, S.Y.; Paek, K.H.; Choi, D.; Park, J.M. Suppression of CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem. Biophys. Res. Commun. 2006, 345, 638–645. [Google Scholar] [CrossRef]
- Geisler, K.; Hughes, R.K.; Sainsbury, F.; Lomonossoff, G.P.; Rejzek, M.; Fairhurst, S.; Olsen, C.E.; Motawia, M.S.; Melton, R.E.; Hemmings, A.M.; et al. Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc. Natl. Acad. Sci. USA 2013, 110, E3360–E3367. [Google Scholar] [CrossRef]
- Bak, S.; Olsen, C.E.; Halkier, B.A.; Møller, B.L. Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol. 2000, 123, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Kahn, R.A.; Nielsen, H.L.; Møller, B.L.; Halkier, B.A. Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 1998, 36, 393–405. [Google Scholar] [PubMed]
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 2000, 24, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Berim, A.; Gang, D.R. The roles of a flavone-6-hydroxylase and 7-O-demethylation in the flavone biosynthetic network of sweet basil. J. Biol. Chem. 2013, 288, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macrì, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef]
- Luo, P.; Wang, Y.H.; Wang, G.D.; Essenberg, M.; Chen, X.Y. Molecular cloning and functional identification of (+)-δ-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 2001, 28, 95–104. [Google Scholar] [CrossRef]
- Li, J.; Deng, C.; Liu, M.; He, Y. Computational identification and systematic classification of cytochrome P450 genes in Pogostemon cablin provide insights into flavonoids biosynthesis. Acta Physiol. Plant. 2023, 45, 46. [Google Scholar] [CrossRef]
- Liu, X.; Gong, Q.; Zhao, C.; Wang, D.; Ye, X.; Zheng, G.; Wang, Y.; Cao, J.; Sun, C. Genome-wide analysis of cytochrome P450 genes in Citrus clementina and characterization of a CYP gene encoding flavonoid 3′-hydroxylase. Hortic. Res. 2023, 10, uhac283. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, H.; Ma, R.; Chang, Y.; Qin, X.; Xu, J.; Fu, Y. Genome-wide transcriptome analysis and characterization of the cytochrome P450 flavonoid biosynthesis genes in pigeon pea (Cajanus cajan). Planta 2022, 255, 120. [Google Scholar] [CrossRef]
- Xia, Y.; He, C.; Yan, S.; Liu, J.; Huang, H.; Li, X.; Su, Q.; Jiang, W.; Pang, Y. New dual functional CYP450 gene involves in isoflavone biosynthesis in Glycine max L. Synth. Syst. Biotechnol. 2023, 8, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Huang, Y.; Tang, Y. Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl. Microbiol. Biotechnol. 2010, 86, 1293–1312. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef]
- Toporkova, Y.Y.; Smirnova, E.O.; Mukhtarova, L.S.; Gorina, S.S.; Grechkin, A.N. Catalysis by allene oxide synthases (CYP74A and CYP74C): Alterations by the Phe/Leu mutation at the SRS-1 region. Phytochemistry 2020, 169, 112152. [Google Scholar] [CrossRef]
- Schuler, M.A. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411–1419. [Google Scholar] [CrossRef]
- Sandermann, H., Jr. Higher plant metabolism of xenobiotics: The ‘green liver’ concept. Pharmacogenetics 1994, 4, 225–241. [Google Scholar] [CrossRef]
- Werck-Reichhart, D. Herbicide metabolism and selectivity: Role of cytochrome P450. In Proceedings of the Brighton Crop Protection Conference Weeds, Brighton, UK, 20–23 November 1995; pp. 813–822. [Google Scholar]
- Riechers, D.E.; Vaughn, K.C.; Molin, W.T. The Role of Plant Glutathione S-transferases in Herbicide Metabolism. In Environmental Fate and Safety Management of Agrochemicals; Clark, M.J., Ohkawa, H., Eds.; Proceedings of the ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2005; pp. 216–232. [Google Scholar]
- Barrett, M.; Polge, N.; Baerg, R.; Bradshaw, R.; Poneleit, C.; Hatzios, K. Role of Cytochrome P450 in Herbicide Metabolism and Selectivity and Multiple Herbicide Metabolizing Cytochrome P450 Activities in Maize. In Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants; Hatzios, K.K., Ed.; Kluwer Academic: Dordrecht, The Netherlands, 1997; pp. 35–50. [Google Scholar]
- Persans, M.W.; Wang, J.; Schuler, M.A. Characterization of maize cytochrome P450 monooxygenases induced in response to safeners and bacterial pathogens. Plant Physiol. 2001, 125, 1126–1138. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, X.; Liu, K.; Zhang, J.; Wu, X.; Zhu, J.; Tu, J. Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol. Biol. 2006, 61, 933–943. [Google Scholar] [CrossRef]
- Nordby, J.N.; Williams, M.M.; Pataky, J.K.; Riechers, D.E.; Lutz, J.D. A common genetic basis in sweet corn inbred Cr1 for cross sensitivity to multiple cytochrome P450-metabolized herbicides. Weed Sci. 2008, 56, 376–382. [Google Scholar] [CrossRef]
- Pataky, J.K.; Meyer, M.D.; Bollman, J.D.; Boerboom, C.M.; Williams, M.M. Genetic basis for varied levels of injury to sweet corn hybrids from three cytochrome P450-metabolized herbicides. J. Am. Soc. Hortic. Sci. 2008, 133, 438–447. [Google Scholar] [CrossRef]
- Duhoux, A.; Delye, C. Reference genes to study herbicide stress response in Lolium sp.: Up-regulation of P450 genes in plants resistant to acetolactate-synthase inhibitors. PLoS ONE 2013, 8, e63576. [Google Scholar] [CrossRef]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef]
- Siminszky, B. Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 2006, 5, 445–458. [Google Scholar] [CrossRef]
- Jang, J.; Khanom, S.; Moon, Y.; Shin, S.; Lee, O.R. PgCYP76B93 docks on phenylurea herbicides and its expression enhances chlorotoluron tolerance in Arabidopsis. Appl. Biol. Chem. 2020, 63, 14. [Google Scholar] [CrossRef]
- Jun, X.; Wang, X.Y.; Gou, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B.; Yan, D.; Dong, W.; Yang, W.; Li, Q.; Zeng, L.; Wang, J.; Wang, L.; Hicks, L.M.; et al. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011, 67, 342–353. [Google Scholar] [CrossRef]
- Millar, A.A.; Jacobsen, J.V.; Ross, J.J.; Helliwell, C.A.; Poole, A.T.; Scofield, G.; Reid, J.B.; Gubler, F. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006, 45, 942–954. [Google Scholar] [CrossRef]
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef]
- Wang, Q.; Hillwig, M.L.; Wu, Y.; Peters, R.J. CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 2012, 158, 1418–1425. [Google Scholar] [CrossRef]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952. [Google Scholar] [CrossRef]
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef]
- Enoki, S.; Tanaka, K.; Moriyama, A.; Hanya, N.; Mikami, N.; Suzuki, S. Grape cytochrome P450 CYP90D1 regulates brassinosteroid biosynthesis and increases vegetative growth. Plant Physiol. Biochem. 2023, 196, 993–1001. [Google Scholar] [CrossRef]
- Godiard, L.; Sauviac, L.; Dalbin, N.; Liaubet, L.; Callard, D.; Czernic, P.; Marco, Y. CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett. 1998, 438, 245–249. [Google Scholar] [CrossRef]
- Höfer, R.; Boachon, B.; Renault, H.; Gavira, C.; Miesch, L.; Iglesias, J.; Ginglinger, J.F.; Allouche, L.; Miesch, M.; Grec, S.; et al. Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 2014, 166, 1149–1161. [Google Scholar] [CrossRef]
- Feldmann, K.A. Cytochrome P450s as genes for crop improvement. Curr. Opin. Plant Biol. 2001, 4, 162–167. [Google Scholar] [CrossRef]
- Singpho, N.L.; Sharma, J.G. Importance of Cytochrome P450 gene family from metabolite biosynthesis to stress tolerance: A review. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Jakarta, Indonesia, 2021; Volume 775, p. 012012. [Google Scholar]
- Shilpashree, H.; Sudharshan, S.; Shasany, A.K.; Nagegowda, D.A. Molecular characterization of three CYP450 genes reveals their role in withanolides formation and defense in Withania somnifera, the Indian Ginseng. Sci. Rep. 2022, 12, 1602. [Google Scholar] [CrossRef]
- George, H.L.; VanEtten, H.D. Characterization of pisatin-inducible cytochrome P450s in fungal pathogens of pea that detoxify the pea phytoalexin pisatin. Fungal Genet. Biol. 2001, 33, 37–48. [Google Scholar] [CrossRef]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems—Biological variations of electron transport chains. Biochim. Biophys. Acta-Gen. Subj. 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Minerdi, D.; Sadeghi, S.J.; Pautasso, L.; Morra, S.; Aigotti, R.; Medana, C.; Gilardi, G.; Gullino, M.L.; Gilardi, G. Expression and role of CYP505A1 in pathogenicity of Fusarium oxysporum f. sp. lactucae. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140268. [Google Scholar] [CrossRef]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef]
- Qi, X.; Bakht, S.; Qin, B.; Leggett, M.; Hemmings, A.; Mellon, F.; Eagles, J.; Werck-Reichhart, D.; Schaller, H.; Lesot, A.; et al. A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 2006, 103, 18848–18853. [Google Scholar] [CrossRef]
- Glauser, G.; Marti, G.; Villard, N.; Doyen, G.A.; Wolfender, J.L.; Turlings, T.C.; Erb, M. Induction and detoxification of maize 1, 4-benzoxazin-3-ones by insect herbivores. Plant J. 2011, 68, 901–911. [Google Scholar] [CrossRef]
- Irmisch, S.; Zeltner, P.; Handrick, V.; Gershenzon, J.; Köllner, T.G. The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol. 2015, 15, 128. [Google Scholar] [CrossRef]
- Liu, Q.; Khakimov, B.; Cárdenas, P.D.; Cozzi, F.; Olsen, C.E.; Jensen, K.R.; Hauser, T.P.; Bak, S. The cytochrome P450 CYP72A552 is key to production of hederagenin-based saponins that mediate plant defense against herbivores. New Phytol. 2019, 222, 1599–1609. [Google Scholar] [CrossRef]
- Rustgi, S.; Springer, A.; Kang, C.; Von Wettstein, D.; Reinbothe, C.; Reinbothe, S.; Pollmann, S. Allene oxide synthase and hydroperoxide lyase, two non-canonical cytochrome P450s in Arabidopsis thaliana and their different roles in plant defense. Int. J. Mol. Sci. 2019, 20, 3064. [Google Scholar] [CrossRef]
- Prince, D.C.; Drurey, C.; Zipfel, C.; Hogenhout, S.A. The leucine-rich repeat receptor-like kinase Brassinosteroid Insensitive1-Associated Kinase1 and the cytochrome P450 Phytoalexin Deficient3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 2014, 164, 2207–2219. [Google Scholar] [CrossRef]
- Wang, E.; Wang, R.; DeParasis, J.; Loughrin, J.H.; Gan, S.; Wagner, G.J. Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat. Biotechnol. 2001, 19, 371–374. [Google Scholar] [CrossRef]
- Paquette, S.M.; Jensen, K.; Bak, S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www.P450.kvl.dk). Phytochemistry 2009, 70, 1940–1947. [Google Scholar] [CrossRef]
- Mafu, S.; Ding, Y.; Murphy, K.M.; Yaacoobi, O.; Addison, J.B.; Wang, Q.; Shen, Z.; Briggs, S.P.; Bohlmann, J.; Castro-Falcon, G.; et al. Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in Maize. Plant Physiol. 2018, 176, 2677–2690. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef]
- Pinot, F.; Beisson, F. Cytochrome P450 metabolizing fatty acids in plants: Characterization and physiological roles. FEBS J. 2011, 278, 195–205. [Google Scholar] [CrossRef]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H.; et al. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef]
- Wang, P.; Ma, L.; Li, Y.; Wang, S.; Li, L.; Yang, R. Transcriptome analysis reveals sunflower cytochrome P450 CYP93A1 responses to high salinity treatment at the seedling stage. Genes Genom. 2017, 39, 581–591. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, Z.; Fu, X.; Liang, C. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 2002, 109, 849–860. [Google Scholar] [CrossRef]
- Trueman, C.L.; Loewen, S.A.; Goodwin, P.H. Can the inclusion of uniconazole improve the effectiveness of acibenzolar-S-methyl in managing bacterial speck (Pseudomonas syringae pv. tomato) and bacterial spot (Xanthomonas gardneri) in tomato? Eur. J. Plant Pathol. 2019, 155, 927–942. [Google Scholar] [CrossRef]
- Tao, X.; Wang, M.X.; Dai, Y.; Wang, Y.; Fan, Y.F.; Mao, P.; Ma, X.R. Identification and expression profile of CYPome in perennial Ryegrass and Tall Fescue in response to temperature stress. Front. Plant Sci. 2017, 8, 1519. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, P.; Udvardi, M.; Peacock, W.; Dennis, E. A prolonged cold treatment-induced cytochrome P450 gene from Arabidopsis thaliana. Plant Cell Environ. 1999, 22, 791–800. [Google Scholar] [CrossRef]
- Chan, Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 2012, 100, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Tian-lai, L.; Miao, L.; Zhou-ping, S. Regulation effect of calcium and salicylic acid on defense enzyme activities in tomato leaves under sub-high temperature stress. Yingyong Shengtai Xuebao 2009, 20, 586–590. [Google Scholar]
- Baron, K.N.; Schroeder, D.F.; Stasolla, C. Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci. 2012, 188, 48–59. [Google Scholar] [CrossRef]
- Robineau, T.; Batard, Y.; Nedelkina, S.; Cabello-Hurtado, F.; LeRet, M.; Sorokine, O.; Didierjean, L.; Werck-Reichhart, D. The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol. 1998, 118, 1049–1056. [Google Scholar] [CrossRef]
- Wedow, J.M.; Yendrek, C.R.; Mello, T.R.; Creste, S.; Martinez, C.A.; Ainsworth, E.A. Metabolite and transcript profiling of Guinea grass (Panicum maximum Jacq) response to elevated [CO2] and temperature. Metabolomics 2019, 15, 51. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.H.; Foster, K.J.; et al. Energy costs of salt tolerance in crop plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, J.; Qin, L.; Shi, W.; Xia, G.; Liu, S. TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol. J. 2020, 18, 791–804. [Google Scholar] [CrossRef]
- Chaurasia, S.; Singh, A.K.; Kumar, A.; Songachan, L.; Yadav, M.C.; Kumar, S.; Kumari, J.; Bansal, R.; Sharma, P.C.; Singh, K. Genome-wide association mapping reveals key genomic regions for physiological and yield-related traits under salinity stress in wheat (Triticum aestivum L.). Genomics 2021, 113, 3198–3215. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Xie, Z.H. Role of abscisic acid in strigolactone-induced salt stress tolerance in arbuscular mycorrhizal Sesbania cannabina seedlings. BMC Plant Biol. 2018, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zheng, D.; Feng, N.; Zhou, H.; Mu, D.; Zhao, L.; Shen, X.; Rao, G.; Meng, F.; Huang, A. Physiological mechanisms of ABA-induced salinity tolerance in leaves and roots of rice. Sci. Rep. 2022, 12, 8228. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 169. [Google Scholar] [CrossRef]
- Balusamy, S.R.; Rahimi, S.; Yang, D.C. Characterization of squalene-induced PgCYP736B involved in salt tolerance by modulating key genes of abscisic acid biosynthesis. Int. J. Biol. Macromol. 2019, 121, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Peng, M.; Luo, Q.; Jiang, M.; Zhang, X.; Zong, X.; Meng, F.; Li, Y. Isolation and detection of transcript-derived fragments (TDFs) in NaCl-stressed black locust (Robinia pseudoacacia L.) using cDNA-AFLP analysis. Acta Physiol. Plant. 2015, 37, 168. [Google Scholar] [CrossRef]
- Wang, X.; Yang, P.; Gao, Q.; Liu, X.; Kuang, T.; Shen, S.; He, Y. Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 2008, 228, 167–177. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Hou, Y.; Xu, Y.; Peng, R.; et al. Knockdown of cytochrome P450 genes Gh_D07G1197 and Gh_A13G2057 on chromosomes D07 and A13 reveals their putative role in enhancing drought and salt stress tolerance in Gossypium hirsutum. Genes 2019, 10, 226. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Wang, H.; Ran, X.; Li, B.; Zhang, J.; Zhang, H. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 2016, 14, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Vishal, B.; Bhal, A.; Kumar, P.P. WRKY9 transcription factor regulates cytochrome P450 genes CYP94B3 and CYP86B1, leading to increased root suberin and salt tolerance in Arabidopsis. Physiol. Plant. 2021, 172, 1673–1687. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Vishal, B.; Ho, W.J.; Lok, F.C.J.; Lee, F.S.M.; Kumar, P.P. Regulation of a cytochrome P450 gene CYP94B1 by WRKY33 transcription factor controls apoplastic barrier formation in roots to confer salt tolerance. Plant Physiol. 2020, 184, 2199–2215. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Huang, Y.; Xian, W.; Wang, J.; Liao, H. Identification and expression analysis of the Glycine max CYP707A gene family in response to drought and salt stresses. Ann. Bot. 2012, 110, 743–756. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Wurtzel, E.T. From epoxycarotenoids to ABA: The role of ABA 8′-hydroxylases in drought-stressed maize roots. Arch. Biochem. Biophys. 2010, 504, 112–117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Lv, Y.; Wan, X.R.; Li, L.M.; Hu, B.; Li, L. Cloning and expression analysis of cDNAs encoding ABA 8′-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE 2014, 9, e97025. [Google Scholar] [CrossRef]
- Chen, J.; Song, Y.; Zhang, H.; Zhang, D. Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol. Biol. Rep. 2013, 31, 946–962. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.C.; Xu, Y.Q.; Li, G.J.; Liao, Y.; Fu, F.L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef]
- Lü, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Xu, Q. CsCYT75B1, a Citrus CYTOCHROME P450 gene, is involved in accumulation of antioxidant flavonoids and induces drought tolerance in transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef]
- Sotelo-Silveira, M.; Cucinotta, M.; Chauvin, A.L.; Chavez Montes, R.A.; Colombo, L.; Marsch-Martínez, N.; de Folter, S. Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiol. 2013, 162, 779–799. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Northey, J.G.; Liang, S.; Jamshed, M.; Deb, S.; Foo, E.; Reid, J.B.; McCourt, P.; Samuel, M.A. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2016, 2, 16114. [Google Scholar] [CrossRef]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.; Song, W. Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signal. Behav. 2018, 13, e1460048. [Google Scholar] [CrossRef]
- Tabrez, S.; Ahmad, M. Cytochrome P450 system as potential biomarkers of certain toxicants: Comparison between plant and animal models. Environ. Monit. Assess. 2013, 185, 2977–2987. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Zhou, Y.; Gong, T.; Wang, J.; Ge, Y. Enhanced phytoremediation of mixed heavy metal (mercury)-organic pollutants (trichloroethylene) with transgenic alfalfa co-expressing glutathione S-transferase and human P450 2E1. J. Hazard. Mater. 2013, 260, 1100–1107. [Google Scholar] [CrossRef]
- Balusamy, S.R.D.; Kim, Y.J.; Rahimi, S.; Senthil, K.S.; Lee, O.R.; Lee, S.; Yang, D.C. Transcript pattern of cytochrome P450, antioxidant and ginsenoside biosynthetic pathway genes under heavy metal stress in Panax ginseng Meyer. Bull. Environ. Contam. Toxicol. 2013, 90, 194–202. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-Target-Site Resistance to Herbicides: Recent Developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef]
- Khanom, S.; Jang, J.; Lee, O.R. Overexpression of ginseng cytochrome P450 CYP736A12 alters plant growth and confers phenylurea herbicide tolerance in Arabidopsis. J. Ginseng Res. 2019, 43, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Hurtado, F.; Batard, Y.; Salaun, J.P.; Durst, F.; Pinot, F.; Werck-Reichhart, D. Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J. Biol. Chem. 1998, 273, 7260–7267. [Google Scholar] [CrossRef]
- Siminszky, B.; Corbin, F.T.; Ward, E.R.; Fleischmann, T.J.; Dewey, R.E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc. Natl. Acad. Sci. USA 1999, 96, 1750–1755. [Google Scholar] [CrossRef]
- Didierjean, L.; Gondet, L.; Perkins, R.; Lau, S.M.; Schaller, H.; O’Keefe, D.; Werck-Reichhart, D. Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke. Plant Physiol. 2002, 130, 179–189. [Google Scholar] [CrossRef]
- Xiang, W.; Wang, X.; Ren, T. Expression of a wheat cytochrome P450 monooxygenase cDNA in yeast catalyzes the metabolism of sulfonylurea herbicides. Pestic. Biochem. Physiol. 2006, 85, 1–6. [Google Scholar] [CrossRef]
- Liu, X.; Bi, B.; Xu, X.; Li, B.; Tian, S.; Wang, J.; Zhang, H.; Wang, G.; Han, Y.; McElroy, J.S. Rapid identification of a candidate nicosulfuron sensitivity gene (Nss) in maize (Zea mays L.) via combining bulked segregant analysis and RNA-seq. Theor. Appl. Genet. 2019, 132, 1351–1361. [Google Scholar] [CrossRef]
- Hamprecht, G.; Witschel, M.; Hawkes, T.R.; Edmunds, A.J.; Morris, J.A.; van Almsick, A. Herbicides with Bleaching Properties. In Modern Crop Protection Compounds; Krämer, W., Schirmer, U., Jeschke, P., Witschel, M., Eds.; ACS Symposium Series; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2011; Volume 58, pp. 197–276. [Google Scholar]
- Paporisch, A.; Rubin, B. Isoxadifen safening mechanism in sweet corn genotypes with differential response to P450-metabolized herbicides. Pestic. Biochem. Physiol. 2017, 138, 22–28. [Google Scholar] [CrossRef]
- Williams, M.M.; Pataky, J.K. Factors affecting differential sensitivity of sweet corn to HPPD-inhibiting herbicides. Weed Sci. 2010, 58, 289–294. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Zhu, W.; Shenoy, A.; Kundrotas, P.; Elofsson, A. Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search. Nat. Commun. 2022, 13, 6028. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).