Clinical Outcome and Molecular Profile in Patients with DDX41 Mutation Hot-Spots
Abstract
1. Introduction
2. Materials and Methods
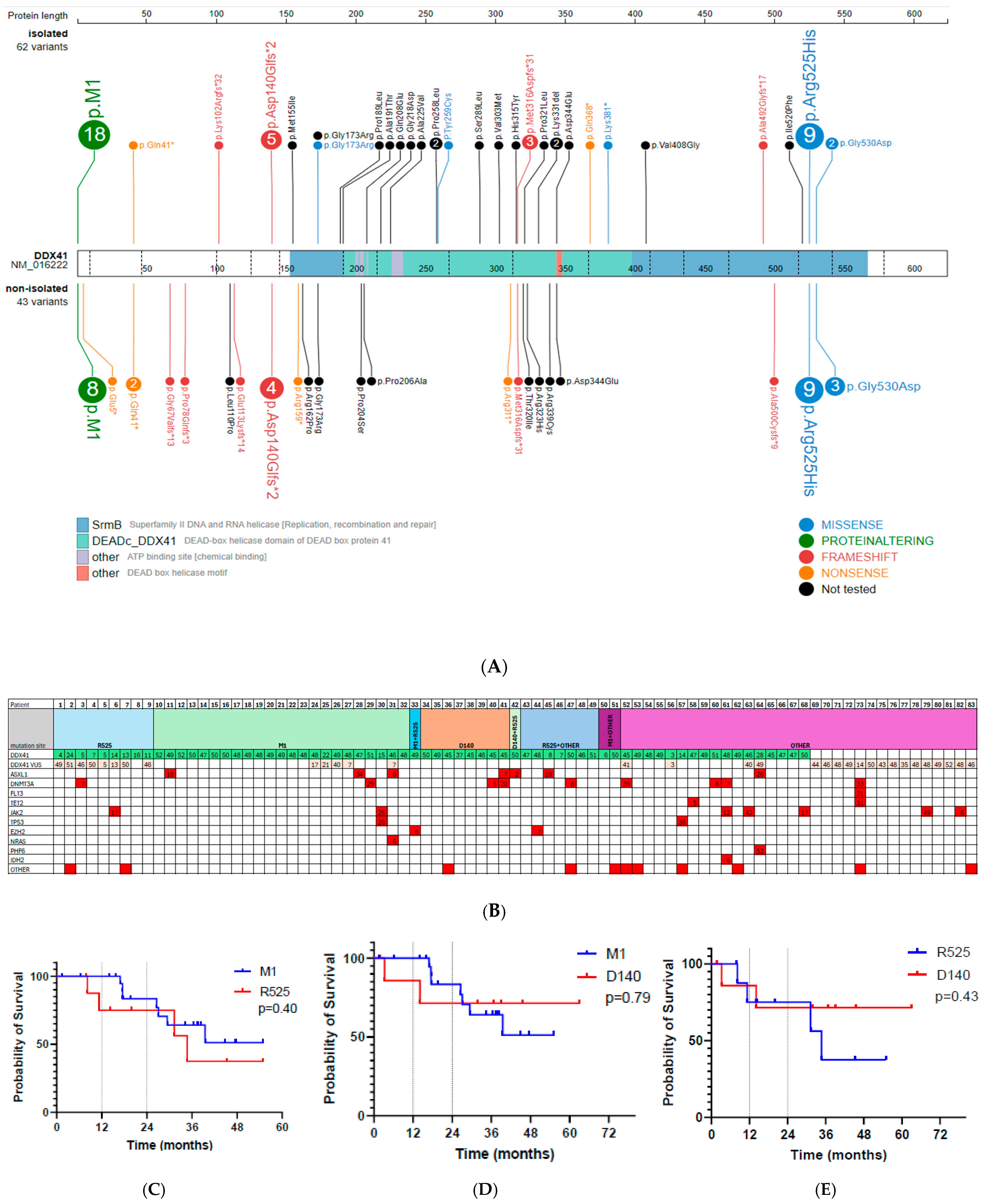
3. Results
4. Discussion
Study Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NGS | Next Generation Sequencing |
| BMT | Bone marrow transplant |
| CHIP | Clonal hematopoiesis of indeterminate potential |
| OS | Overall survival |
| MDS | Myelodysplastic syndrome |
| AML | Acute Myeloid leukemia |
| MPN | Myeloproliferative neoplasm |
| CCUS | Clonal cytopenia of undetermined significance |
References
- Winstone, L.; Jung, Y.; Wu, Y. DDX41: Exploring the roles of a versatile helicase. Biochem. Soc. Trans. 2024, 52, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y. Molecular pathophysiology of germline mutations in acute myeloid leukemia. Int. J. Hematol. 2024, 120, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Bataller, A.; Loghavi, S.; Gerstein, Y.; Bazinet, A.; Sasaki, K.; Chien, K.S.; Hammond, D.; Montalban-Bravo, G.; Borthakur, G.; Short, N.; et al. Characteristics and clinical outcomes of patients with myeloid malignancies and DDX41 variants. Am. J. Hematol. 2023, 98, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ong, F.; Sasaki, K. Current Understanding of DDX41 Mutations in Myeloid Neoplasms. Cancers 2023, 15, 344. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, H.B.; Nanaa, A.; Viswanatha, D.S.; Foran, J.M.; Badar, T.; Sproat, L.; He, R.; Nguyen, P.; Jevremovic, D.; Salama, M.E.; et al. Genetic features and clinical outcomes of patients with isolated and comutated DDX41-mutated myeloid neoplasms. Blood Adv. 2022, 6, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Duployez, N.; Largeaud, L.; Duchmann, M.; Kim, R.; Rieunier, J.; Lambert, J.; Bidet, A.; Larcher, L.; Lemoine, J.; Delhommeau, F.; et al. Prognostic impact of DDX41 germline mutations in intensively treated acute myeloid leukemia patients: An ALFA-FILO study. Blood 2022, 140, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.E.; Routbort, M.J.; DiNardo, C.D.; Bueso-Ramos, C.E.; Kanagal-Shamanna, R.; Khoury, J.D.; Thakral, B.; Zuo, Z.; Yin, C.C.; Loghavi, S.; et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am. J. Hematol. 2019, 94, 757–766. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Qin, T.; Xu, Z.; Qu, S.; Pan, L.; Li, B.; Wang, H.; Zhang, P.; Yan, X.; et al. Comparison of the revised 4th (2016) and 5th (2022) editions of the World Health Organization classification of myelodysplastic neoplasms. Leukemia 2022, 36, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Makishima, H.; Saiki, R.; Nannya, Y.; Korotev, S.C.; Gurnari, C.; Takeda, J.; Momozawa, Y.; Best, S.; Krishnamurthy, P.; Yoshizato, T.; et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasms. Blood 2023, 141, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Badar, T.; Nanaa, A.; Foran, J.M.; Viswanatha, D.; Al-Kali, A.; Lasho, T.; Finke, C.; Alkhateeb, H.B.; He, R.; Gangat, N.; et al. Clinical and molecular correlates of somatic and germline DDX41 variants in patients and families with myeloid neoplasms. Haematologica 2023, 108, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Al-Kali, A.; Nanaa, A.; Viswanatha, D.; He, R.; Nguyen, P.; Jevremovic, D.; Foran, J.M.; Yi, C.A.; Greipp, P.T.; Gangat, N.; et al. Observation and treatment in DDX41-mutated acute myeloid leukemia and myelodysplastic syndrome. Blood Cancer J. 2023, 13, 49. [Google Scholar] [CrossRef] [PubMed]
| Variable | Patients Identified to Have DDX41 Mutation (N = 68) |
|---|---|
| Median age, yrs (range) | 72 (30–93) |
| Male sex, N (%) | 48 (70%) |
| Diagnosis | |
| MDS, N (%) | 30 (44%) |
| AML, N (%) | 23 (34%) |
| MPN, N (%) | 4 (6%) |
| DDX41 carrier, N(%) | 6 (8%) |
| CCUS, N (%) | 4 (6%) |
| CHIP, N (%) | 1 (1%) |
| Abnormal cytogenetics, N (%) | 20 (29%) |
| Median DDX41 VAF % | 15% |
| AML progression, N (%) | 8 (16%) |
| Underwent BM transplantation | 20 (29%) |
| Confirmed germline mutation | 16 (24%) |
| Median BM blast% | 10% |
| Variable | DDX41 p.M1|, N = 23 | DDX41 p.Arg525His, N = 9 | DDX41 p.Asp140GlyFs*2, N = 8 | p Value |
|---|---|---|---|---|
| Median age, yrs (range) | 72 (30–83) | 69 (59–90) | 63.5 (60–82) | 0.77 |
| Male sex, N (%) | 15 (65%) | 7 (78%) | 7 (88%) | 0.56 |
| Diagnosis | ||||
| MDS, N (%) | 9 (39%) | 5 (56%) | 7 (88%) | |
| AML, N (%) | 6 (26%) | 2 (22%) | 1 (12.5%) | |
| MPN, N (%) | 1 (4%) | 1 (11%) | 0 (0%) | |
| DDX41 carrier, N(%) | 3 (13%) | 0 (0%) | 0 (0%) | |
| CCUS, N (%) | 3 (13%) | 0 (0%) | 0 (0%) | |
| Mean number of co-mutations, (range) | 0.3 (0–2) | 0.6 (0–2) | 0.5 (0–2) | 0.63 |
| Co-mutations | ||||
| ASXL1, N (%) | 3 (13%) | 0 (0%) | 1 (13%) | 0.79 |
| JAK2, N (%) | 1 (4%) | 1 (11%) | 0 (0%) | 0.69 |
| DNMT3A, N (%) | 1 (4%) | 1 (11.1%) | 2 (29%) | 0.27 |
| TP53, N (%) | 1 (4%) | 0 (0%) | 0 (0%) | 1.00 |
| NRAS, N (%) | 1 (4%) | 0 (0%) | 0 (0%) | 0.59 |
| Abnormal cytogenetics, N (%) | 2 (9%) | 3 (33%) | 2 (25%) | 0.17 |
| Median DDX41 VAF % (range) | 48 (15–52) | 11 (4–24) | 45 (37–50) | <0.001 |
| AML progression, N (%) | 1 (5.9%) | 2 (25%) | 0 (0%) | 0.28 |
| Labs | ||||
| Median hemoglobin (g/dL) | 11.2 | 10.2 | 10.4 | 0.68 |
| Median platelet (×109/L) | 94 | 96 | 153 | 0.64 |
| Median white blood count (×109/L) | 2.8 | 2.5 | 2.3 | 0.24 |
| Median BM blast % | 12 | 11 | 7 | 0.57 |
| Overall survival (OS) | ||||
| 12-month OS (%) | 100% | 75% | 86% | Cohort 1 vs. 2: 0.41 Cohort 1 vs. 3: 0.79 Cohort 2 vs. 3: 0.43 |
| 24-month OS (%) | 83% | 75% | 71% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toumeh, N.; Jabban, Y.; Nanaa, A.; He, R.; Viswanatha, D.; Jevremovic, D.; Foran, J.M.; Arana Yi, C.Y.; Saliba, A.N.; Hefazi Torghabeh, M.; et al. Clinical Outcome and Molecular Profile in Patients with DDX41 Mutation Hot-Spots. Hematol. Rep. 2025, 17, 26. https://doi.org/10.3390/hematolrep17030026
Toumeh N, Jabban Y, Nanaa A, He R, Viswanatha D, Jevremovic D, Foran JM, Arana Yi CY, Saliba AN, Hefazi Torghabeh M, et al. Clinical Outcome and Molecular Profile in Patients with DDX41 Mutation Hot-Spots. Hematology Reports. 2025; 17(3):26. https://doi.org/10.3390/hematolrep17030026
Chicago/Turabian StyleToumeh, Nadia, Yazan Jabban, Ahmad Nanaa, Rong He, David Viswanatha, Dragan Jevremovic, James M. Foran, Cecilia Y. Arana Yi, Antoine N. Saliba, Mehrdad Hefazi Torghabeh, and et al. 2025. "Clinical Outcome and Molecular Profile in Patients with DDX41 Mutation Hot-Spots" Hematology Reports 17, no. 3: 26. https://doi.org/10.3390/hematolrep17030026
APA StyleToumeh, N., Jabban, Y., Nanaa, A., He, R., Viswanatha, D., Jevremovic, D., Foran, J. M., Arana Yi, C. Y., Saliba, A. N., Hefazi Torghabeh, M., Hogan, W. J., Shah, M. V., Mangaonkar, A. A., Patnaik, M. M., Alkhateeb, H. B., & Al-Kali, A. (2025). Clinical Outcome and Molecular Profile in Patients with DDX41 Mutation Hot-Spots. Hematology Reports, 17(3), 26. https://doi.org/10.3390/hematolrep17030026








