Abstract
In recent years, CRISPR-Cas9 technology has become a powerful and indispensable tool for targeted mutagenesis in plants, including applications such as gene knockout, prime-editing, multiplex gene editing, and regulation of gene transcription. As the number of potential genome editing approaches expands at a very fast pace, rapid, efficient, and cost-effective analytical strategies are needed to screen large numbers of mutants, including the detection of off-target events. In this study, we reported a detection method based on High-Resolution Melting (HRM) analysis to discriminate between wild-type (wt) and edited lines of different varieties of Vitis vinifera and grapevine rootstocks. Those edited lines were obtained through Agrobacterium tumefaciens mediated transformation of embryogenic calli using the CRISPR/SpCas9 system and targeting VviEPFL9-1 and VviEPFL9-2, two paralogous genes involved in stomata cell fate induction. The method clearly distinguished between the wt allele and the mutated one and was partially effective in distinguishing different types of mutation. Moreover, HRM data elaboration based on a Principal Component Analysis (PCA) allowed one to group populations of lines which originated from the same transformation event. Our study demonstrates the reliability of HRM as a fast and cost-effective diagnostic tool for the screening of edited lines and the evaluation of off-target events.
1. Introduction
Several biotic (bacteria, viruses, fungi, insects, parasites, etc.) and abiotic stresses (drought, salinity, heat, cold, etc.) can affect agriculture production around the world and one strategy to develop climate-resilient, stress-tolerant crops with better quality and increased production relies on the targeted mutagenesis of key genes, known to have a crucial role in determining agronomic, nutritional, or qualitative traits of particular interest [1,2,3,4]. During the past years, several new genomic techniques (NGT), including targeted mutagenesis methodologies called genome editing (GE), have been widely employed in plants [5,6]. These techniques allow one to site-specifically modify a DNA sequence of one or a few selected genes, either in a homozygous or heterozygous state, by introducing single nucleotide variants (SNV) or nucleotide insertions or deletions (InDels). Among genome editing approaches, the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas9) is the simplest and the most versatile and has become the favorite tool for genetic manipulation [7,8]. CRISPR/Cas9 system is designed from a component of the known bacterial immune system and consists of a single guide RNA (sgRNA) that identifies a specific DNA sequence and the Cas9 protein [9]. This complex produces double-strand breaks (DSBs) at the targeted site. By designing the specific 20mer sequence of the sgRNA, complementary to a specific site in the genome and close to a protospacer adjacent motif (PAM) site, any desired genomic region can be targeted. This aspect makes it simpler to handle in comparison to other GE techniques such as transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs) which require any specific target to modify complex and large elements of the vector [7,8,9,10,11].
Because of its versatility, efficiency, and simplicity, the CRISPR/Cas9 system is used both for functional genomic research, to clearly prove the function of a gene in a specific organism, but also for those genes whose function has been deeply deciphered, to generate plant prototypes to be tested in the field (when the national regulation allows field trials). To this purpose, over recent years, protocols have been developed to deliver Cas9 and sgRNA in the form of a ribonucleoprotein complex directly to plant protoplasts and to regenerate entire plants from protoplasts both in herbaceous crops [12,13,14,15,16] and in vegetatively propagated tree species [17,18,19,20,21]. According to the European Commission’s recent proposal aimed to update European GMO legislation, which is still in the amendment process, in most cases edited plants regenerated from protoplasts may be considered as a category 1 NGT. Therefore, if this proposal is approved by the European Parliament and the Council, NGT1 plants will be exempted from the requirements of Directive 2001/18 on the deliberate release into the environment of GMOs [22]. The process of transfecting protoplasts with ribonucleoproteins (RNPs) is therefore a highly sought-after technology, particularly for vegetatively propagated plants such as woody fruit trees, like grapevine, for which it is important to maintain the identical genetic make-up in order to guarantee the cultivar typicity of fruits and their derivatives. However, this methodology lacks a selection phase that would allow for the regeneration of plants only from edited protoplasts, while excluding the non-edited ones. The challenge lies in the lack of established protocols to reliably isolate and propagate the edited cells from a mixed population and to precisely detect the resulting genetic changes. Therefore, the availability of a rapid, high-throughput and low-cost method to screen the regenerated plants, identifying those with a mutation in the target site, is essential.
While single-bp insertions or deletions (indels) have an enormous effect on the target gene, knocking out its function through the frameshift of its coding region, analytical detection may be challenging due to the small size of the mutation. Traditionally, screening for the identification of mutations generated by the CRISPR-Cas9 system has been achieved using several techniques such as Sanger and Illumina sequencing, TaqMan® assays, and restriction enzyme analysis [23]. Regarding the first two methods, amplicon-sequencing outputs can require quite a long processing time while a low sensitivity in the detection of small indels is reported for the last two methods [24].
HRM analysis is a fluorescence-based technique that measures the melting temperature of amplified double-stranded DNA, allowing one to discriminate between amplicons with different melting temperatures [25]. The melting temperature depends on GC content, length, and nucleotide sequence [26]. A HRM curve analysis has been widely used to identify mutations and single nucleotide polymorphisms in various genes [27,28,29,30] and in plants too, including Vitis vinifera [31]. In addition it has recently been used to reliably identify CRISPR-Cas9-induced indels in plants [32,33,34]
Although HRM analysis was previously applied in grapevine [31] and used to verify both the quality [35,36] and the authenticity of wine [37], in this study we report a simple and low-cost strategy to perform molecular genotyping in order to discriminate edited grapevine by using the High-Resolution Melting analysis data. Such implemented workflow is intended as a screening process for the selection of edited lines. Several grapevine lines of different varieties and rootstocks, putatively edited in two paralogous genes, VviEPFL9-1 and VviEPFL9-2, involved in the leaf stomata formation were analyzed. The effect of VviEPFL9 paralogs knockout (KO) on leaf stomatal density was reported in detail by Shahbaz et al. [38], together with its influence on physiological traits (e.g., stomatal conductance, photosynthesis, and plant survival rate under water-stressed conditions). The present study, conversely, aimed to emphasize the significant effort required for the selection of the most promising edited lines among the huge amount of putatively edited lines and the valuable contribution of a HRM method in discriminating between wt and mutated alleles during the initial stage of the molecular characterization or in subsequent phases to confirm line identity.
2. Materials and Methods
2.1. Plant Material (Gene Transfer Experiments, In Vitro and Greenhouse Cultivation)
Details regarding plant generation are described in Clemens et al. [11] (for the editing of VviEPFL9-1) and in Shahbaz et al. [38] (for the editing of VviEPFL9-2). Three binary vectors were used for gene transfer experiments (Cloning Service, Hamburg, Germany). In addition to the common CRISPR/Cas9 elements and nptII gene, they carried a specific guide RNA designed with CRISPR-P 2.0 software (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR accessed on 1 November 2025) [39] and validated for RNA secondary structure (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi accessed on 1 November 2025): (1) sgEPFL9-1 GCACATACAATGAATGCAAA, recognizing a 20 bp region in the third exon of VviEPFL9-1; (2) sg937EPFL9-2 CATTCATTGTAAGTGCAGGT, recognizing a 20 bp region in the third exon of VviEPFL9-2 (reverse strand); and (3) sg938EPFL9-2 GACTGATGATTGGATCCACC, recognizing a 20 bp region in the third exon of VviEPFL9-2 (forward strand, upstream and adjacent to sg937). Gene transfer was carried out by means of Agrobacterium tumefaciens strain EHA105 on the embryogenic calli of the varieties ‘Sugraone’, ‘Syrah’, and of the rootstock Kober 5BB according to the procedure described by Dalla Costa et al. [40]. Actively proliferating Agrobacterium cultures, pre-induced with 100 μM acetosyringone (AS) for 3 h, were re-suspended in liquid GS1CA medium containing AS to an optical density at 600 nm (OD600) of 0.3–0.45. These bacterial suspensions were then combined with approximately 5 g of embryogenic callus in a total volume of 20 mL. The mixture was gently agitated at 60 rpm for 10 min at 25 °C, subsequently centrifuged, blotted on sterile Whatman paper, and transferred onto solid GS1CA medium. Co-cultivation was carried out in the dark at 25 °C for 48 h. Following co-cultivation, the callus was washed with liquid GS1CA medium supplemented with 1 g/L Timentin, centrifuged, blotted dry, and transferred onto solid GS1CA medium containing 1 g/L Timentin. Cultures were maintained in the dark at 25 °C for 4 weeks. After this period, calli were subcultured monthly onto fresh GS1CA medium supplemented with 1 g/L Timentin and 150 mg/L Kanamycin, under the same dark conditions at 25 °C, for a total duration of 8 months. Mature embryos at the torpedo stage were then transferred to an NN medium supplemented with 25 mg/L Kanamycin and incubated under a 16 h photoperiod to promote differentiation and germination. Regenerated plants were analyzed by PCR for the initial screening of SpCas9 integration into the 20 µL final volume containing 1× PCR BIO (Resnova, Rome, Italy), 0.5 µM of each primer (SpCas9_Fw 5′-CTTCAGAAAGGACTTCCAATTC-3′ and SpCas9_Rv, 5′-ATGATCAAGTCCTTCTTCACTT-3′), and 30 ng of genomic DNA. In vitro edited lines and the wt control were propagated in sterile baby jars in WP medium (McCown and Lloyd, 1981) and were grown in a climatic chamber at 100 photosynthetic photon flux density (PPFD) ± 20 (µmol m−2 s−1), 24 °C, and a 16/8 light/dark photoperiod. In vitro Kober 5BB edited plants knocked out in VviEPFL9-1 and in vitro ‘Syrah’ edited plants knocked out in VviEPFL9-2 were sequentially subjected to Sanger sequencing, Illumina sequencing as described in Clemens et al. [11], and HRM analysis during in vitro propagation, two or three months after the first screening. Regarding ‘Sugraone’ edited plants knocked out in VviEPFL9-1, they were subjected to Illumina sequencing after the initial screening and then, after one year of in vitro propagation, were acclimatized in the greenhouse for further experiments as reported in Clemens et al. [11]. Subsequently, approximately 2 years after the initial screening, these plants were re-analyzed by Illumina sequencing and evaluated by the HRM method. The CRISPResso software version 1.0.13 (https://crispresso.rocks/ accessed on 1 November 2025) was used to process (with default parameters) the raw paired-end reads and to visualize the mutation profiles in the target sequences.
2.2. DNA Extraction
Genomic DNA from ‘Sugraone’, ‘Syrah’, and Kober 5BB samples was extracted from freshly frozen leaf tissue (approximately 100 mg) using the NucleoSpin Plant II kit (Macherey–Nagel, Düren, Germany) according to the manufacturer’s instruction, quantified using Nanodrop 8800 (Thermo Fischer Scientific, Waltham, MA, USA). In addition, fluorometric measurements were also performed for each extracted DNA, using the QuantiFluor® dsDNA System (Promega, Madison, WI, USA). To improve DNA quality, a re-extraction step was performed for selected ‘Syrah’ lines. In these cases, small, healthy, fully green leaves were used, as this tissue typically contains lower levels of polysaccharides and phenolic compounds. Approximately 100 mg of leaf material was processed using the NucleoSpin Plant II kit according to the manufacturer’s protocol, with careful attention to complete tissue homogenization and thorough membrane washing to remove contaminants. DNA was eluted in nuclease-free water, maintained on ice during handling, and immediately stored at −20 °C.
2.3. Detection of T DNA Integration Point
T-DNA integration points of the putative ‘Syrah’ clones were identified according to the method described in Dalla Costa et al. [40]. The library was sequenced by Illumina MiSeq (PE300) platform at the Sequencing Platform Facility of Fondazione Edmund Mach (San Michele all’Adige, Italy) and the genomic region was subsequently validated by PCR amplification. PCR was performed in a final volume of 20 µL containing 1× PCR BIO (Resnova, Rome, Italy), 40 ng of genomic DNA, and 0.5 µM of the following: Chr13fw8581line880 primer (AACTTCGGCCGGTCTAGAGC) annealed on the T-DNA closed to the LB and Chr13rv8581_line880 (ACAAACAAGTGGTGGAGATGAGT) annealed on chromosome 13 of the grapevine reference genome. Amplification products were checked on an agarose gel, purified using PureLink Quick Gel Extraction (Invitrogen, Carlsbad, CA, USA) and sequenced by Sanger sequencing (FEM Sequencing Platform Facility). The sequencing results were analyzed with the Blast tool using the Vitis vinifera PN40024 reference genome v5 available on the Grapegenomics.com web portal (https://www.grapegenomics.com/pages/PN40024/blast.php, accessed on 1 November 2025).
2.4. qPCR-HRM Assay
The HRM experiments were performed on the Rotor-Gene Q 5plex Platform (Qiagen, GmbH, Hilden, Germany) instrument in a final reaction volume of 20 μL, including 1X FAST Eva Green qPCR Master Mix (Fisher Molecular Biology, Allentown, PA, USA), 1 ng of genomic DNA, 300 nM of primers in order to amplify EPFL9-1 (primer EPFL9-1_fw and EPFL9-1_rv, see Table S1), and EPFL9-2 (primer EPFL9-2_fw and EPFL9-2_rv, see Table S1). At least two technical replicates were performed for each sample.
The PCR thermocycling conditions were as follows: initial denaturation of 2 min at 95 °C, followed by 35 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 20 s, with one fluorescence reading per annealing step. The program of HRM was as follows: denaturation at 95 °C for 1 min, renaturation at 40 °C for 1 min, and ramp from 75 °C to 85 °C increasing at 0.1 °C per second. Fluorescence data were acquired at a frequency of 20 readings per °C increment.
2.5. HRM Data Analysis
The HRM curve data were analyzed after normalization of the melting curve using the Rotor-Gene Q series software version 2.3.5 (Qiagen, Venlo, The Netherlands https://www.qiagen.com/gb/products/instruments-and-automation/pcr-instruments/rotor-gene-q-mdx, accessed on 1 November 2025).
The normalization regions were chosen as near as possible to but not overlapping the region of pre-melting and post-melting in order to eliminate the fluorescence variance. This software can characterize samples using the auto-calling function to auto-call the genotypes that match the controls (reference genotype) with the highest confidence percentage, clustering similar curve shapes automatically into groups representing different genotypes with high accuracy. Wild-type DNA was included in each experiment and chosen as a reference genotype. For the calculation of the scores (confidence values expressed as percentage confidence) of the HRM analysis, a threshold of 80% (confidence percentage) was set up, below that value the samples are classified as variants. This threshold was applied during curve normalization, as it offered an optimal compromise between minimizing noise and retaining meaningful variation among samples. This value was determined based on preliminary analyses and is in line with thresholds reported in previous HRM studies.
High-Resolution Melting normalized data were exported to a .csv file from Rotor-Gene Q Series software version 2.3.5. Data are organized with samples on columns and temperature values on rows. Each cell value describes the normalized value (from 0 to 100) of fluorescence intensity. The files were then imported to R-studio [41] and handled with R functions of tibble and dyplr R packages. Predicted melt curves were obtained from the nucleotide sequence reported in Table S3 with the DECIPHER R package. When available two or more Illumina profiles and the correspondent percentage contribution was summed to achieve the combination of the contributions [42,43]. In case of residue from the sum of correspondent contributions, it was equally divided and added to each of them. Curves were plotted with the ggplot R package.
2.6. Statistical Analysis
Principal Component Analysis has been performed to our data. Firstly, a covariance matrix is computed from the input data showing the degree of correlation between two variables, which in our case are emission of fluorescence and time series temperature [44,45]. Initially, our data were filtered considering temperature values from 75.85 to 84.00, so as to include only the melting domain, and the PCA was performed on a covariance matrix of our data by the R function prcomp() (factoextra R package) with default parameters. In order to compare our groups, defined by colors, an ANOVA test (adonis() R function) was performed on each PC used to show our data. When a PC result was statistically significant, a post hoc Tukey test (TukeyHSD() R function) was carried out to see which separations were statistically significant among pair comparison. PCA plots are shown with R functions which are part of the ggplot2, ggpubr and paletteer R packages.
2.7. Droplet Digital PCR Quantification
The Syrah DNA samples analyzed by HRM were tested in droplet digital PCR (ddPCR) using the BioRad QX200 Droplet Digital PCR System. Each sample (8 lines, 2 biological replicates per line) was analyzed to measure the copy number of grapevine endogenous gene VviCHI and Cas9 (Dalla Costa et al. [22]). The reaction volume was 20 μL containing 2X ddPCR Supermix for probes (BioRad, Pleasanton, CA, USA), 300 nM of primers (see Table S2), and 200 nM of probes and (should be indicated as a quantity, in ng, not a volume) of template DNA for the ddPCR singleplex assay. Droplet generation was carried out in DG8 cartridges (BioRad, Pleasanton, CA, USA) loaded on a QX200 droplet generator (Bio-Rad Laboratories, Inc.).
Droplets were then transferred to 96-well plates amplified in a PCR thermocycler GeneAmp PCR System 9700 (Thermo Fisher Scientific, Waltham, MA, USA) with the following thermal profile: 10 min DNA polymerase activation at 95 °C, 45 cycles of a two-step thermal profile for 30 s at 94 °C for denaturation, 60 s at 55 °C for annealing and extension, and droplets stabilization at 98 °C for 10 min followed by an infinite hold at 4 °C. After thermal cycling, the 96-well plates were transferred to a QX200 droplet reader (Bio-Rad Laboratories, Inc.) and data were analyzed with QX Manager 1.2 Standard Edition (Bio-Rad Laboratories, Inc. https://www.bio-rad.com/sites/default/files/2023-10/10000167263.pdf, accessed on 1 November 2025). According to the manufacturer’s instructions and documents of the Joint Research Centre (JRC) [46,47], data were accepted for the subsequent analyses if the number of droplets was more than 10,000 per 20 μL reaction with a clear discrimination between positive and negative signals, if negative controls (NTC) were ≤2 fluorescent droplets and positive control and sample were >2 fluorescent droplets.
3. Results
3.1. Set Up of a HRM Method to Evaluate Specific Target Gene Sequences in Grapevine
Two HRM methods were set up to evaluate the presence of multiple alleles or nucleotide mutations in two grapevine genes, VviEPFL9-1 (Vitvi05g01370) and VviEPFL9-2 (Vitvi07g04390) which were the target of a biotechnological application aimed at knocking them out through the CRISPR/Cas9 system [38]. Different genotypes of grapevine plants were tested belonging to different Vitis spp.: the Vitis vinifera wine varieties ‘Syrah’ and ‘Cabernet Sauvignon’, the Vitis vinifera table grape variety ‘Sugraone’, and the rootstock Kober 5BB (Vitis berlandieri × Vitis riparia). As shown in Figure 1 (Figure 1a refers to VviEPFL9-1 and Figure 1b refers to VviEPFL9-2), for each gene the melting curves of the four genotypes were overlapping, thus proving that no polymorphism was present among the four genotypes in the amplified region, nor different alleles within the genotype. Moreover, melting peaks were single and sharp indicating that the primers chosen to amplify the selected regions were very specific and were suitable to discriminate between different alleles or the presence of nucleotide mutations.
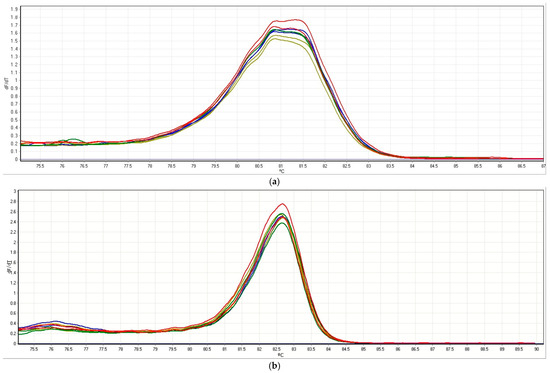
Figure 1.
Normalized melting peaks of qPCR-HRM assay employing the primers VviEPFL9-1 (a) and VviEPFL9-2 (b). A different color has been used per each grapevine genotype, Sugraone (n = 2) is shown in blue, ’Syrah’ (n = 2) in dark yellow, Cabernet Sauvignon (n = 2) in green, and Kober 5 BB (n = 2) in red.
3.2. Screening of the Mutations In Vitro and Greenhouse-Cultivated Edited Lines
Transgenic plants regenerated from Agrobacterium tumefaciens co-cultured calli were analyzed with the HRM method to screen the presence of mutations caused by the CRISPR/Cas9 system in the genes VviEPFL9-1 and VviEPFL9-2. The screening was carried out in three different situations. In the first, VviEPFL9-1 was assessed on a set of in vitro Kober edited plants, while in the second on biological replicates of ‘Sugraone’ wt and edited lines cultivated in a greenhouse. In the third, the gene VviEPFL9-2 was assessed in KO lines of the variety ‘Syrah’ and of the rootstock Kober 5BB. The results between the predicted melt curves from the Illumina and Sanger sequences and those obtained experimentally (Figure S1) show a clear separation among variants with mutations and wt samples. In the second situation, the melting phase deduced from the nucleotide sequence (S1M, S1N) is quite similar between those gained experimentally (S1O, S1P), which have a more gradual decrease in fluorescence intensity. On the other hand, in the first and third situations the predicted melting phases (S1A- S1E, S1Q- S1R) start at a lower temperature than the analytical ones (S1F- S1L, S1S- S1T).
A covariance matrix was considered for the Principal Component Analysis (PCA). In addition, for each sample, an Illumina profile of the target site was produced to have a precise picture of the mutations generated after the Cas9 nuclease cleavage.
Figure 2a shows the PCA of HRM data for the gene VviEPFL9-1 on a set of in vitro Kober edited plants. For each sample, the Illumina mutation profiles in the target site were summarized on the right and reported in detail in Supplementary Table S3. According to such data, some lines resulted in having the same mutation pattern and were classified in cluster 1, 2, and 3.

Figure 2.
(a) The PCA of VviEPFL9-1 edited lines and wt Kober, (b) the PCA of VviEPFL9-1 edited lines and wt Sugraone, and (c) the PCA of VviEPFL9-2 edited lines and wt plants. For each panel, on the left, there is a table with the significant (significance level 0.05) pair comparisons among the classes of samples for each PC. On the right, Illumina results are described in a table by specifying the modifications and the percentage of valid reads from the CRISPResso software. (a) The Illumina clusters were circled (“Ko” stands for Kober variety, “1” for EPFL9-1 gene, and the last number corresponds to the sample name (Ko_1_samplename)). (b) “Su” stands for Sugraone variety, “1” for EPFL9-1 gene, “LA/LC” represents a specific line, and the last number corresponds to the sample name (Su_1_LA/LC_samplename). (c) “Ko/Sy” stands for the variety, “2” for EPFL9-2 gene, “sgs1/sgs2” represents the 937/938 guide RNA, and the last number corresponds to the sample name (Ko/Sy_2_sgs1/sgs2_samplename).
The wt samples and the mutant lines were clearly separated along the first component, which explained almost 88% of the variance. Along this axis, it was also possible to distinguish a well-defined group, indicated as cluster 2. According to the Illumina profile, those samples shared the same mutation, namely a 12 bp deletion. Moreover, samples from clusters 1 and 3 were located next to each other in the PCA. In contrast, the distribution of the remaining samples, which exhibited a diverse range of mutation, is scattered over the graph. The unique significant pair comparison in both PCs is between cluster 3 and cluster 2. An interesting case is represented by sample ‘790’ which resulted in being transgenic but not mutated in the target site, as shown by its Sanger profile in the Supplementary Table S3. Indeed, in the PCA graph this sample can be classified within the group of ‘wt’ samples, confirming the absence of any mutations.
The HRM method was also employed for the screening of biological replicates of ‘Sugraone’ wild-type (wt) and of two lines (line A and line C) edited in the VviEPFL9-1 gene (Figure 2b; the associated Illumina profiles are reported in detail in Supplementary Table S3). These plants were already selected by the developer and cultivated in the greenhouse for a phenotypic characterization. The three groups of samples were clearly separated along the horizontal axis of the PCA as well as verified by the corresponding pair comparison table, where the distinction among each pair is significant. Interestingly, the two samples of line A, which maintain 30% of the wt allele, are closer to the wt group compared to samples of line C. A third set of HRM data related to the analysis of the mutations of the gene VviEPFL9-2 in KO lines of the variety ‘Syrah’ and of the rootstock Kober 5BB is shown in Figure 2c. Wild-type samples were grouped together and a well-defined cluster was also observed for the ‘Syrah’ edited lines, which showed the same pattern of mutation according to Illumina data. Along the x axis, which shows nearly 77% of explained variance, all the pair comparisons among the corresponding classes are significant. As expected, Kober edited lines with a different mutation profile did not cluster in the PCA.
3.3. Case Study: Clones Discrimination
To test the ability of HRM analysis to identify putative clones, the VviEPFL9-2 edited lines of the ‘Syrah’ sample displaying an identical mutation profile and a deletion of a Cytosine (Supplementary Table S3) were selected for an in-depth analysis. The PCA showed that the edited ‘Syrah’ samples result in a defined cluster localized in a specific, continuous, and exclusive area of the plot (Figure 2c), and this is in line with Sanger sequencing data. Also, the melt curve analysis showed a similar trend for the ‘Syrah’ edited sample, with a clear shift in the dissociation curve compared to ‘Syrah’ wt (Figure 3A). However, six ‘Syrah’ samples (Sy_2sg1_798, Sy_2sg1_800, Sy_2sg1_801, Sy_2sg1_802, Sy_2sg1_803, Sy_2sg1_805) showed a confidence value, which is calculated to provide a level of confidence in the result, lower than the others (Figure 3A). In order to investigate if the DNA quality may impact on the confidence value, the DNA of selected ‘Syrah’ samples was extracted again and the HRM assay repeated. The re-extracted DNA samples exhibited higher quantity and better quality compared to the first ones (Figure 3B) and their melt curve appeared tighter with similar scoring of confidence percentage, comparable to the rest of the group (Figure 3C). According to these results, the DNA quality appears to be crucial to guarantee the correct confidence value in HRM analysis.
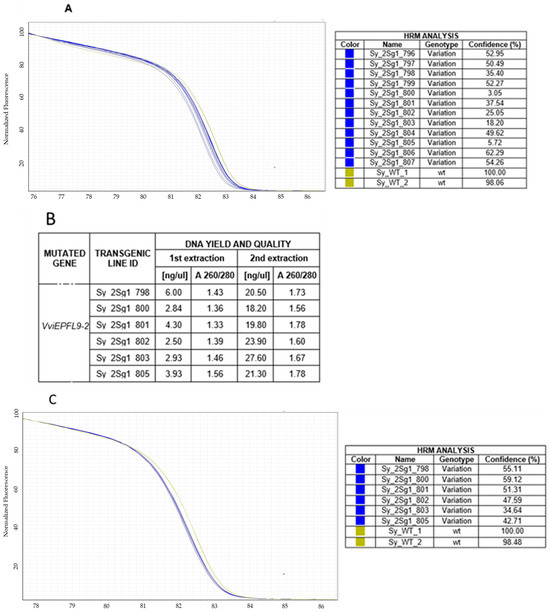
Figure 3.
Normalized melting curves of ‘Syrah’ samples. (A) DNA sample variants were isolated using a commercial isolation kit. ‘Syrah’ edited line shown in blue and ‘Syrah’ wt in dark yellow. (B) DNA yield (ng/µL) and quality parameters of ‘Syrah’ samples from two different DNA extraction sessions. Values were assessed using the spectrophotometric method. (C) Melting curves of re-extracted samples are shown.
To understand whether ‘Syrah’ samples were different lines with the same profile in the target site or, on the contrary, were clones deriving from the same transformation event, a T-DNA copy number assay was carried out through the quantification of SpCas9 with the droplet digital PCR technique (ddPCR). The analysis of a set of ‘Syrah’ lines showed that the mean copy number value for each line was around two copies (Figure S2).
Additionally, to definitively discriminate if ‘Syrah’ lines derive from the same transformation event and should be more correctly classified as clones, a method based on high-throughput sequencing was performed to investigate T-DNA genomic integration points in two of the ‘Syrah’ samples, 801 and 802. According to the analytical output, in samples 801 and 802 T-DNA integration occurred in the same genomic region, namely an intergenic region in chromosome 13. Specifically, two copies of T-DNA integrated in an inverted tandem, respectively, in positions 3,826,701 and 3,826,813 of chromosome 13 as reported in the scheme of Figure 4.
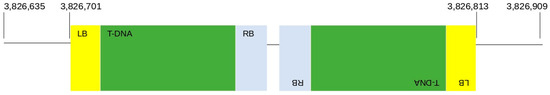
Figure 4.
Schematic representation of the T-DNA integration profile in ‘Syrah’ lines.
The genomic position was validated on intact genomic DNA of the two analyzed samples, 801 and 802, and on the remaining ‘Syrah’ lines. Figure 4 showed that the junction region between the genomic position 3,826,701 in chromosome 13 and the left border (LB) of the first T-DNA cassette was amplified in all the analyzed samples, confirming that they are clones deriving from the same T-DNA transformation event (Figure S3). Moreover, another amplification confirmed the second T-DNA integration point in all the samples.
3.4. Off-Target Analysis
Analysis of potential off-target events is required when the CRISPR/Cas9 system is used. The search for possible off-target sites is performed using online tools and is based on the identification of the sites most similar to the target site in the reference genome of the organism under study. In the case of the VviEPFL9-1 target site, as expected, the most similar off-target in the Kober 5BB genome was on the paralogous VviEPFL9-2 gene (see Figure S4), where the mismatch between the two consists of three nucleotides. The HRM analysis of the off-target site in the gene VviEPFL9-2 showed that amplicons from wt Kober 5BB, as well as VviEPFL9-1 KO mutants, clustered together into a group with a similar melting curve profile (Figure S4). The Rotor-Gene Q Series software version 2.3.5 classified these samples as wt, excluding the occurrence of off-target effects in the similar site in the paralogous gene. These results indicate that the HRM assay can efficiently be used to evaluate the presence of potential off-target events.
4. Discussion
In recent years, innovative techniques have emerged to accelerate the genetic improvement of crops without the use of transgenesis, the most important of which are cisgenesis and gene editing (using a DNA-free approach). Once putatively edited lines have been regenerated, they need to be screened to identify those with the desired mutation [48,49,50]. Razzaq et al. [51] reported several strategies that have been used in the past or are currently used to screen mutants, such as annealing at critical temperature polymerase chain reaction (ACT-PCR), polyacrylamide gel electrophoresis (PAGE)-mediated genotyping, T7 endonuclease I (T7EI) approach, restriction enzyme site loss technique, and direct sequencing of cloned PCR products. Each technique has its advantages and disadvantages. For example, the method based on direct sequencing of cloned PCR products can be time-consuming and prohibitively expensive for high-throughput screening. In particular, Illumina sequencing of the target sequence is realistically the only method capable of fully and accurately characterizing the mutation profile in a putative mutant line, but it remains a relatively expensive technique when applied to tens or hundreds of samples needing to be screened. When analyzing large numbers of samples, the need for a rapid and efficient workflow to discriminate between mutants and non-mutants is critical. This step is important for the developer to initially select modified plants for subsequent experimentation. Consequently, reducing the length and cost of the process is of paramount importance.
Streamlining the screening step is particularly important in the case of lines regenerated from protoplasts transfected with CRISPR/Cas9 ribonucleoproteins to knock out specific candidate genes. In fact, since the protoplasts are not subjected to any post-transfection selection regime, after a few months of culture, plants are regenerated that originate both from protoplasts that have incorporated a mutation in the target site, as well as from wt protoplasts. In this last case, plants must be identified and discarded. For example, in grapevine, the proportion of edited plants among the total number of regenerated plants is around 10% [18], although this percentage can vary depending on the efficiency of the sgRNA [17] and the plant variety [17,19,20]. The method proposed here, based on real-time PCR followed by High-Resolution DNA Melting analysis (qPCR-HRM), proved to be very fast and reliable in distinguishing the mutant lines from the non-mutant lines even when the mutation profile is reduced to single nucleotide variants (SNVs) or small insertions or deletions (InDels), whilst many other molecular diagnostic techniques depend on the DNA quality of the sample [52]. In fact, by exploiting the detection of melting temperature (TM) and differences in amplicons ranging in size from approximately 70 bp to 130 bp, this method is efficient in identifying single nucleotide polymorphisms (SNPs). The approach chosen to search sequence-variants by HRM was “reference curve-based directed genotyping”, where one of possible genotypes is selected as the reference, which in the reported examples corresponded to “wild-type”, and specific deviations from it are subsequently identified and classified as mutants [33,51]. It has been reported that the choice of reference genotype can affect the samples classification accuracy [53], suggesting that a larger number of reference samples can improve genotype classification. We selected the appropriate reference for each target gene and sample set (Figure 2) in order to decrease the assignment error. Overall, the experimental melting curves showed good discrimination between wt and edited lines and were able to identify lines that are transgenic but not mutated in the target site. In addition, a Principal Component Analysis (PCA) was implemented in order to explore the potential power of the dissociation curves to discriminate among mutation profiles. As shown in PCA plots (Figure 2), this analysis was able to cluster populations of lines with the same mutation (see the Syrah clones in Figure 2c), or biological replicates of a line (Figure 2b).
The use of this rapid and low-cost screening technique could also be useful for re-examining plant material long after the initial molecular characterization. In fact, several months elapse between the initial analysis of the in vitro plants and their phenotyping in the greenhouse, during which the plant must be propagated to obtain an adequate number of biological replicates. In the case of particularly complex experiments, involving many plant lines and biological replicates, it may be important to carry out a round of screening to confirm the identity of the plants, in order to identify any errors that may have occurred during the in vitro propagation phase.
Furthermore, despite the increasing specificity of new versions of nucleases [54] there may be cases of gene families whose members are very similar in sequence, making it difficult to design very specific sgRNAs. In other cases, the aim of the study may be to knock out more than one member of the family to avoid the very common functional redundancy of genes belonging to the same family. In these situations the HRM technique can be employed for the identification of off-target events in regions of the genome with a high degree of sequence similarity to the on-target, thereby reducing the cost of this mandatory assessment.
Finally, regarding the technical features of the technique, according to the literature, a relevant source of inaccuracy was low DNA quality of the plant sample [55,56,57,58]. DNA degradation or impurities can alter melting curve profiles, decrease fluorescence stability, and reduce the ability to distinguish between closely related genotypes. In this study, we observed that high-quality DNA obtained through optimized tissue selection and verified using both spectrophotometric and fluorometric quantification resulted in sharper and more reproducible melting curves. As shown in Figure 4 in the Syrah lines, an improvement in the quality of samples’ DNA may reduce the variation in melt profiles, increasing discriminatory power of the technique. These findings highlight the necessity of incorporating rigorous DNA quality control into HRM workflows, especially when small sequence differences must be resolved.
5. Conclusions
The experience presented in this paper has strengthened the fact that the HRM technique, when optimized appropriately, is capable of discriminating between mutated and wild-type profiles with sufficient accuracy. It is evident that the HRM technique, in contrast to sequencing, is unable to precisely ascertain the mutation profile or the presence of multiple patterns within a single sample. Nevertheless, it has confirmed a degree of reliability when employed for screening applications. This could serve as a viable and cost-effective alternative to DNA sequencing for screening purposes, with the prospect of subsequent confirmation of the result through sequencing to definitively validate the mutation profile in the initially selected samples. Our findings confirm that HRM analysis, when properly optimized, is a reliable and efficient tool for the preliminary screening of edited grapevine lines. The experience presented in this study reinforces that HRM can effectively discriminate between mutated and wild-type profiles with satisfactory accuracy. However, unlike sequencing, HRM cannot precisely determine the exact mutation or detect the presence of multiple mutation patterns within a single sample. Despite this limitation, it remains a cost-effective strategy for large-scale screening, allowing one to focus sequencing efforts only on selected samples. The application of HRM with PCA can finally provide an overview of the data, limiting the impact of expensive analyses that may not be accessible to all laboratories.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijpb16040126/s1, Figure S1: Comparison between predicted and analytical normalised melt curves for the three scenarios; Figure S2: Measure of Sp Cas9 copy number through ddPCR; Figure S3: Validation of the T-DNA integration point analysis through a qualitative PCR in Syrah lines; Figure S4: Off-target evaluation; Table S1: Primer sequences of VviEPFL9-1 and VviEPFL9-2 for HRM; Table S2: Primer and probe sequences of VvChi and SpCas9 for ddPCR; Table S3: Illumina and Sanger sequencing data for edited grapevine lines.
Author Contributions
Conceptualization, K.S. and L.D.C.; methodology, K.S., S.C., U.S., P.V. and O.Z.; software, D.L.R.; validation, U.M., D.V. and L.D.C.; formal analysis, K.S., L.D.C., S.C., U.S., P.V. and O.Z.; writing—original draft preparation, K.S.; writing—review and editing, U.M., L.D.C., O.Z., P.V., D.V., K.S. and D.L.R.; supervision, U.M. and L.D.C.; funding acquisition, U.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Italian Ministry of Health, in the current research framework, research code IZS LT0720.
Data Availability Statement
The datasets presented in this study can be found in online repositories: NCBI Sequence Read Archive, BioProject accession number: PRJNA820619.
Conflicts of Interest
Authors P.V. and O.Z. are employed by Mercier Novatech company. The remaining authors declare that there are no business or financial relationships that could be considered a potential conflict of interest in conducting this research.
References
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel Alleles of Rice eIF4G Generated by CRISPR/Cas9-Targeted Mutagenesis Confer Resistance to Rice Tungro Spherical Virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a Bacterial Speck Resistant Tomato by CRISPR/Cas9-Mediated Editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef]
- Zhang, Z.; Ge, X.; Luo, X.; Wang, P.; Fan, Q.; Hu, G.; Xiao, J.; Li, F.; Wu, J. Simultaneous Editing of Two Copies of GH14-3-3D Confers Enhanced Transgene-Clean Plant Defense against Verticillium Dahliae in Allotetraploid Upland Cotton. Front. Plant Sci. 2018, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Grape in the First Generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome Editing in Plants: Exploration of Technological Advancements and Challenges. Cells 2019, 8, 1386. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S. A Guide to Genome Engineering with Programmable Nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef]
- Clemens, M.; Faralli, M.; Lagreze, J.; Bontempo, L.; Piazza, S.; Varotto, C.; Malnoy, M.; Oechel, W.; Rizzoli, A.; Dalla Costa, L. VvEPFL9-1 Knock-Out via CRISPR/Cas9 Reduces Stomatal Density in Grapevine. Front. Plant Sci. 2022, 13, 878001. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Jeff Xi, J.; Qiu, J.-L.; et al. Targeted Genome Modification of Crop Plants Using a CRISPR-Cas System. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-Free Genome Editing of Bread Wheat Using CRISPR/Cas9 Ribonucleoprotein Complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-Free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Yang, S.H.; Kim, S.W.; Lee, S.; Koo, Y. Optimized Protocols for Protoplast Isolation, Transfection, and Regeneration in the Solanum Genus for the CRISPR/Cas-Mediated Transgene-Free Genome Editing. Appl. Biol. Chem. 2024, 67, 21. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-Free Genome Editing of Brassica Oleracea and B. Rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 2018, 871, 1594. [Google Scholar] [CrossRef]
- Gambino, G.; Nuzzo, F.; Moine, A.; Chitarra, W.; Pagliarani, C.; Petrelli, A.; Boccacci, P.; Delliri, A.; Velasco, R.; Nerva, L.; et al. Genome Editing of a Recalcitrant Wine Grape Genotype by Lipofectamine-Mediated Delivery of CRISPR/Cas9 Ribonucleoproteins to Protoplasts. Plant J. 2024, 119, 404–412. [Google Scholar] [CrossRef]
- Najafi, S.; Bertini, E.; D’Incà, E.; Fasoli, M.; Zenoni, S. DNA-Free Genome Editing in Grapevine Using CRISPR/Cas9 Ribonucleoprotein Complexes Followed by Protoplast Regeneration. Hortic. Res. 2023, 10, uhac240. [Google Scholar] [CrossRef]
- Scintilla, S.; Salvagnin, U.; Giacomelli, L.; Zeilmaker, T.; Malnoy, M.A.; Rouppe van der Voort, J.; Moser, C. Regeneration of Non-Chimeric Plants from DNA-Free Edited Grapevine Protoplasts. Front. Plant Sci. 2022, 13, 1078931. [Google Scholar] [CrossRef]
- Tricoli, D.M.; Debernardi, J.M. An Efficient Protoplast-Based Genome Editing Protocol for Vitis Species. Hortic. Res. 2024, 11, uhad266. [Google Scholar] [CrossRef] [PubMed]
- DIRECTIVE 2001/18/EC of the European Parliament and of the council of 12 March 2001 on the Deliberate Release into the Environment of Genetically Modified Organisms and Repealing Council Directive 90/220/EEC. Off. J. L 2001, 106, 1–39.
- European Network of GMO Laboratories (ENGL). Detection of Food and Feed Plant Products Obtained by New Mutagenesis Techniques; European Union Reference Laboratory for GM Food and Feed: Ispra, Italy, 2019. [Google Scholar]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of On-Target and off-Target Mutations Generated by CRISPR/Cas9 and Other Sequence-Specific Nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Druml, B.; Cichna-Markl, M. High Resolution Melting (HRM) Analysis of DNA-Its Role and Potential in Food Analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Hemmert, A.C.; Kent, J.O.; Rejali, N.A. DNA Melting Analysis. Mol. Asp. Med. 2024, 97, 101268. [Google Scholar] [CrossRef]
- Dufresne, S.D.; Belloni, D.R.; Wells, W.A.; Tsongalis, G.J. BRCA1 and BRCA2 Mutation Screening Using SmartCycler II High-Resolution Melt Curve Analysis. Arch. Pathol. Lab. Med. 2006, 130, 185–187. [Google Scholar] [CrossRef]
- Reed, G.H.; Wittwer, C.T. Sensitivity and Specificity of Single-Nucleotide Polymorphism Scanning by High-Resolution Melting Analysis. Clin. Chem. 2004, 50, 1748–1754. [Google Scholar] [CrossRef]
- Thomas, H.R.; Percival, S.M.; Yoder, B.K.; Parant, J.M. High-Throughput Genome Editing and Phenotyping Facilitated by High Resolution Melting Curve Analysis. PLoS ONE 2014, 9, e114632. [Google Scholar] [CrossRef]
- Distefano, G.; La Malfa, S.; Gentile, A.; Wu, S.-B. EST-SNP Genotyping of Citrus Species Using High-Resolution Melting Curve Analysis. Tree Genet. Genomes 2013, 9, 1271–1281. [Google Scholar] [CrossRef]
- Pereira, L.; Martins-Lopes, P. Vitis vinifera L. Single-Nucleotide Polymorphism Detection with High-Resolution Melting Analysis Based on the UDP-Glucose:Flavonoid 3-O-Glucosyltransferase Gene. J. Agric. Food Chem. 2015, 63, 9165–9174. [Google Scholar] [CrossRef]
- Denbow, C.J.; Lapins, S.; Dietz, N.; Scherer, R.; Nimchuk, Z.L.; Okumoto, S. Gateway-Compatible CRISPR-Cas9 Vectors and a Rapid Detection by High-Resolution Melting Curve Analysis. Front. Plant Sci. 2017, 8, 1171. [Google Scholar] [CrossRef]
- Li, R.; Ba, Y.; Song, Y.; Cui, J.; Zhang, X.; Zhang, D.; Yuan, Z.; Yang, L. Rapid and Sensitive Screening and Identification of CRISPR/Cas9 Edited Rice Plants Using Quantitative Real-Time PCR Coupled with High Resolution Melting Analysis. Food Control 2020, 112, 107088. [Google Scholar] [CrossRef]
- Denbow, C.; Ehivet, S.; Okumoto, S. High Resolution Melting Temperature Analysis to Identify CRISPR/Cas9 Mutants from Arabidopsis. Bio-Protocol 2018, 8, e2944. [Google Scholar] [CrossRef]
- Sanzani, S.; Miazzi, M.; Di Rienzo, V.; Fanelli, V.; Gambacorta, G.; Taurino, M.; Montemurro, C. A Rapid Assay to Detect Toxigenic Penicillium Spp. Contamination in Wine and Musts. Toxins 2016, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Samaras, A.; Madesis, P.; Karaoglanidis, G.S. Detection of sdhB Gene Mutations in SDHI-Resistant Isolates of Botrytis Cinerea Using High Resolution Melting (HRM) Analysis. Front. Microbiol. 2016, 7, 1815. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Gomes, S.; Castro, C.; Eiras-Dias, J.E.; Brazão, J.; Graça, A.; Fernandes, J.R.; Martins-Lopes, P. High Resolution Melting (HRM) Applied to Wine Authenticity. Food Chem. 2017, 216, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, U.; Videau, P.; Coulonnier, E.; Papon, C.; Navarro-Payá, D.; Valenzuela, A.V.; Matus, J.T.; Malnoy, M.; Zekri, O.; Fiorani, F.; et al. Reduced Stomatal Density Improves Water-Use Efficiency in Grapevine under Climate Scenarios of Decreased Water Availability. Plant Cell Rep. 2025, 44, 195. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol Plant. 2017, 10, 530–532. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to Produce T-DNA Free CRISPRed Fruit Trees via Agrobacterium Tumefaciens Stable Gene Transfer. Sci. Rep. 2020, 10, 20155. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of Single-Nucleotide Polymorphisms by High-Resolution Melting of Small Amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-Resolution DNA Melting Analysis for Simple and Efficient Molecular Diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal Component Analysis. WIREs Comp. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Groth, D.; Hartmann, S.; Klie, S.; Selbig, J. Principal Components Analysis. Methods Mol. Biol. 2013, 930, 527–547. [Google Scholar] [CrossRef] [PubMed]
- Gatto, F.; Savini, C.; Sacco, M.G.; Vinciguerra, D.; Buttinger, G.; Corbisier, P.; Mazzara, M.; Emons, H. Single and Multi-Laboratory Validation of a Droplet Digital PCR Method. Food Control 2022, 140, 109117. [Google Scholar] [CrossRef]
- Pecoraro, S.; Berben, G.; Burns, M.; Corbisier, P.; De Giacomo, M.; De Loose, M.; Dagand, E.; Dobnik, D.; Eriksson, R.; Holst-Jensen, A.; et al. Overview and Recommendations for the Application of Digital PCR; European Network of GMO Laboratories (ENGL): Ispra, Italy, 2019. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Vinciguerra, D.; Giacomelli, L.; Salvagnin, U.; Piazza, S.; Spinella, K.; Malnoy, M.; Moser, C.; Marchesi, U. Integrated Approach for the Molecular Characterization of Edited Plants Obtained via Agrobacterium Tumefaciens-Mediated Gene Transfer. Eur. Food Res. Technol. 2022, 248, 289–299. [Google Scholar] [CrossRef]
- Guertler, P.; Pallarz, S.; Belter, A.; Eckermann, K.N.; Grohmann, L. Detection of Commercialized Plant Products Derived from New Genomic Techniques (NGT)-Practical Examples and Current Perspectives. Food Control 2023, 152, 109869. [Google Scholar] [CrossRef]
- Shillito, R.D.; Whitt, S.; Ross, M.; Ghavami, F.; De Vleesschauwer, D.; D’halluin, K.; Van Hoecke, A.; Meulewaeter, F. Detection of Genome Edits in Plants-from Editing to Seed. Vitr. Cell. Dev. Biol.-Plant 2021, 57, 595–608. [Google Scholar] [CrossRef]
- Razzaq, A.; Saleem, F.; Kanwal, M.; Mustafa, G.; Yousaf, S.; Arshad, H.M.I.; Hameed, M.K.; Khan, M.S.; KhanJoyia, F.A. Modern Trends in Plant Genome Editing: An Inclusive Review of the CRISPR/Cas9 Toolbox. Int. J. Mol. Sci. 2019, 20, 4045. [Google Scholar] [CrossRef]
- A Guide to High Resolution Melting (HRM) Analysis. Available online: https://documents.thermofisher.com/TFS-Assets/LSG/manuals/cms_070283.pdf (accessed on 1 November 2025).
- Carrubba, A.; Abbate, L.; Sarno, M.; Sunseri, F.; Mauceri, A.; Lupini, A.; Mercati, F. Characterization of Sicilian Rosemary (Rosmarinus officinalis L.) Germplasm through a Multidisciplinary Approach. Planta 2020, 251, 37. [Google Scholar] [CrossRef]
- Lee, B.B.; Schott, E.J.; Behringer, D.C.; Bojko, J.; Kough, A.; Plough, L.V. Rapid Genetic Identification of the Blue Crab Callinectes Sapidus and Other Callinectes spp. Using Restriction Enzyme Digestion and High Resolution Melt (HRM) Assays. Front. Mar. Sci. 2020, 7, 633. [Google Scholar] [CrossRef]
- Graham, N.; Patil, G.B.; Bubeck, D.M.; Dobert, R.C.; Glenn, K.C.; Gutsche, A.T.; Kumar, S.; Lindbo, J.A.; Maas, L.; May, G.D.; et al. Plant Genome Editing and the Relevance of Off-Target Changes. Plant Physiol. 2020, 183, 1453–1471. [Google Scholar] [CrossRef]
- Garritano, S.; Gemignani, F.; Voegele, C.; Nguyen-Dumont, T.; Le Calvez-Kelm, F.; De Silva, D.; Lesueur, F.; Landi, S.; Tavtigian, S.V. Determining the Effectiveness of High Resolution Melting Analysis for SNP Genotyping and Mutation Scanning at the TP53 Locus. BMC Genet. 2009, 10, 5. [Google Scholar] [CrossRef]
- Fernandes, T.J.R.; Costa, J.; Oliveira, M.B.P.P.; Mafra, I. COI Barcode-HRM as a Novel Approach for the Discrimination of Hake Species. Fish. Res. 2018, 197, 50–59. [Google Scholar] [CrossRef]
- Fitzcharles, E.M. Rapid Discrimination between Four Antarctic Fish Species, Genus Macrourus, Using HRM Analysis. Fish. Res. 2012, 127–128, 166–170. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).