lncRNA-mRNA-miRNA Networks in Arabidopsis thaliana Exposed to Micro-Nanoplastics
Abstract
1. Introduction
2. Materials and Methods
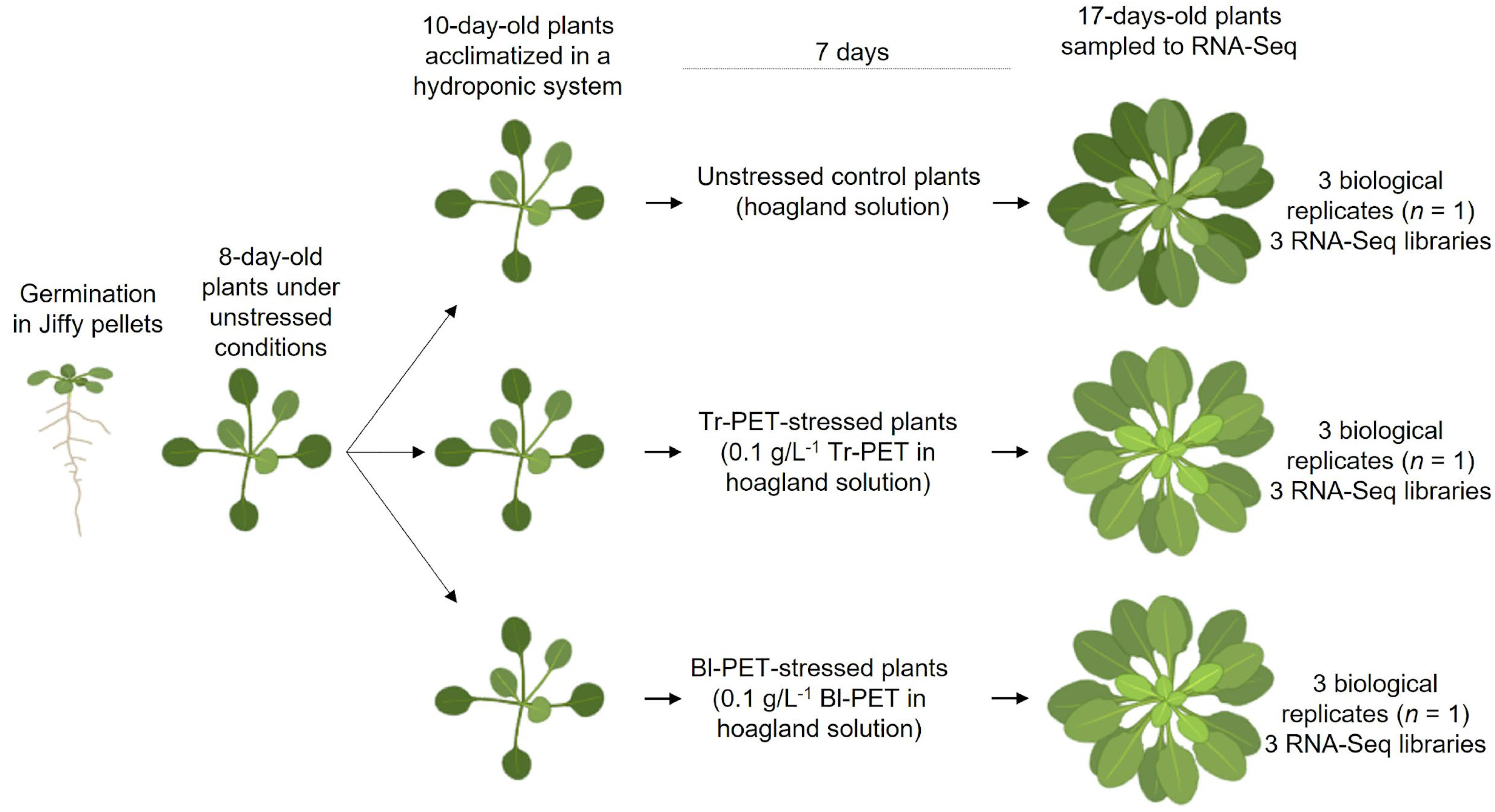
2.1. Plant Material and Stress Treatment
2.2. RNA Isolation, Library Sequencing, and Transcript Expression
2.3. Identification of Novel Unannotated lncRNAs, Read Mapping, and Expression Profiling
2.4. In Silico Prediction of Regulatory Function of Novel lncRNAs
2.5. Gene Set Enrichment Analysis
2.6. Network Predictive Protein–Protein Interaction Analysis
2.7. Chromosomal Localization and Cis-Acting Regulatory Elements in Promoter Sequences
3. Results
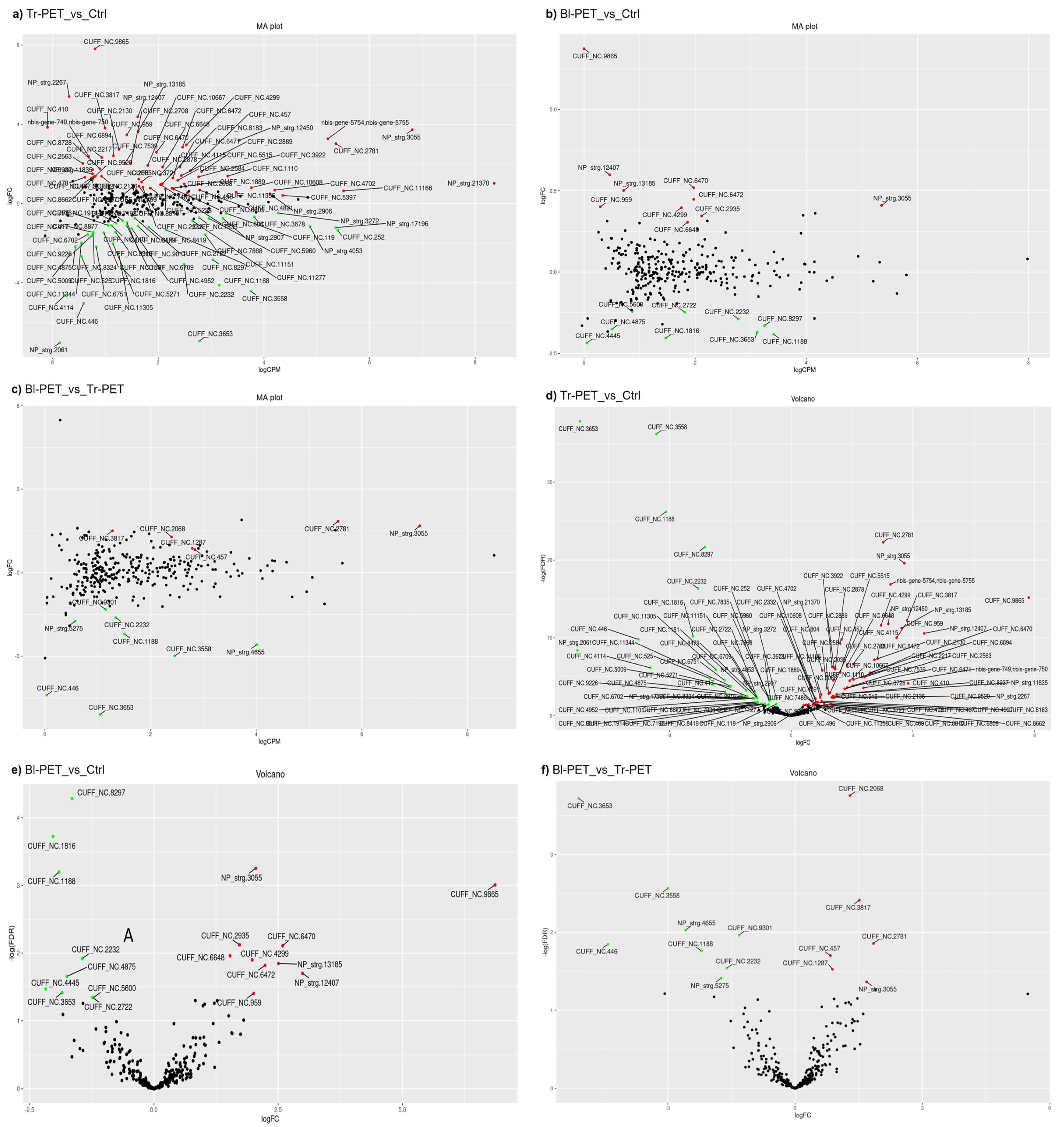
3.1. Global Characterization and Identification of lncRNAs and Their Target Protein-Coding Genes in A. thaliana Response to Micro-Nanoplastics
3.2. Functional Analysis of Target Protein-Coding Genes and Splice Variants
3.2.1. Identification of mRNAs Interacting with PET-Modulated lncRNAs
3.2.2. Tr-PET Versus Unstressed Control
3.2.3. Bl-PET Versus Unstressed Control
3.3. Identification of miRNAs Interacting with PET-Modulated lncRNAs
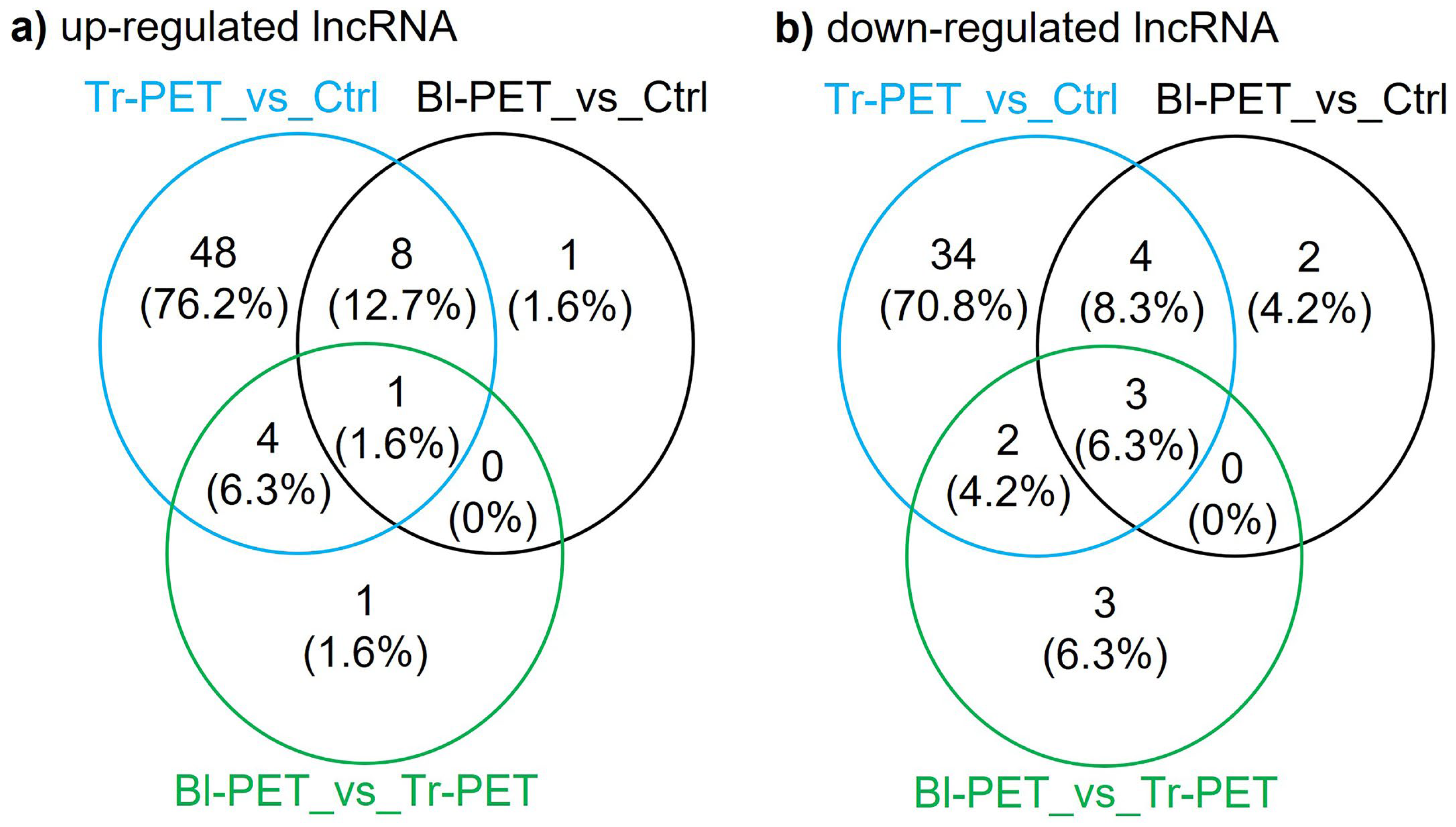
3.4. LncRNAs Up- or Down-Regulated by Micro-Nanoplastics
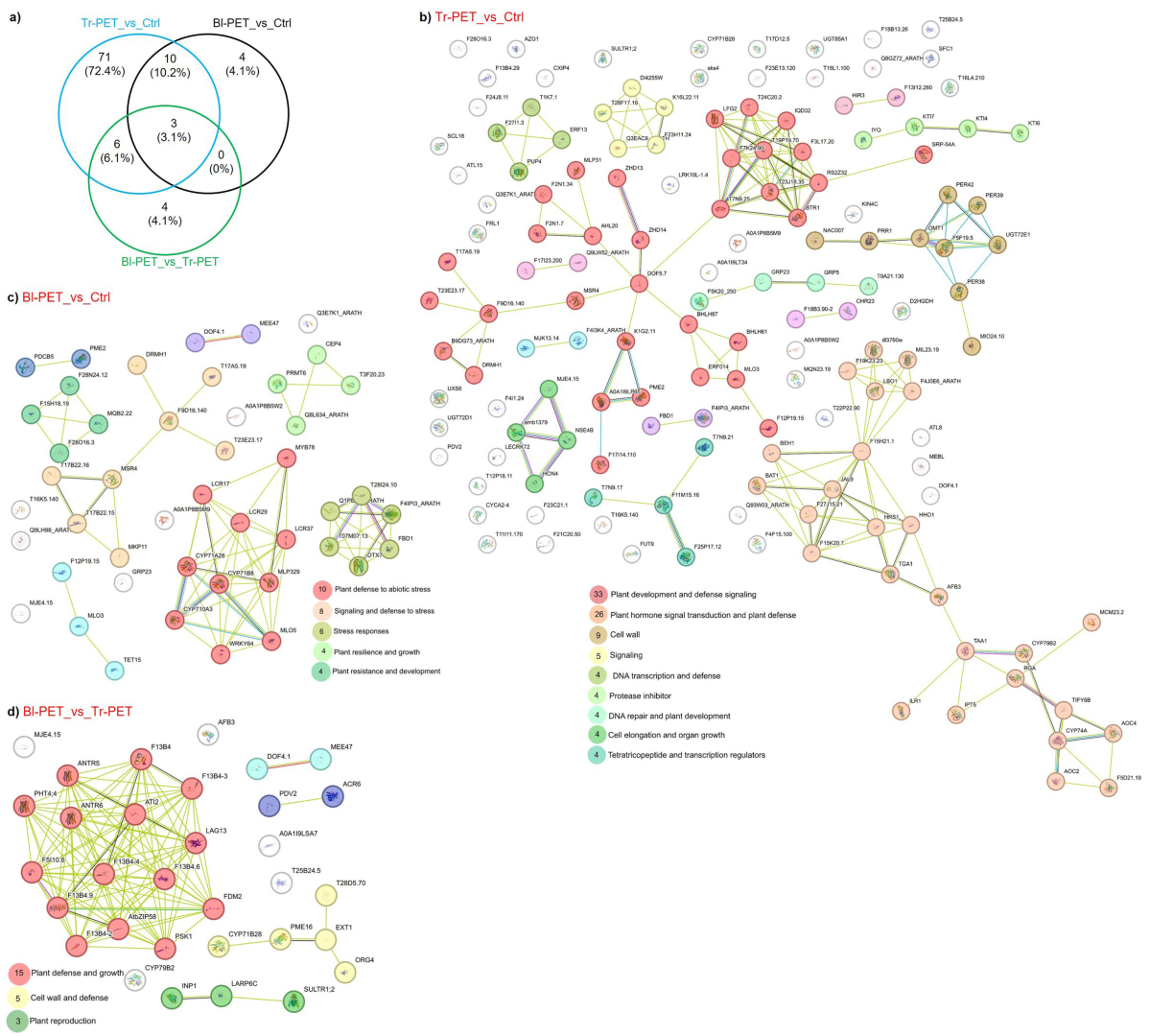
3.5. Functional Analysis of Protein-Coding Genes Targeted by lncRNAs
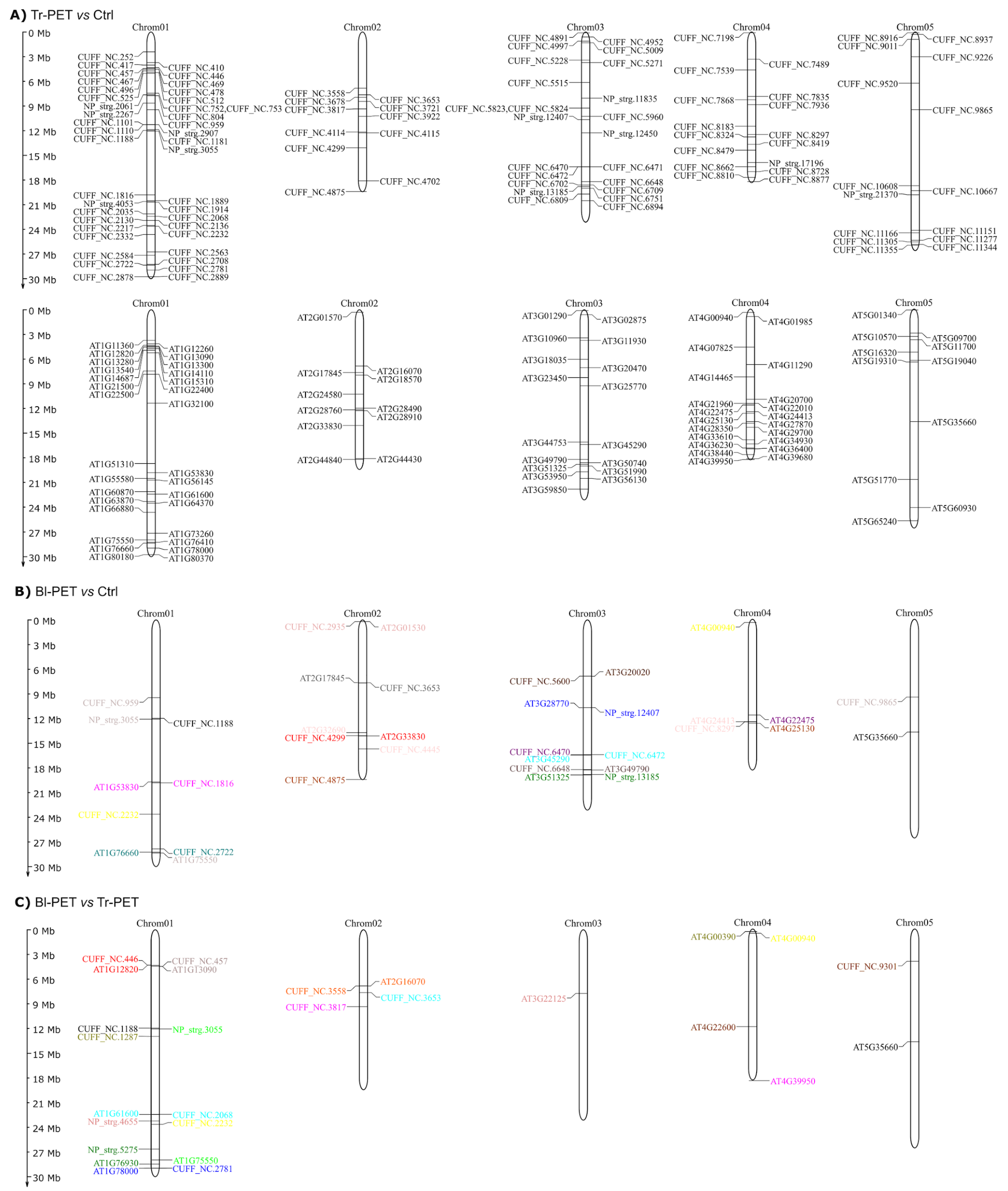
3.6. Chromosomal Localization of lncRNAs and Cis-Acting Regulatory Elements in Their Promoter Sequences
4. Discussion
4.1. lncRNA Roles in Micro-Nanoplastics Stress
4.1.1. Tr-PET Versus Unstressed Control
4.1.2. Bl-PET Versus Unstressed Control
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hovhannisyan, H.; Gabaldón, T. The Long Non-Coding RNA Landscape of Candida Yeast Pathogens. Nat. Commun. 2021, 12, 7317. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Jardim Poli, P.; Fischer-Carvalho, A.; Tahira, A.C.; Chan, J.D.; Verjovski-Almeida, S.; Murilo Sena, A. Long Non-Coding RNA Levels Are Modulated in Schistosoma Mansoni Following in Vivo Praziquantel Exposure. Non-Coding RNA 2024, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Nizhynska, V.; Morales, A.M.; Pisupati, R.; Nordborg, M. Population-level annotation of lncRNAs in Arabidopsis reveals extensive expression variation associated with transposable element-like silencing. Plant Cell 2023, 36, 85–111. [Google Scholar] [CrossRef]
- Rahman, M.-M.; Omoto, C.; Kim, J. Genome-Wide Exploration of Long Non-Coding RNAs of Helicoverpa armigera in Response to Pyrethroid Insecticide Resistance. Insects 2024, 15, 146. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Niu, M.; Wang, Y.; Xu, R.; Guo, Y.; Zhang, C. Roles of Long Noncoding RNAs in Human Inflammatory Diseases. Cell Death Discov. 2024, 10, 235. [Google Scholar] [CrossRef]
- Gendrel, A.-V.; Heard, E. Noncoding RNAs and Epigenetic Mechanisms during X-Chromosome Inactivation. Annu. Rev. Cell Dev. Biol. 2014, 30, 561–580. [Google Scholar] [CrossRef]
- Meller, V.H.; Joshi, S.S.; Deshpande, N. Modulation of Chromatin by Noncoding RNA. Annu. Rev. Genet. 2015, 49, 673–695. [Google Scholar] [CrossRef]
- Zhao, X.; Li, F.; Ali, M.; Li, X.; Fu, X.; Zhang, X. Emerging Roles and Mechanisms of LncRNAs in Fruit and Vegetables. Hortic. Res. 2024, 11, uhae046. [Google Scholar] [CrossRef]
- Gonzales, L.R.; Blom, S.; Henriques, R.; Bachem, C.W.B.; Immink, R.G.H. LncRNAs: The Art of Being Influential without Protein. Trends Plant Sci. 2024, 29, 770–785. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global Identification of Arabidopsis LncRNAs Reveals the Regulation of MAF4 by a Natural Antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [PubMed]
- Palos, K.; Nelson, A.C.; Yu, L.; Brock, J.R.; Railey, C.E.; Wu, H.-Y.L.; Sokołowska, E.; Skirycz, A.; Hsu, P.Y.; Gregory, B.D.; et al. Identification and Functional Annotation of Long Intergenic Non-Coding RNAs in Brassicaceae. Plant Cell 2022, 34, 3233–3260. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhu, T.; Shi, Y.; Chen, Y.; Li, W.J.; Chan, R.J.; Chen, D.; Zhang, W.; Yuan, Y.A.; Xu, J.; et al. An Antisense Intragenic LncRNA SEAIRa Mediates Transcriptional and Epigenetic Repression of SERRATE in Arabidopsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2216062120. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Lan, Y.; Zhou, X.; Yu, B.; Zhu, T.; Yang, F.; Fu, L.-Y.; Chao, H.; Wang, J.; Feng, R.-X.; et al. Single-Cell Transcriptome Analysis Dissects LncRNA-Associated Gene Networks in Arabidopsis. Plant Commun. 2023, 5, 100717. [Google Scholar] [CrossRef]
- Liu, J.; Zhong, X. Epiallelic Variation of Non-Coding RNA Genes and Their Phenotypic Consequences. Nat. Commun. 2024, 15, 1375. [Google Scholar] [CrossRef]
- Wang, Y.; Folimonova, S.Y. Long Noncoding RNAs in Plant–Pathogen Interactions. Phytopathology 2023, 113, 1380–1386. [Google Scholar] [CrossRef]
- Cui, C.; Wan, H.; Li, Z.; Ai, N.; Zhou, B. Long noncoding RNA TRABA suppresses β-glucosidase-encoding BGLU24 to promote salt tolerance in cotton. Plant Physiol. 2024, 194, 1120–1138. [Google Scholar] [CrossRef]
- Fahad, M.; Tariq, L.; Muhammad, S.; Wu, L. Underground Communication: Long Non-Coding RNA Signaling in the Plant Rhizosphere. Plant Commun. 2024, 5, 100927. [Google Scholar] [CrossRef]
- Jin, X.; Wang, Z.; Li, X.; Ai, Q.; Wong, D.C.J.; Zhang, F.; Yang, J.; Zhang, N.; Si, H. Current Perspectives of LncRNAs in Abiotic and Biotic Stress Tolerance in Plants. Front. Plant Sci. 2023, 14, 1334620. [Google Scholar] [CrossRef]
- Numan, M.; Sun, Y.; Li, G. Exploring the Emerging Role of Long Non-Coding RNAs (LncRNAs) in Plant Biology: Functions, Mechanisms of Action, and Future Directions. Plant Physiol. Biochem. 2024, 212, 108797. [Google Scholar] [CrossRef]
- Yadav, A.; Mathan, J.; Dubey, A.K.; Singh, A. The Emerging Role of Non-Coding RNAs (NcRNAs) in Plant Growth, Development, and Stress Response Signaling. Non-Coding RNA 2024, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhao, H.; Niu, Y.; Wang, Y. Long Noncoding RNA from Betula platyphylla, BplncSIR1, Confers Salt Tolerance by Regulating BpNAC2 to Mediate Reactive Oxygen Species Scavenging and Stomatal Movement. Plant Biotechnol. J. 2023, 22, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Saeid, B.; Bhalla, P.L.; Singh, M.B. Identifying Long Non-Coding RNAs Involved in Heat Stress Response during Wheat Pollen Development. Front. Plant Sci. 2024, 15, 1344928. [Google Scholar] [CrossRef]
- Bai, Y.; He, J.; Yao, Y.; An, L.; Cui, Y.; Li, X.; Yao, X.; Xiao, S.; Wu, K. Identification and Functional Analysis of Long Non-Coding RNA (LncRNA) and Metabolites Response to Mowing in Hulless Barley (Hordeum vulgare L. Var. Nudum Hook. F.). BMC Plant Biol. 2024, 24, 666. [Google Scholar] [CrossRef]
- Magar, N.D.; Shah, P.; Barbadikar, K.M.; Bosamia, T.C.; Madhav, M.S.; Mangrauthia, S.K.; Pandey, M.K.; Sharma, S.; Shanker, A.K.; Neeraja, C.N.; et al. Long Non-Coding RNA-Mediated Epigenetic Response for Abiotic Stress Tolerance in Plants. Plant Physiol. Biochem. 2024, 206, 108165. [Google Scholar] [CrossRef]
- Weng, M.; Zhang, D.; Wang, H.; Yang, C.; Lin, H.; Pan, Y.; Lin, Y. Long Non-Coding RNAs and Their Potential Function in Response to Postharvest Senescence of Sparassis latifolia during Cold Storage. Sci. Rep. 2024, 14, 747. [Google Scholar] [CrossRef]
- Wang, X.; Fan, H.; Wang, B.; Yuan, F. Research Progress on the Roles of LncRNAs in Plant Development and Stress Responses. Front. Plant Sci. 2023, 14, 1138901. [Google Scholar] [CrossRef]
- Ackah, M.; Jin, X.; Zhang, Q.; Amoako, F.K.; Wang, L.; Attaribo, T.; Zhao, M.; Yuan, F.; Herman, R.A.; Qiu, C.; et al. Long Noncoding RNA Transcriptome Analysis Reveals Novel LncRNAs in Morus Alba “Yu-711” Response to Drought Stress. Plant Genome 2022, 17, e2027. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, Y.; He, R.; Jiang, L.; Qu, Z.; Gu, J.; Yang, J.; Legascue, M.F.; Wang, Z.-Y.; Ariel, F.; et al. LncRNA DANA1 Promotes Drought Tolerance and Histone Deacetylation of Drought Responsive Genes in Arabidopsis. EMBO Rep. 2024, 25, 796–812. [Google Scholar] [CrossRef]
- Cao, W.; Yang, L.; Zhuang, M.; Lv, H.; Wang, Y.; Zhang, Y.; Ji, J. Plant Non-Coding RNAs: The New Frontier for the Regulation of Plant Development and Adaptation to Stress. Plant Physiol. Biochem. PPB 2024, 208, 108435. [Google Scholar] [CrossRef]
- Imaduwage, I.; Hewadikaram, M. Predicted Roles of Long Non-Coding RNAs in Abiotic Stress Tolerance Responses of Plants. Mol. Hortic. 2024, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Traubenik, S.; Charon, C.; Blein, T. From Environmental Responses to Adaptation: The Roles of Plant LncRNAs. Plant Physiol. 2024, 195, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Lamin-Samu, A.T.; Zhuo, S.; Ali, M.; Lu, G. Long Non-Coding RNA Transcriptome Landscape of Anthers at Different Developmental Stages in Response to Drought Stress in Tomato. Genomics 2022, 114, 110383. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2010, 331, 76–79. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-Loop Stabilization Represses Antisense Transcription at the Arabidopsis FLC Locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef]
- Whittaker, C.; Dean, C. The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef]
- Wang, T.-Z.; Liu, M.; Zhao, M.-G.; Chen, R.; Zhang, W.-H. Identification and Characterization of Long Non-Coding RNAs Involved in Osmotic and Salt Stress in Medicago truncatula Using Genome-Wide High-Throughput Sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Y.; Mou, F.; Tian, Y.; Chen, L.; Zhang, S.; Qiong, J.; Li, X. Genome-Wide Small RNA Analysis of Soybean Reveals Auxin-Responsive MicroRNAs That Are Differentially Expressed in Response to Salt Stress in Root Apex. Front. Plant Sci. 2016, 6, 1273. [Google Scholar] [CrossRef]
- Khandal, H.; Parween, S.; Roy, R.; Meena, M.K.; Chattopadhyay, D. MicroRNA Profiling Provides Insights into Post-Transcriptional Regulation of Gene Expression in Chickpea Root Apex under Salinity and Water Deficiency. Sci. Rep. 2017, 7, 4632. [Google Scholar] [CrossRef]
- Unver, T.; Tombuloglu, H. Barley Long Non-Coding RNAs (LncRNA) Responsive to Excess Boron. Genomics 2019, 112, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Qian, N.; Gao, X.; Lang, X.; Deng, H.; Bratu, T.M.; Chen, Q.; Stapleton, P.; Yan, B.; Min, W. Rapid Single-Particle Chemical Imaging of Nanoplastics by SRS Microscopy. Proc. Natl. Acad. Sci. USA 2024, 121, e2300582121. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Barbosa, A. Chemical Reactivity Theory to Analyze Possible Toxicity of Microplastics: Polyethylene and Polyester as Examples. PLoS ONE 2024, 19, e0285515. [Google Scholar] [CrossRef] [PubMed]
- Ortega, D.E.; Cortés-Arriagada, D. Atmospheric Microplastics and Nanoplastics as Vectors of Primary Air Pollutants—A Theoretical Study on the Polyethylene Terephthalate (PET) Case. Environ. Pollut. 2023, 318, 120860. [Google Scholar] [CrossRef]
- Patil, P.B.; Maity, S.; Sarkar, A. Potential Human Health Risk Assessment of Microplastic Exposure: Current Scenario and Future Perspectives. Environ. Monit. Assess. 2022, 194, 898. [Google Scholar] [CrossRef]
- Dal Yöntem, F.; Aydoğan Ahbab, M. Mitochondria as a Target of Micro- and Nanoplastic Toxicity. Camb. Prism. Plast. 2024, 2, e6. [Google Scholar] [CrossRef]
- Djouina, M.; Loison, S.; Body-Malapel, M. Recent Progress in Intestinal Toxicity of Microplastics and Nanoplastics: Systematic Review of Preclinical Evidence. Microplastics 2024, 3, 217–233. [Google Scholar] [CrossRef]
- Eom, H.-J.; Nam, S.-E.; Rhee, J.-S. Polystyrene Microplastics Induce Mortality through Acute Cell Stress and Inhibition of Cholinergic Activity in a Brine Shrimp. Mol. Cell. Toxicol. 2020, 16, 233–243. [Google Scholar] [CrossRef]
- Naz, S.; Manan, A.; Khan, N.A.; Ullah, Q.; Zaman, F.; Qadeer, A.; Khan, I.B.; Danabas, D.; Kiran, A.; Skalickova, S.; et al. Unraveling the Ecotoxicological Effects of Micro and Nano-Plastics on Aquatic Organisms and Human Health. Front. Environ. Sci. 2024, 12, 1390510. [Google Scholar] [CrossRef]
- Aardema, H.; Vethaak, A.D.; Kamstra, J.H.; Legler, J. Farm Animals as a Critical Link between Environmental and Human Health Impacts of Micro-and Nanoplastics. Microplastics Nanoplastics 2024, 4, 5. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, J.; Zou, M.; Jia, Z.; Zhou, S.; Li, Y. Microplastics in Soils: A Review of Methods, Occurrence, Fate, Transport, Ecological and Environmental Risks. Sci. Total Environ. 2020, 748, 141368. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chen, H.; Liao, Y.; Ye, Z.; Li, M.; Klobučar, G. Ecotoxicity and Genotoxicity of Polystyrene Microplastics on Higher Plant Vicia faba. Environ. Pollut. 2019, 250, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Karalija, E.; Carbó, M.; Coppi, A.; Colzi, I.; Dainelli, M.; Gašparović, M.; Grebenc, T.; Gonnelli, C.; Papadakis, V.; Pilić, S.; et al. Interplay of Plastic Pollution with Algae and Plants: Hidden Danger or a Blessing? J. Hazard. Mater. 2022, 438, 129450. [Google Scholar] [CrossRef] [PubMed]
- Dainelli, M.; Castellani, M.B.; Pignattelli, S.; Falsini, S.; Ristori, S.; Papini, A.; Colzi, I.; Coppi, A.; Gonnelli, C. Growth, Physiological Parameters and DNA Methylation in Spirodela polyrhiza (L.) Schleid Exposed to PET Micro-Nanoplastic Contaminated Waters. Plant Physiol. Biochem. 2024, 207, 108403. [Google Scholar] [CrossRef]
- Falsini, S.; Colzi, I.; Chelazzi, D.; Dainelli, M.; Schiff, S.; Papini, A.; Coppi, A.; Gonnelli, C.; Ristori, S. Plastic Is in the Air: Impact of Micro-Nanoplastics from Airborne Pollution on Tillandsia usneoides (L.) L. (Bromeliaceae) as a Possible Green Sensor. J. Hazard. Mater. 2022, 437, 129314. [Google Scholar] [CrossRef]
- Dainelli, M.; Pignattelli, S.; Bazihizina, N.; Falsini, S.; Papini, A.; Baccelli, I.; Mancuso, S.; Coppi, A.; Castellani, M.B.; Colzi, I.; et al. Can Microplastics Threaten Plant Productivity and Fruit Quality? Insights from Micro-Tom and Micro-PET/PVC. Sci. Total Environ. 2023, 895, 165119. [Google Scholar] [CrossRef]
- Roy, T.; Dey, T.K.; Jamal, M. Microplastic/Nanoplastic Toxicity in Plants: An Imminent Concern. Environ. Monit. Assess. 2022, 195, 27. [Google Scholar] [CrossRef]
- Dainelli, M.; Colzi, I.; Giosa, D.; Gargiulo, G.; Passo, C.L.; Pernice, I.; Falsini, S.; Ristori, S.; Pignattelli, S.; Miniati, A.; et al. Coding and Non-Coding Transcripts Modulated by Transparent and Blue PET Micro-Nanoplastics in Arabidopsis thaliana. Plant Physiol. Biochem. 2025, 220, 109409. [Google Scholar] [CrossRef]
- Ciriminna, R.; Pagliaro, M. Biodegradable and Compostable Plastics: A Critical Perspective on the Dawn of Their Global Adoption. ChemistryOpen 2019, 9, 8–13. [Google Scholar] [CrossRef]
- Li, S.; Wang, T.; Guo, J.; Dong, Y.; Wang, Z.; Gong, L.; Li, X. Polystyrene Microplastics Disturb the Redox Homeostasis, Carbohydrate Metabolism and Phytohormone Regulatory Network in Barley. J. Hazard. Mater. 2021, 415, 125614. [Google Scholar] [CrossRef]
- Luo, H.; Liu, C.; He, D.; Sun, J.; Li, J.; Pan, X. Effects of Aging on Environmental Behavior of Plastic Additives: Migration, Leaching, and Ecotoxicity. Sci. Total Environ. 2022, 849, 157951. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.K.; Choudhary, D.; Vishwakarma, A.; Mudgal, M.; Srivastava, A.K.; Singh, A. Microplastics in Freshwater Environment: Occurrence, Analysis, Impact, Control Measures and Challenges. Int. J. Environ. Sci. Technol. 2022, 20, 6865–6896. [Google Scholar] [CrossRef]
- Ahmed, A.S.S.; Billah, M.M.; Ali, M.M.; Bhuiyan, M.K.A.; Guo, L.; Mohinuzzaman, M.; Hossain, M.B.; Rahman, M.S.; Islam, M.S.; Yan, M.; et al. Microplastics in Aquatic Environments: A Comprehensive Review of Toxicity, Removal, and Remediation Strategies. Sci. Total Environ. 2023, 876, 162414. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Li, T.; Li, M.; Yang, S.; Shen, M.; Liu, H. Research Advances on the Toxicity of Biodegradable Plastics Derived Micro/Nanoplastics in the Environment: A Review. Sci. Total Environ. 2024, 916, 170299. [Google Scholar] [CrossRef]
- Tsochatzis, E.D.; Gika, H.; Theodoridis, G.; Maragou, N.; Thomaidis, N.; Corredig, M. Microplastics and Nanoplastics: Exposure and Toxicological Effects Require Important Analysis Considerations. Heliyon 2024, 10, e32261. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Liang, X.; Lu, S.; Ren, J.; Zhang, Y.; Han, Z.; Gao, B.; Sun, K. Effects of Microplastics on Soil Carbon Pool and Terrestrial Plant Performance. Carbon Res. 2024, 3, 37. [Google Scholar] [CrossRef]
- Iqbal, S.; Xu, J.; Gui, H.; Bu, D.; Alharbi, S.A.; Khan, S.; Nadir, S. Interactive Effects of Microplastics and Typical Pollutants on the Soil-Plant System: A Mini-Review. Circ. Agric. Syst. 2024, 4, e007. [Google Scholar] [CrossRef]
- Bosker, T.; Bouwman, L.J.; Brun, N.R.; Behrens, P.; Vijver, M.G. Microplastics Accumulate on Pores in Seed Capsule and Delay Germination and Root Growth of the Terrestrial Vascular Plant Lepidium sativum. Chemosphere 2019, 226, 774–781. [Google Scholar] [CrossRef]
- Lian, J.; Wu, J.; Xiong, H.; Zeb, A.; Yang, T.; Su, X.; Su, L.; Liu, W. Impact of Polystyrene Nanoplastics (PSNPs) on Seed Germination and Seedling Growth of Wheat (Triticum aestivum L.). J. Hazard. Mater. 2020, 385, 121620. [Google Scholar] [CrossRef]
- Giorgetti, L.; Spanò, C.; Muccifora, S.; Bottega, S.; Barbieri, F.; Bellani, L.; Ruffini Castiglione, M. Exploring the Interaction between Polystyrene Nanoplastics and Allium cepa during Germination: Internalization in Root Cells, Induction of Toxicity and Oxidative Stress. Plant Physiol. Biochem. 2020, 149, 170–177. [Google Scholar] [CrossRef]
- De Silva, Y.; Rajagopalan, U.; Kadono, H. Microplastics on the Growth of Plants and Seed Germination in Aquatic and Terrestrial Ecosystems. Glob. J. Environ. Sci. Manag. 2021, 7, 347–368. [Google Scholar] [CrossRef]
- Colzi, I.; Renna, L.; Bianchi, E.; Castellani, M.B.; Coppi, A.; Pignattelli, S.; Loppi, S.; Gonnelli, C. Impact of Microplastics on Growth, Photosynthesis and Essential Elements in Cucurbita pepo L. J. Hazard. Mater. 2022, 423, 127238. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Engeseth, N.J. Comparison of Growth and Quality between Hydroponically Grown and Soil-Grown Lettuce under the Stress of Microplastics. ACS EST Water 2022, 2, 1182–1194. [Google Scholar] [CrossRef]
- Rillig, M.C.; Lehmann, A.; Souza Machado, A.A.; Yang, G. Microplastic Effects on Plants. New Phytol. 2019, 223, 1066–1070. [Google Scholar] [CrossRef]
- Azeem, I.; Adeel, M.; Ahmad, M.A.; Shakoor, N.; Jiangcuo, G.D.; Azeem, K.; Ishfaq, M.; Shakoor, A.; Ayaz, M.; Xu, M.; et al. Uptake and Accumulation of Nano/Microplastics in Plants: A Critical Review. Nanomaterials 2021, 11, 2935. [Google Scholar] [CrossRef]
- Ekner-Grzyb, A.; Duka, A.; Grzyb, T.; Lopes, I.; Chmielowska-Bąk, J. Plants Oxidative Response to Nanoplastic. Front. Plant Sci. 2022, 13, 1027608. [Google Scholar] [CrossRef]
- Ma, J.; Aqeel, M.; Khalid, N.; Nazir, A.; Alzuaibr, F.M.; Al-Mushhin, A.A.M.; Hakami, O.; Iqbal, M.F.; Chen, F.; Alamri, S.; et al. Effects of Microplastics on Growth and Metabolism of Rice (Oryza sativa L.). Chemosphere 2022, 307, 135749. [Google Scholar] [CrossRef]
- Sun, X.-D.; Yuan, X.-Z.; Jia, Y.; Feng, L.-J.; Zhu, F.-P.; Dong, S.-S.; Liu, J.; Kong, X.; Tian, H.; Duan, J.-L.; et al. Differentially Charged Nanoplastics Demonstrate Distinct Accumulation in Arabidopsis thaliana. Nat. Nanotechnol. 2020, 15, 755–760. [Google Scholar] [CrossRef]
- Lian, J.; Liu, W.; Sun, Y.; Men, S.; Wu, J.; Zeb, A.; Yang, T.; Ma, L.Q.; Zhou, Q. Nanotoxicological Effects and Transcriptome Mechanisms of Wheat (Triticum aestivum L.) under Stress of Polystyrene Nanoplastics. J. Hazard. Mater. 2021, 423, 127241. [Google Scholar] [CrossRef]
- Teng, L.; Zhu, Y.; Li, H.; Song, X.; Shi, L. The Phytotoxicity of Microplastics to the Photosynthetic Performance and Transcriptome Profiling of Nicotiana tabacum Seedlings. Ecotoxicol. Environ. Saf. 2022, 231, 113155. [Google Scholar] [CrossRef]
- Pehlivan, N.; Gedik, K. Coping with the Un-Natural: Tracking Transcriptional Activation and Macromolecular Profiles in Arabidopsis under Microplastic Exposure. Environ. Exp. Bot. 2022, 199, 104870. [Google Scholar] [CrossRef]
- Martín, C.; Pirredda, M.; Fajardo, C.; Costa, G.; Sánchez-Fortún, S.; Nande, M.; Mengs, G.; Martín, M. Transcriptomic and Physiological Effects of Polyethylene Microplastics on Zea mays Seedlings and Their Role as a Vector for Organic Pollutants. Chemosphere 2023, 322, 138167. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Han, Z.; Wang, Q.; Wang, T.; Sun, Z.; Yu, Y.; Han, X.; Yu, H. Toxicity Effects of Nanoplastics on Soybean (Glycine max L.): Mechanisms and Transcriptomic Analysis. Chemosphere 2023, 313, 137571. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, Y.; Yin, S.; Xiao, K.; Xiong, Q.; Bian, S.; Liang, S.; Hou, H.; Hu, J.; Yang, J. Metabolomics Revealing the Response of Rice (Oryza sativa L.) Exposed to Polystyrene Microplastics. Environ. Pollut. 2020, 266, 115159. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, X.; Wang, F.; Wang, Z.; Bian, Y.; Gu, C.; Wen, X.; Kengara, F.O.; Schäffer, A.; Jiang, X.; et al. Positively Charged Microplastics Induce Strong Lettuce Stress Responses from Physiological, Transcriptomic, and Metabolomic Perspectives. Environ. Sci. Technol. 2022, 56, 16907–16918. [Google Scholar] [CrossRef]
- Wang, Y.; Xiang, L.; Wang, F.; Redmile-Gordon, M.; Bian, Y.; Wang, Z.; Gu, C.; Jiang, X.; Schäffer, A.; Xing, B. Transcriptomic and Metabolomic Changes in Lettuce Triggered by Microplastics-Stress. Environ. Pollution 2023, 320, 121081. [Google Scholar] [CrossRef]
- Yu, Z.; Xu, X.; Guo, L.; Yuzuak, S.; Lu, Y. Physiological and Biochemical Effects of Polystyrene Micro/Nano Plastics on Arabidopsis thaliana. J. Hazard. Mater. 2024, 469, 133861. [Google Scholar] [CrossRef]
- Conn, S.J.; Hocking, B.; Dayod, M.; Xu, B.; Athman, A.; Henderson, S.; Aukett, L.; Conn, V.; Shearer, M.K.; Fuentes, S.; et al. Protocol: Optimising Hydroponic Growth Systems for Nutritional and Physiological Analysis of Arabidopsis thaliana and Other Plants. Plant Methods 2013, 9, 4. [Google Scholar] [CrossRef]
- Ekvall, M.T.; Lundqvist, M.; Kelpsiene, E.; Šileikis, E.; Gunnarsson, S.B.; Cedervall, T. Nanoplastics Formed during the Mechanical Breakdown of Daily-Use Polystyrene Products. Nanoscale Adv. 2019, 1, 1055–1061. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: www.bioinformatics.babraham.ac.uk (accessed on 1 September 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dainat, J. AGAT: Another Gff Analysis Toolkit to Handle Annotations in Any GTF/GFF Format (Version v1.4.0) Zenodo. 2024. Available online: https://www.doi.org/10.5281/zenodo.3552717 (accessed on 1 September 2024).
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene Structure and Function Annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Amatria-Barral, I.; González-Domínguez, J.; Touriño, J. PRIblast: A Highly Efficient Parallel Application for Comprehensive LncRNA–RNA Interaction Prediction. Future Gener. Comput. Syst. 2022, 138, 270–279. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 46. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, gkad347. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Chao, J.; Kong, Y.; Wang, Q.; Sun, Y.; Gong, D.; Lv, J.; Liu, G. MapGene2Chrom, a Tool to Draw Gene Physical Map Based on Perl and SVG Languages. PubMed 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.K.; Bhanu, D.; Maheswari, M. Epigenetics and Transgenerational Memory in Plants under Heat Stress. Plant Physiol. Rep. 2020, 25, 583–593. [Google Scholar] [CrossRef]
- Mladenov, V.; Fotopoulos, V.; Kaiserli, E.; Karalija, E.; Maury, S.; Baranek, M.; Segal, N.; Testillano, P.S.; Vassileva, V.; Pinto, G.; et al. Deciphering the Epigenetic Alphabet Involved in Transgenerational Stress Memory in Crops. Int. J. Mol. Sci. 2021, 22, 7118. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Bhukya, D.P.N.; Sunkar, R.; Chavali, S.; Allu, A.D. Molecular Basis of Priming-Induced Acquired Tolerance to Multiple Abiotic Stresses in Plants. J. Exp. Bot. 2022, 73, 3355–3371. [Google Scholar] [CrossRef]
- Gao, S.; Wang, J.; Jiang, N.; Zhang, S.; Wang, Y.; Zhang, J.; Li, N.; Fang, Y.; Yang, L.; Chen, S.; et al. Hyponastic Leaves 1 Protects Pri-MiRNAs from Nuclear Exosome Attack. Proc. Natl. Acad. Sci. USA 2020, 117, 17429–17437. [Google Scholar] [CrossRef]
- Gao, F.; Chen, J.; Ma, T.; Li, H.; Wang, N.; Li, Z.; Zhang, Z.; Zhou, Y. The Glutathione Peroxidase Gene Family in Thellungiella salsuginea: Genome-Wide Identification, Classification, and Gene and Protein Expression Analysis under Stress Conditions. Int. J. Mol. Sci. 2014, 15, 3319–3335. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Wang, B.; Hou, Y.; Qin, L.; Hou, B.-K. The ArabidopsisUGT87A2, a Stress-Inducible Family 1 Glycosyltransferase, Is Involved in the Plant Adaptation to Abiotic Stresses. Physiol. Plant. 2016, 159, 416–432. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Mühlenbock, P.; Van Breusegem, F. Stress Homeostasis—The Redox and Auxin Perspective. Plant Cell Environ. 2011, 35, 321–333. [Google Scholar] [CrossRef]
- Husar, S.; Berthiller, F.; Fujioka, S.; Rozhon, W.; Khan, M.; Kalaivanan, F.; Elias, L.; Higgins, G.; Li, Y.; Schuhmacher, R.; et al. Overexpression of the UGT73C6 Alters Brassinosteroid Glucoside Formation in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Islam, S.; Griffiths, C.A.; Blomstedt, C.K.; Le, T.-N.; Gaff, D.F.; Hamill, J.D.; Neale, A.D. Increased Biomass, Seed Yield and Stress Tolerance Is Conferred in Arabidopsis by a Novel Enzyme from the Resurrection Grass Sporobolus stapfianus That Glycosylates the Strigolactone Analogue GR24. PLoS ONE 2013, 8, e80035. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.; Ross, A.R.S.; Bowles, D.J. Use of the Glucosyltransferase UGT71B6 to Disturb Abscisic Acid Homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Qiao, Z.; Wang, H.; Zhu, L.; Zhang, L. Flavonoids: Promising Anticancer Agents. Med. Res. Rev. 2003, 23, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Meßner, B.; Thulke, O.; Schäffner, A.R. Arabidopsis Glucosyltransferases with Activities toward Both Endogenous and Xenobiotic Substrates. Planta 2003, 217, 138–146. [Google Scholar] [CrossRef] [PubMed]

| Differentially Expressed Genes | Tr-PET vs. Ctrl | Bl-PET vs. Ctrl | Bl-PET vs. Tr-PET | ||||||
| Up | Down | Total | Up | Down | Total | Up | Down | Total | |
| Total | 1152 | 950 | 2102 | 266 | 161 | 427 | 219 | 334 | 553 |
| Kornienko plus novel identified lncRNAs | 61 | 43 | 104 | 10 | 9 | 19 | 6 | 8 | 14 |
| Novel lncRNAs | 7 | 4 | 11 | 3 | 0 | 3 | 1 | 2 | 3 |
| Differentially Expressed Isoforms | Tr-PET vs. Ctrl | Bl-PET vs. Ctrl | Bl-PET vs. Tr-PET | ||||||
| Up | Down | Total | Up | Down | Total | Up | Down | Total | |
| Total | 2328 | 1994 | 4322 | 821 | 598 | 1419 | 784 | 813 | 1597 |
| Kornienko plus novel identified lncRNAs | 156 | 114 | 270 | 50 | 43 | 93 | 54 | 54 | 108 |
| Novel lncRNAs | 10 | 8 | 18 | 3 | 4 | 7 | 1 | 2 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galbo, R.; Giosa, D.; Gargiulo, G.; Bonomo, A.; Basso, M.F.; Negussu, M.; Giovino, A.; Vergata, C.; Colzi, I.; Gonnelli, C.; et al. lncRNA-mRNA-miRNA Networks in Arabidopsis thaliana Exposed to Micro-Nanoplastics. Int. J. Plant Biol. 2025, 16, 70. https://doi.org/10.3390/ijpb16020070
Galbo R, Giosa D, Gargiulo G, Bonomo A, Basso MF, Negussu M, Giovino A, Vergata C, Colzi I, Gonnelli C, et al. lncRNA-mRNA-miRNA Networks in Arabidopsis thaliana Exposed to Micro-Nanoplastics. International Journal of Plant Biology. 2025; 16(2):70. https://doi.org/10.3390/ijpb16020070
Chicago/Turabian StyleGalbo, Roberta, Domenico Giosa, Gaetano Gargiulo, Andrea Bonomo, Marcos Fernando Basso, Miriam Negussu, Antonio Giovino, Chiara Vergata, Ilaria Colzi, Cristina Gonnelli, and et al. 2025. "lncRNA-mRNA-miRNA Networks in Arabidopsis thaliana Exposed to Micro-Nanoplastics" International Journal of Plant Biology 16, no. 2: 70. https://doi.org/10.3390/ijpb16020070
APA StyleGalbo, R., Giosa, D., Gargiulo, G., Bonomo, A., Basso, M. F., Negussu, M., Giovino, A., Vergata, C., Colzi, I., Gonnelli, C., Dainelli, M., Martinelli, F., & Giuffrè, L. (2025). lncRNA-mRNA-miRNA Networks in Arabidopsis thaliana Exposed to Micro-Nanoplastics. International Journal of Plant Biology, 16(2), 70. https://doi.org/10.3390/ijpb16020070












