Abstract
Wheat quality and quantity are challenged by increasing global temperature, which poses an urgent need for heat tolerance breeding in wheat. The identification of seedling-stage factors highly associated with reproductive-stage performance can enable early-stage selection and enhance the efficiency and effectiveness of breeding. This study investigated the myeloblastosis (MYB) gene family, one of the largest transcription factor (TF) gene families in plants, for its response to seedling- and reproductive-stage heat stress in wheat. Genome-wide analysis of MYB TF genes identified 876 TaMYB genes, and 48 genes were selected for qRT-PCR expression analysis based on in silico expression analysis under abiotic stresses. Correlation analysis of the quantitative real-time polymerase chain reaction (qRT-PCR) expression pattern of selected TaMYB genes in a heat-tolerant genotype (Perenjori) and two heat-sensitive genotypes (Brazil32 and Yitpi) at the seedling stage and grain-filling stage identified five TaMYB genes (TaMYB-327, TaMYB-049, TaMYB-030, TaMYB-226, and TaMYB-023) for the early-stage selection of heat tolerance and four TaMYB genes (TaMYB-232, TaMYB-343, TaMYB-305, and TaMYB399) for the early-stage selection of heat sensitivity in wheat. As important stress-responsive genes, these MYB genes showed similar expression patterns between early and late developmental stages, indicating the existence of a correlation for heat tolerance at the two stages, and therefore providing the theoretical basis for the early selection of heat tolerance in wheat.
1. Introduction
Wheat production is largely affected by heat stress (HS) at both seedling and grain-filling stages. Heat stress significantly impacts plant growth and development through damage in photosynthetic machinery, membrane permeability, protein dysfunction, ROS production, and increased oxidative stress in plant cells [,]. At the same time, a 2 °C increase in average temperature during the wheat-growing season is estimated to reduce grain production by 50% in Australia []. To mitigate the problem of rising temperature in wheat production, heat tolerance breeding in wheat is needed. Early-stage selection (ESS) can increase the efficiency and effectiveness of the breeding process. A good understanding of the correlation between the traits at the seedling stage and reproductive stage is crucial for determining the opportunity of ESS in plant breeding []. Morphologically heat-tolerant seedlings of wheat produced a higher yield at the adult stage compared to susceptible seedlings []. ESS is also effective in indirect selection at the seedling stage for difficult-to-measure traits at the mature stage. At the seedling stage, HS affects seedling establishment due to higher seedling mortality [], restricted nutrient absorption [], disruption in leaf growth, which limits the photosynthetic area [], and damage of vascular tissues, which results in stunted growth and subsequent stem collapse in plants []. Heat-tolerant genotypes can be distinguished from sensitive ones using various phenotypic traits, such as canopy temperature, biomass, chlorophyll content, chlorophyll fluorescence, and their heat tolerance index [,]. However, seedling root length showed significant variation under HS in contrasting wheat genotypes []. Similarly, HS at the grain-filling stage is critical for grain production. It causes oxidative damage to photosynthetic machinery, reduces photosynthesis and ATP production, and accelerates the grain-filling rate, reducing grain yield [,]. Heat-tolerant genotypes can be distinguished using traits that have strong correlation with grain yield, such as chlorophyll content, canopy temperature, flag leaf senescence, etc. The grain yield is the major economic product; thus, measuring the effect of HS on grain yield itself can be a suitable phenotypic selection trait for adult-stage heat tolerance. A strong positive correlation was reported between root length at the seedling stage and grain yield at the adult stage under HS in wheat based on the evaluation of the seedling damage index and adult damage index in wheat genotypes [], which paves the way for using the morphological trait ESS for heat tolerance breeding in wheat. However, no genetic factors for ESS have been reported in wheat for heat tolerance breeding. Plants undergo multiple cellular, physiological, and molecular changes in response to HS by signal transduction, osmolyte synthesis, antioxidant defense, and expression of the HS-associated gene []. These responses are produced by activating different stress regulators, such as transcription factors (TFs), which regulate rapid signaling pathways to enhance stress acclimation in plants []. The induction of the signaling cascade by TFs leading to profound changes in the expression of stress-related genes is considered to play an important role in HS adaptation []. The advancement in omics-based approaches has facilitated the understanding of mechanism of HS in plants, and further research on stress-adaptive transcription factors and HSPs is required to develop resistant cultivars []. MYB TFs play a crucial role in the heat tolerance mechanism by regulating the calcium signaling pathway, where the absence of MYB TF increases the [Ca2+]cyst under HS and enhances HS tolerance []. In addition, the mechanism of MYB TF in regulating different biotic and abiotic stresses has broadly been discussed [,]. The MYB TF family gene is one of the largest TF family genes controlling the plant response to several biotic and abiotic stresses, development, differentiation, defense, and metabolism [,]. The heat stress response is regulated by a complex network of TFs in plants, and MYB TFs are integral components of the heat-induced signaling pathway and hold great potential for developing heat tolerance []. The first plant MYB gene was isolated from maize, which encoded c-MYB-like TF and was functionally associated with anthocyanin biosynthesis [], and was studied in different species, including model plants Arabidopsis and rice []. The transcriptome analysis of the heat-tolerant genotype Raj3765 revealed an abundance of MYB TF genes playing a role in thermotolerance []. The heat-induced expression of MYB TF genes has also been reported in several plant species: AtMYB68, AtMYB30, and LiMYB305 in transgenic Arabidopsis [,,]; Os-MYB55 and OsMYB1 in rice [,]; LeAN2 in transgenic tomato []; BnaMYB111L in tobacco []; and BdMYB056 and BdMYB091 in Brachypodium [].
In wheat (Triticum aestivum L.), ref. [] identified 66 TaMYB genes, and overexpression of TaMYB32 and TaMYB80 genes enhanced salt, heat, and drought tolerance []. Enhanced expression of MYB TF genes due to HS at 37 °C and 42 °C for 2 h was observed in developing seeds of a moderately heat-tolerant wheat genotype []. Similarly, ref. [] identified 393 R2R3-MYB and 12 R1R2R3-MYB genes in wheat, and the overexpression of the TaMYB344 gene enhanced drought, heat, and salt tolerance. Recently, ref. [] identified 719 putative TaMYB genes in wheat, with nine target genes regulated by TaMYB for salt tolerance. Also, previous transcriptomic research conducted in our lab found that, among 60 TF families, MYB and MYB-related TFs were the top TF families responsible for abiotic stress defense in wheat []. However, a comparative analysis of the heat-induced expression of TaMYB TF genes in contrasting wheat genotypes has not been studied so far.
Early-stage selection for physiological traits has been proved to be cost-effective for increasing genetic gain in breeding populations []. Several studies have focused on morphological and physiological traits, such as canopy temperature [], whereas minimal effort has been reported in the identification and evaluation of the molecular and genetic traits for ESS in wheat. Therefore, investigation for feasible TF genes in wheat seedlings showing strong association with adult-stage performance under HS can increase the precision and speed of selection for a higher genetic gain in wheat breeding.
In this study, MYB TF genes were identified and characterized in the wheat genome and the qRT-PCR expression of TaMYB genes was analyzed in contrasting wheat genotypes at seedling and grain-filling stages under HS. The objectives of this study were (a) to evaluate the performance of heat-tolerant and -sensitive genotypes at the seedling and grain-filling stage under HS; (b) to understand the TaMYB gene expression pattern between contrasting genotypes; (c) to evaluate the correlation of expression patterns of TaMYB TF genes at seedling and grain-filling stages; and (d) to identify candidate genes for ESS for heat tolerance and sensitivity in wheat.
2. Materials and Methods
2.1. Identification and Characterization of MYB TF Genes in Wheat
The wheat reference genome and protein sequences were downloaded from the International Wheat Genome Sequencing Consortium website (IWGSC RefSeq v1.1, https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies, accessed on 17 July 2020) []. The latest hidden Markov model (HMM) profile of the MYB and MYB-related domains (PF00249, PF13921, PF14215, PF11831, and PF14379), downloaded from the protein family (Pfam) database (http://pfam.xfam.org/family/, accessed on 17 July 2020) [], were used as a query sequence for HMM search using HMMER3.1b2 software (http://hmmer.org/) in the wheat reference protein sequence version 1.1 (http://plants.ensembl.org/Triticum_aestivum/Info/Index), with an E-value threshold of E < 1 × 10−5.
2.2. Selection of Key TaMYB Genes for Expression Analysis
Putative MYB genes identified were grouped into different sub-families based on the NCBI-CD search (https://www.ncbi.nlm.nih.gov/cdd, accessed on 17 July 2020), and the number of genes was reduced using the Treetrimmer principle []. The reduced number of MYB genes were characterized by their phylogenetic relationship (MEGAX) [], conserved motif composition (MEME online tool) [], gene structure (GSDS) [], chromosomal localization (MapChart 2.32) [], and gene duplication (sequence identity and similarity program) followed by the selection of abiotic-stress-responsive genes based on in silico expression analysis (wheat expression database, https://www.wheat-expression.com/) []. Gene-based primers were validated for specificity using agarose gel electrophoresis for single bands and single melting curves in RT-qPCR.
2.3. Plant Materials and Heat Stress Treatment
2.3.1. Seedling Stage Heat Treatment
A heat treatment experiment was conducted using a completely randomized design with three biological replicates. The seeds of three wheat genotypes—one heat-tolerant genotype (Perenjori) and two heat-sensitive genotypes (Brazil32 and Yitpi) identified by []—were obtained from Australian Winter Cereals Collection. Surface-sterilized seeds were germinated in room temperature (25 ± 1 °C) under dark conditions for 36 h, and germinated seeds with a protruding coleoptile and radicle were transferred to a plastic folder lined with sterile black calico cloth at 35 ± 1 °C for HS treatment and at 25 ± 1 °C for the control, in a semi-hydroponic system []. The seedlings after three days of heat treatment were used for morphological trait (root depth) measurement followed by sampling the whole seedling (snap frozen) for storage at −80 °C for RNA extraction.
2.3.2. Post-Anthesis Heat Treatment
The three wheat genotypes used for the seedling stage experiment were grown in pots (8 cm × 8 cm × 16 cm) containing a potting mixture (5:2:3 compost: peat: sand, pH 6.0) in a naturally lit glasshouse at The University of Western Australia, Crawley, Western Australia (31°59′ S, 115°49′ E). Twelve pots per genotype (three biological replicates, two time points, and two treatments (one as the control and another as HS)) were grown in two sets (set A for flag leaf sampling and set B for harvesting to measure yield parameters at maturity) using a completely randomized block design. A single plant per pot was maintained and regularly watered and fertilized fortnightly with “Dimond red” (Campbells Fertilisers Australasia Pty Ltd., Laverton North, VIC, Australia) from four weeks after sowing until the end of the grain-filling period.
At anthesis, each plant was tagged on wheat heads to record the anthesis date. At the 10th day post-anthesis (DPA), 12 pots were shifted to a control environment room (CER) for heat treatment set at 37/27 °C (day/night) with a 14 h photoperiod and 420 mmol m-2s-1 light intensity until 11DPA and 13DPA, while the other 12 pots were left in a glasshouse to grow as the control. The flag leaf was sampled and snap frozen in liquid nitrogen for molecular analysis after heat treatment at 11DPA and 13DPA. The pots containing plants from set B, one with the flag leaf intact, were returned to the glasshouse after each treatment and grown until physiological maturity. Yield parameters such as the grain number of the main tiller and grain weight of the main tiller (g) were measured at harvest.
2.3.3. Statistical Analysis
Morphological data were analyzed by using an analysis of variance (ANOVA) test for both root depth (RD) at the seedling stage and thousand-kernel weight (TKW) at the reproductive stage followed by the Tukey HSD test. The heat damage index (HDI) was used to compare measurements under HS and under control conditions. The damage index (DI) due to HS was calculated as follows:
- (1)
- Root depth heat damage index (RD_HDI) = (root depth under control condition − root depth under HS condition)/root depth under control condition.
- (2)
- Thousand-kernel weight heat damage index (TKW_HDI) = (TKW under control condition -TKW under HS condition)/TKW under control condition.
2.4. RT-qPCR Expression Analysis at Seedling and Grain-Filling Stage
The total RNA was extracted from whole seedlings and the flag leaf using the RNeasy Plant Mini Kit (Qiagen) with an on-column DNase digestion with RNase-free DNase I (Qiagen). The extracted total RNA was quantified using Qubit RNA BR Assay kit in a Qubit 3.0 Fluorometer (Invitrogen by life technologies Ref: Q33216) and the integrity was tested using 1.5% agarose gel. The quantified RNA was used for the cDNA synthesis using a SensiFAST cDNA Synthesis Kit from Meridian Bioscience (BIO-65054) following the kit protocol.
A real-time quantitative polymerase chain reaction (RT-qPCR) for validated gene-linked primers (Supplementary Table S7) was conducted with a SensiFAST SYBR Lo-ROX mix on an Applied Biosystem 7500/7500 Fast Real-Time PCR System using the same protocol from our previous experiment []. Each sample was analyzed in three biological replications and two technical repeats with β-actin as a housekeeping gene. Gene expression values calculated by using the 2−∆∆CT method [] were used to calculate the fold change (FC) for each gene by comparing the expression under treatment with the control for both tolerant and sensitive genotypes. Genes were considered significantly up- or downregulated by two folds when their Log2FC values were ≥1 and ≤−1, respectively [].
3. Results
3.1. Identification and Characterization of TaMYB TF Genes
A total of 1896 putative homologs of MYB genes (1082 from PF00249; 620 from PF13921; 97 from PF14215; 89 from PF14379; and 8 from PF11831) were identified in the wheat genome. Among these 1896, 876 (825 from PF00249, 48 from PF14215, and 3 from PF14379) non-redundant putative homologous genes were categorized into four sub-families based on the gene annotation and number of domains identified from the NCBI-CD search. They are as follows: (1) TaMYB-1R sub-family: 252 TaMYB genes with one domain; (2) TaMYB-2R sub-family: 542 TaMYB genes with two domains, also known as TaMYB-R2R3 genes; (3) TaMYB-3R sub-family: 62 TaMYB genes with three domains, also known as TaMYB-R1R2R3 genes; and (4) TaMYB-4R sub-family: 20 TaMYB genes with four domains (Supplementary Table S1). We also identified 159 TaMYB (21 TaMYB-1R, 75 TaMYB-2R, 45 TaMYB-3R, and 18 TaMYB-4R) unique genes compared to the findings of [] (Supplementary Table S2). Among the 876 TaMYB TF genes, 409 MYB TF genes representing the whole MYB gene family were selected using Treetrimmer for further analysis (Supplementary Table S3). Phylogenetic analysis categorized these genes into four distinct clusters of TaMYB-1R (42 genes), TaMYB-2R (332 genes), TaMYB-3R (31 genes), and TaMYB-4R (4 genes) sub-families (Figure 1).
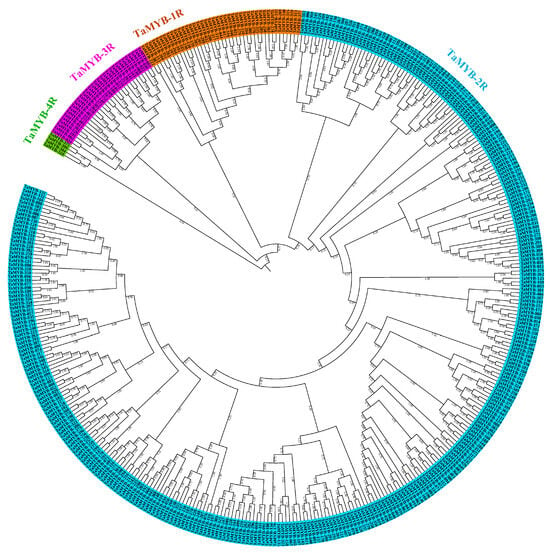
Figure 1.
Phylogenetic tree of 409 TaMYB TF genes in wheat. The unrooted neighbor-joining tree was constructed using a full-length protein sequence of 409 TaMYB TF genes in MEGA X software with a bootstrap test of 1000 replications, which classified these genes into four sub-families: TaMYB-1R (42 TF genes in orange color); TaMYB-2R (332 TF genes in blue color); TaMYB-3R (31 TF genes in purple color); and TaMYB-4R sub-family (4 TF genes in green color).
Further, gene structure analysis with the Gene Structure Display Server (GSDS) online server identified 41 intronless TF genes and 4 TF genes (TaMYB-250, TaMYB-375, TaMYB-304, and TaMYB-272) with the highest number (14) of introns. Two genes, TaMYB-272 and TaMYB-399, were found to have the highest number of exons (14 CDS), while fifty TF genes contained only a single exon (Supplementary Figure S1). Similarly, conserved motif analysis using the MEME online tool showed that the number of motifs ranged from 1 to 13 per TF gene. Motif 10 (200 amino acids) was the longest motif, while motif 7 and motif 8 were the shortest, with 12 amino acids. Motif 3 (16 amino acids long) was the most frequent among the 369 sites, while motif 10 (200 amino acids long) and motif 15 (182 amino acids long) were the least frequent, present only in three sites as shown in Supplementary Figure S1. Details of the conserved motifs discovered in the 409 TaMYB TF genes are given in Supplementary Table S4.
3.2. Chromosomal Distribution and Gene Duplication of TaMYB TF Genes
The selected 409 TaMYB genes (137 genes in sub-genome A, 134 genes in sub-genome B, and 134 genes in sub-genome D) were unevenly distributed among all 21 chromosomes of wheat, ranging from 25 on chromosome 1B and 2D to 14 on chromosome 5D and 1 gene on an unknown position, as visualized with MapChart (Figure 2). Gene duplication analysis showed that 66 pairs of genes were segmental duplicates and that no tandem duplicates were present. The highest sequence similarity and identity of 99.7% was present between segmental duplicated gene pair TaMYB-386 and TaMYB-371, present on chromosome 7B and 7A, respectively, closely followed by the similarity of 99.6% and identity of 98.6% between the two gene pairs of TaMYB-287 and TaMYB-267 and TaMYB-295 and TaMYB-281, both present on chromosome 5 (Figure 2 and Supplementary Table S5).

Figure 2.
Chromosomal distribution of 409 TaMYB TF genes on 21 wheat chromosomes and an unknown chromosome generated in MapChart software. The relative positions of genes are displayed on the left side of each chromosome and the name of the genes is on the right. The number of genes per chromosome ranged from 25 on chromosomes 1B and 2D to 1 on an unknown position.
3.3. In Silico Expression Profiling of TaMYB TF Genes
An in silico gene expression pattern analysis of 409 TaMYB genes under abiotic stress using the wheat expression database showed that the genes were grouped into nine major clusters (Supplementary Figure S2). The number of genes grouped in the clusters ranged from 2 genes in cluster 7 to 254 genes in cluster 9. The details of the genes in each group are given in Supplementary Table S6.
The exon-spanning primers developed for 48 TaMYB TF genes representing all four sub-families (6 from TaMYB-1R, 31 from TaMYB-2R, 8 from TaMYB-3R, and 3 from TaMYB-4R sub-families) are given in Supplementary Table S7.
3.4. Morphological Response of Wheat Genotypes Under HS
The morphological assessment of heat-treated wheat seedlings showed a significant interaction effect in Anova analysis, with a p-value of 0.000, indicating significant damage in the root growth of both heat-tolerant (Perenjori) and heat-sensitive genotypes (Brazil32 and Yitpi) (Supplementary Figure S3). However, the root depth heat damage index (RD_HDI) calculated in Perenjori was 41%, showing less damage compared to the 59% in Yitpi and 60% in Brazil32 under the HS condition, as shown in Figure 3a. The RD_HDI was almost similar in both the sensitive genotypes Brazil32 and Yitpi at seedling-stage HS, showing a similar level of susceptibility.
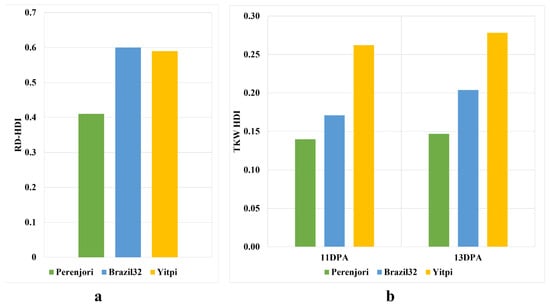
Figure 3.
Differences in heat damage index (HDI) between heat-tolerant genotype (Perenjori in green color) and heat-susceptible genotypes (Brazil32 in blue color and Yitpi in yellow color) at (a) seedling and (b) grain-filling stages. The root depth heat damage index (RD_HDI) at seedling stage shows a similar level of damage in Brazil32 and Yitpi (a), while thousand-kernel weight heat damage index (TKW_HDI) at 11DPA and 13DPA shows a difference in the level of damage (b). The RD_HDI and TKW_HDI are lower in Perenjori both at seedling and grain-filling stages under heat stress compared to Brazil32 and Yitpi.
Similarly, yield parameters measured as grain number and grain weight at 11DPA and 13DPA were used to calculate thousand-kernel weight (TKW) at the reproductive stage (Supplementary Figure S4). Anova analysis showed significant main effects with a p-value lower than 0.000 and a non-significant interaction effect with a p-value of 0.723, while the Tukey HSD test revealed that TKW was significantly reduced in Brazil32 and Yitpi due to HS, while a non-significant difference was observed in Perenjori between HS and the control. A reduction in TKW due to HS showed a slightly higher reduction at 13DPA compared to 11DPA in all three genotypes, while the extent of damage was higher in sensitive genotypes compared to the tolerant genotype as shown in Figure 3b. The TKW heat damage index (TKW_HDI) at 11DPA was 14% in Perenjori, 17% in Brazil32, and 26% in Yitpi, which increased to 15% in Perenjori, 20% in Brazil32, and 28% in Yitpi at 13DPA HS. The sensitive genotype Yitpi showed a higher TKW_HDI compared to Brazil 32 at both 11DPA and 13DPA, showing a higher susceptibility to HS.
3.5. Quantitative Gene Expression Analysis Using RT-qPCR
RT-qPCR expression analysis of 48 TaMYB TF gene-linked markers (Supplementary Table S7) under the HS condition showed a differential expression pattern of genes with growth stages and genotypes.
3.5.1. Differential Gene Expression at Seedling Stage
RT-qPCR expression analysis of 48 TaMYB TF genes in the heat-stressed wheat seedlings showed 40 differential expressed genes (DEG), while the remaining 8 TF genes were not significantly expressed. Some genes were significantly differentially expressed only in Perenjori or Brazil32 or Yitpi, while some were commonly expressed in all three genotypes. The total number of DEGs in Perenjori, Brazil32, and Yitpi was 20, 24, and 14, respectively. There were 14 DEGs (TaMYB-023, TaMYB-014, TaMYB-206, TaMYB049, TaMYB-300, TaMYB-267, TaMYB-030, TaMYB-405, TaMYB-327, TaMYB-157, TaMYB-211, TaMYB-296, TaMYB-226, and TaMYB-304) uniquely expressed in the heat-tolerant genotype (Perenjori), showing heat tolerance at the seedling stage. Among these 14 DEGs, 5 were upregulated and 11 were downregulated. Ten DEGs (TaMYB-286, TaMYB-232, TaMYB-343, TaMYB-375, TaMYB-305, TaMYB-399, TaMYB-363, TaMYB-369, TaMYB-356, and TaMYB-272) were commonly expressed in the sensitive genotypes (Brazil32 and Yitpi), showing heat sensitivity at the seedling stage. Among these 10 DEGs, 2 were upregulated and 8 were downregulated in both genotypes. In addition, nine DEGs were uniquely expressed in Brazil32 and one in Yitpi. Three DEGs were common between Perenjori and Brazil32, one between Perenjori and Yitpi, and two among all three genotypes (Figure 4a).
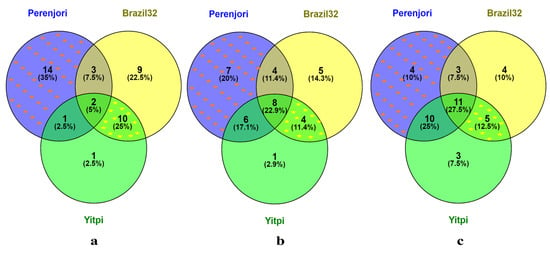
Figure 4.
Venn diagram of DEGs showing heat-tolerant genotype (Perenjori) and two heat-susceptible genotypes (Brazil32 and Yitpi) at (a) seedling, (b) 11DPA, and (c) 13DPA. At the seedling stage, 14 DEGs (TaMYB-023, TaMYB-014, TaMYB-206, TaMYB049, TaMYB-300, TaMYB-267, TaMYB-030, TaMYB-405, TaMYB-327, TaMYB-157, TaMYB-211, TaMYB-296, TaMYB-226, and TaMYB-304) were uniquely expressed in Perenjori (highlighted with orange stars), and 10 DEGs (TaMYB-286, TaMYB-232, TaMYB-343, TaMYB-375, TaMYB-305, TaMYB-399, TaMYB-363, TaMYB-369, TaMYB-356, and TaMYB-272) were commonly expressed between Brazil32 and Yitpi (highlighted with yellow stars). At 11DPA, seven DEGs (TaMYB-292, TaMYB-049, TaMYB-308, TaMYB-030, TaMYB-327, TaMYB-299, and TaMYB-226) were uniquely expressed in Perenjori (highlighted with orange stars), and four DEGs (TaMYB-232, TaMYB-343, TaMYB-305, and TaMYB-140) were commonly expressed between Brazil32 and Yitpi (highlighted with yellow stars). At 13DPA, four DEGs (TaMYB-209, TaMYB-023, TaMYB-232, and TaMYB-327) were uniquely expressed in Perenjori (highlighted with orange stars), and five DEGs (TaMYB-300, TaMYB-382, TaMYB-399, TaMYB-377, and TaMYB-245) were commonly expressed between Brazil32 and Yitpi (highlighted with yellow stars).
3.5.2. Differential Gene Expression at Grain-Filling Stage
RT-qPCR expression analysis of 48 TaMYB TF genes at 11DPA showed a differential expression of 34 TF genes, while the remaining 14 TF genes were not expressed significantly. The total number of DEGs in Perenjori, Brazil32, and Yitpi were 25, 21, and 19, respectively. Seven DEGs (TaMYB-292, TaMYB-049, TaMYB-308, TaMYB-030, TaMYB-327, TaMYB-299, and TaMYB-226) were uniquely expressed in Perenjori, showing heat tolerance at 11DPA. Among the seven DEGs, four were upregulated and three were downregulated. On the other hand, four DEGs (TaMYB-232, TaMYB-343, TaMYB-305, and TaMYB-140) were commonly expressed between Brazil32 and Yitpi, showing heat sensitivity at 11DPA. Among these four DEGs, two DEGs were downregulated and one DEG was upregulated in both Brazil32 and Yitpi, while one DEG was upregulated in Brazil32 and downregulated in Yitpi. Five unique DEGs were shown in Brazil32 and one in Yitpi. Four DEGs were common between Perenjori and Brazil32, six between Perenjori and Yitpi, and eight among all three genotypes (Figure 4b).
At 13DPA, 40 DEGs out of 48 TaMYB genes were detected, while the remaining 8 genes were not expressed significantly. The total number of DEGs in Perenjori, Brazil32, and Yitpi was 28, 23, and 29, respectively. Four DEGs (TaMYB-209, TaMYB-023, TaMYB-232, and TaMYB-327) were uniquely expressed in Perenjori, showing heat tolerance at 13DPA. Among them, three DEGs were upregulated and one DEG was downregulated. Five DEGs (TaMYB-300, TaMYB-382, TaMYB-399, TaMYB-377, and TaMYB-245) were commonly expressed between Brazil32 and Yitpi, showing heat sensitivity at 13DPA. Among them, two DEGs were upregulated in both Brazil32 and Yitpi while three DEGs were upregulated in Brazil32 and downregulated in Yitpi. Four DEGs were unique to Brazil32 and three to Yitpi. Three DEGs were common between Perenjori and Brazil32, ten between Perenjori and Yitpi, and eleven among all three genotypes (Figure 4c).
3.5.3. Differential Expression Pattern of TaMYB TF Genes in Contrasting Wheat Genotypes
There were different numbers of DEGs in Perenjori, Brazil32, and Yitpi. Some genes were commonly expressed between all three genotypes, while some were uniquely expressed only in one genotype or between two or three genotypes. In Perenjori, the total number of DEGs was 20 at the seedling stage compared to 25 at 11DPA and 28 at 13DPA, and the number of uniquely expressed DEGs was 14 (TaMYB-023, TaMYB-014, TaMYB-206, TaMYB049, TaMYB-300, TaMYB-267, TaMYB-030, TaMYB-405, TaMYB-327, TaMYB-157, TaMYB-211, TaMYB-296, TaMYB-226, and TaMYB-304) at the seedling stage, which dropped to 7 (TaMYB-292, TaMYB-049, TaMYB-308, TaMYB-030, TaMYB-327, TaMYB-299, and TaMYB-226) at 11DPA and to 4 (TaMYB-209, TaMYB-023, TaMYB-232, and TaMYB-327) at 13DPA (Figure 4a). The expression profile of DEGs unique to Perenjori at the seedling stage, 11DPA, and 13DPA showed that a single DEG (TaMYB-327) was expressed at all three time points, three DEGs (TaMYB-049, TaMYB-030, and TaMYB-226) were expressed both at the seedling stage and 11DPA, and one DEG (TaMYB-023) was expressed both at the seedling stage and 13DPA (Figure 5). The remaining DEGs were unique to either the seedling stage, 11DPA or 13DPA.
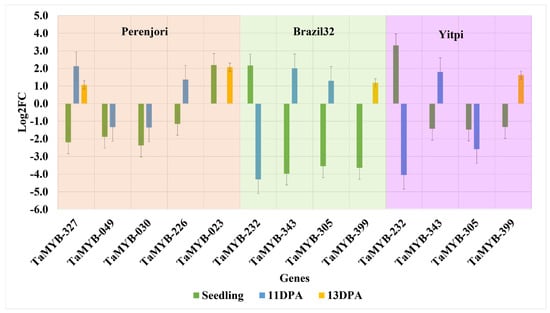
Figure 5.
Expression profile of DEGs in the heat-tolerant genotype (Perenjori) and two heat-susceptible genotypes (Brazil32 and Yitpi) at three heat treatment time points. At the seedling stage, five DEGs were expressed only in Perenjori and four DEGs in both Brazil32 and Yitpi; at 11DPA, four DEGs were expressed in Perenjori and three DEGs in both Brazil32 and Yitpi; and at 13DPA, two DEGs were expressed in Perenjori and one DEG each in both Brazil32 and Yitpi. The highlighted color pattern differentiates the genes significantly upregulated/downregulated in all three genotypes: orange for Perenjori, green for Brazil32, and purple for Yitpi.
In the sensitive genotype, Brazil32, the number of DEGs decreased from 24 at the seedling stage to 21 at 11DPA and slightly increased to 23 at 13DPA. The number of uniquely expressed DEGs was nine (TaMYB-288, TaMYB-287, TaMYB-292, TaMYB-281, TaMYB-295, TaMYB-301, TaMYB-382, TaMYB-273, and TaMYB-361) at the seedling stage, five (TaMYB-295, TaMYB-138, TaMYB-382, TaMYB-369, and TaMYB-152) at 11DPA, and four (TaMYB-281, TaMYB-295, TaMYB-138, and TaMYB-157) at 13DPA. The expression profile of uniquely expressed DEGs in Brazil32 showed that one DEG (TaMYB-295) was expressed at all three time points, one DEG (TaMYB-382) was expressed both at the seedling stage and 11DPA, one DEG (TaMYB-281) was expressed both at the seedling stage and 13DPA, and one DEG (TaMYB-138) was expressed both at 11DPA and 13DPA. The remaining DEGs were unique to either the seedling stage, 11DPA, or 13DPA. Similarly, in Yitpi, the number of DEGs increased from 14 at the seedling stage to 19 at 11DPA and 29 at 13DPA. The number of uniquely expressed DEGs was one (TaMYB-161) at the seedling stage, one DEG (TaMYB-267) at 11DPA, and three DEGs (TaMYB-352, TaMYB-273, and TaMYB-356) at 13DPA. There were no DEGs commonly expressed between time points.
The number of DEGs commonly expressed between the two sensitive genotypes Brazil32 and Yitpi was 10 (TaMYB-286, TaMYB-232, TaMYB-343, TaMYB-375, TaMYB-305, TaMYB-399, TaMYB-363, TaMYB-369, TaMYB-356, and TaMYB-272) at the seedling stage, 4 (TaMYB-232, TaMYB-343, TaMYB-305, and TaMYB-140) at 11DPA, and 5 DEGs (TaMYB-300, TaMYB-382, TaMYB-399, TaMYB-377, and TaMYB-245) at 13DPA. The expression profile of commonly expressed DEGs between Brazil32 and Yitpi showed that three DEGs (TaMYB-232, TaMYB-343, and TaMYB-305) were expressed both at the seedling stage and 11DPA and that one DEG (TaMYB-399) was expressed at both the seedling stage and 13DPA (Figure 5).
4. Discussion
The ESS for adult-stage traits such as yield and yield-related traits is important in crop breeding for minimizing the cost, time, and effort of growing crops to maturity. The morpho-physiological traits, such as canopy temperature, biomass, chlorophyll content, chlorophyll fluorescence, and their heat tolerance index, have been considered in the potential for the ESS of heat-tolerant genotypes in wheat [,]. However, morphological indices are prone to variability due to the influence of environmental factors and genotypes, so it is difficult to make an accurate assessment of heat tolerance []. Therefore, exploring potential genetic traits associated with heat tolerance and yield for ESS can speed up the wheat breeding cycle and increase genetic gain. The MYB TF genes have been studied in several crop species along with microorganisms. They are involved in a diverse range of functions in plants, from plant growth and development and the response to biotic and abiotic stress to the production of secondary metabolites and cell cycle regulation []. In wheat, previous studies have been mainly focused on the identification and characterization of MYB-2R sub-family genes under salt, drought, and HS. This study identified the TaMYB genes from all four sub-families in the wheat genome, established phylogenetic relationships, examined conserved motifs and gene structure, and analyzed the in silico expression pattern to enable the selection of 48 TaMYB genes as putative candidate genes related to heat-tolerant and heat-sensitive phenotypes in wheat. The morphological analysis of the seedling root depth heat damage index (RD_HDI) and thousand-kernel weight heat damage index (TKW_HDI) confirmed Perejori as heat-tolerant and Brazil32 and Yitpi as heat-sensitive genotypes at both seedling and reproductive stages. The qRT-PCR gene expression analysis of TaMYB genes in contrasting wheat genotypes at seedling and grain-filling stages revealed candidate genes for ESS for heat tolerance and for heat sensitivity in wheat.
In this study, 876 TaMYB TF genes were identified in the wheat genome and classified into four sub-families, with the highest number of genes in the TaMYB-2R sub-family and smallest number of genes in TaMYB-4R. Similarly, four MYB sub-families, with the highest number of genes in the MYB-2R sub-family, were reported in rice and Arabidopsis []. This number is higher than the number of MYB genes identified in the wheat genome in previous studies: 60 TaMYB by [], 405 TaMYB by [], and 719 TaMYB by []. We also identified 159 TaMYB (21 TaMYB-1R, 75 TaMYB-2R, 45 TaMYB-3R, and 18 TaMYB-4R) genes unique to the TaMYB genes reported by []. The number of TaMYB genes identified in wheat was also much higher than the TaMYB genes identified in model plants rice (155 OsMYB) and Arabidopsis (197 AtMYB) []. This might be due to the larger genome size of hexaploid wheat, with three sub-genomes, compared to diploid plants. Gene duplication is also considered as a major factor for the progressive gene evolution and expansion of gene families. The 409 TaMYB genes analyzed in this study were unevenly distributed in all 21 wheat chromosomes in the three sub-genomes A, B, and D, with 66 pairs of segmental duplicated genes (Supplementary Table S4 and Figure S2). The segmental duplication of MYB genes has been reported in several species, including 176 in pearl millet [], 29 in Potato [], and 36 in sunflower []. The analysis of conserved motifs indicated that motif 3 is the most frequently observed motif across 369 sites, suggesting its potential significance in shaping the functional characteristics of this gene. Similar evidence of MYB genes with conserved motifs sharing similar functional characteristics has been reported by []. However, some motifs are uniquely present only in a few genes, which might be responsible for the gene-specific roles in the plant.
The seedling-stage heat tolerance was determined based on the RD_HDI. The seedling root development was severely affected in Brazil32 and Yitpi compared to Perenjori under HS (Figure 3a). The performance of these genotypes was consistent with the previous results []. The MYB genes play a significant role in root development, evident from the high expression of HvMYB5 in the root tissue of 15-day-old barley seedlings []. The Arabidopsis MYB gene, AtMYB68, was expressed in the root pericycle cells at the base of lateral roots and primordia under high-temperature stress, indicating their role in root development under the HS condition []. In addition, MYB genes were responsible for heat-responsive gene expression and seedling-stage heat tolerance in potato [] and transgenic tobacco [].
The grain-filling stage heat tolerance was measured with the TKW_HDI in three genotypes of wheat in this experiment (Figure 3). The TKW is directly associated with grain development and yield and it is a useful indicator for heat tolerance []. MYB genes play important roles in grain development through transcription regulation or are directly involved in the biosynthesis of metabolites; for example, TaTCL2 was involved in anthocyanin accumulation during grain development, contributing to seed color in wheat through the MBW complex [], and the overexpression of the TaMYB13-1 gene in transgenic wheat enhanced the accumulation of fructan in the leaves and stem, eventually contributing to a higher spike and grain weight under drought stress []. In maize, the ZmMYB14 gene acts as a key transcription regulator of ZmBT1 for the biosynthesis of starch in developing endosperm []. In rice, OsMYB55 enhanced heat tolerance through the activation of genes related to amino acid metabolism and maintained a higher grain yield under HS [].
The RT-qPCR expression analysis of TaMYB genes showed a variation in the total number of DEGs at the seedling stage (40 DEGs), 11DPA (35 DEGs), and 13DPA (40DEGs) under heat stress (Figure 4). These changes in the number of DEGs were also accompanied by changes in the genes in each genotype, which showed that the heat-induced expression of some TaMYB genes were specific to developmental stages. The developmental-stage-specific expression of PoMYB genes has also been reported in Pleurotus osteratus mushroom in RNA sequencing and qPCR expression analysis [].
At the same time, the variation in the number of DEGs between 11DPA and 13DPA at grain-filling stage HS reveals that the expression of TaMYB genes is also affected by the duration of heat treatment within the same developmental stage. The variation in the heat-responsive expression of MYB TF genes under HS applied at 40 °C for 0, 10, 20, 60 min, and 2h was also reported in rice panicle in a microarray experiment []. The expression difference of MYB genes is also explained by the difference in their gene structure. Introns are a non-coding region in a gene; however, they may play significant roles in gene regulation and expression. The gene structure analysis of TaMYB genes in this study revealed a significant variation in the number of intron–exon compositions. Some TaMYB genes contained a higher number (14) of introns while some were intronless (Supplementary Figure S1). The genes with short and fewer introns show early expression, while genes with longer introns exhibit delayed expression []. This might be the reason for the time-point-specific expression of some TaMYB genes at 11DPA and 13DPA.
There were different numbers of DEGs in Perenjori, Brazil32, and Yitpi under HS (Figure 4). The unique DEGs expressed only in Perenjori were associated with the heat-tolerant phenotype and were considered as heat-tolerant TaMYB genes. However, the commonly expressed DEGs between Brazil32 and Yitpi and those not expressed in Perenjori under HS were associated with the heat-sensitive phenotype of Brazil32 and Yitpi and were considered as TaMYB genes for heat sensitivity in this study. The different expression patterns of the MYB73 gene in two contrasting wheat genotypes, Suan and Joeun seedlings, were reported after 50 days of HS []. Also, the abundance of MYB and MYB-related genes was reported in an awn transcript analysis of the heat-sensitive wheat genotype PBW343 under heat stress []. Similarly, a relatively higher fold expression of heat shock proteins (sHsp-1, Hsp17, and HsfA4) in the heat-tolerant genotypes (WH730 and WH1218) compared to heat-sensitive genotypes (WH711 and WH157) under HS at 15DAA was responsible for heat tolerance in wheat []. The difference in the expression of 35 transcripts was also reported between the heat-tolerant and heat-sensitive rice genotypes under HS at the early milky stage []. These results indicate that the HS-induced expression of TaMYB genes in wheat genotypes differs with heat tolerance and sensitivity.
The selection of genotypes at early growth stages is more convenient in terms of time, cost, and effort compared to the adult stage. Therefore, identifying seedling factors that have a strong association with adult-stage performance is essential for the effective implementation of ESS in crop breeding. The genetic basis for the ESS of heat tolerance can be a reliable method in heat tolerance breeding. In this study, common DEGs of TaMYB genes between the seedling and grain-filling stage were selected as candidate genes for the ESS of wheat genotypes for heat tolerance breeding. Five TaMYB genes were identified as candidate genes in Perenjori for heat tolerance. One DEG (TaMYB-327) exhibited expression across all three time points of heat treatment. Therefore, it was considered as a candidate gene for both long-term heat tolerance (13DPA) and short-term heat tolerance (11DPA). In addition, the three DEGs (TaMYB-030, TaMYB-049, and TaMYB-226) expressed both at the seedling stage and 11DPA were candidate genes for short-term heat tolerance, and one DEG (TaMYB-023) expressed both at the seedling stage and 13DPA was a candidate gene for long-term heat tolerance. Similarly, DEGs commonly expressed between two sensitive genotypes, Brazil32 and Yitpi, at the seedling stage and 11DPA or/and 13DPA were candidate genes for selecting heat-sensitive genotypes for ESS. The three DEGs (TaMYB-232, TaMYB-343, and TaMYB-305) expressed both at the seedling stage and 11DPA were candidate genes for short-term heat sensitivity, and one DEG (TaMYB-399) expressed both at the seedling stage and 13DPA was a candidate gene for long-term heat sensitivity. These genes were negative regulators of heat tolerance in wheat and can be utilized to identify heat-sensitive genotypes in wheat breeding. Similar evidence of MYB genes as negative regulators of HS tolerance has been reported in blueberry, where overexpression of the VcMYB4a gene enhanced heat sensitivity []. The negative regulatory roles of MYB TF genes have also been reported in the regulation of anthocyanin accumulation in lily by LhR3MYB2 [] and in tobacco flower by GtMYB1R1 and GtMYB1R9 [], as well as the salt tolerance of Arabidopsis by AtDIV2 [].
In addition, some DEGs were commonly expressed between the tolerant and sensitive genotypes, where one DEG (TaMYB-299) at the seedling stage, six DEGs (TaMYB-352, TaMYB-273, TaMYB-296, TaMYB-377, TaMYB-363, and TaMYB-304) at 11DPA, and 10 DEGs (TaMYB-288, TaMYB-287, TaMYB-286, TaMYB-292, TaMYB-375, TaMYB-053, TaMYB-161, TaMYB-369, TaMYB-140, and TaMYB-304) at 13DPA were commonly expressed between Perenjori and Yitpi. Similarly, three DEGs (TaMYB-261, TaMYB-138, and TaMYB-275) at the seedling stage, four DEGs (TaMYB-287, TaMYB-375, TaMYB-399, and TaMYB-361) at 11DPA, and three DEGs (TaMYB-014, TaMYB-206, and TaMYB-226) at 13DPA were commonly expressed between Perenjori and Brazil32 (Figure 4). These commonly expressed genes between tolerant and sensitive genotypes might be associated with other common developmental functions characteristic to the MYB gene family.
5. Conclusions
Heat tolerance breeding is a complex procedure in wheat and requires a significant amount of time, cost, and effort. The identification of seedling-stage factors with a strong correlation with reproductive-stage performance and employing ESS in breeding can enhance the efficiency and effectiveness in wheat heat tolerance breeding. In this study, we have identified TaMYB TF genes demonstrating a strong correlation in their RT-qPCR expression at seedling and grain-filling stages. Five TaMYB genes (TaMYB-327, TaMYB-049, TaMYB-030, TaMYB-226, and TaMYB-023) can be utilized for the early-stage selection of heat tolerance and four TaMYB genes (TaMYB-232, TaMYB-343, TaMYB-305, and TaMYB-399) for the early-stage selection of heat sensitivity in wheat. The identified genes can be used for further investigation of target genes and proteins in the regulatory network directly expressed due to HS. They provide an idea of how MYB may regulate HS tolerance in wheat, which helps in sustainable wheat breeding for heat tolerance. These genes can also be further validated in field trials with multiple breeding lines to ensure the consistency in gene expression across lines.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijpb16020041/s1, Supplementary Tables: Supplementary Table S1: Domain and annotation details of 876 non-redundant TaMYB TF genes identified using hmm profile of MYB protein domain (PF00249, PF13921, and PF14215) against the International Wheat Genome Sequencing Consortium website (IWGSC) RefSeq v1.1. Supplementary Table S2: Domain information of 159 unique TaMYB TF genes identified in this analysis compared to the TaMYB TF genes reported by Sukumaran et. al., 2023 []. There were 21 TaMBYB-1R, 75 TaMYB-2R, 45 TaMYB-3R, and 18 TaMYB-4R genes unique in the four sub-families. Supplementary Table S3: The 409 TaMYB TF genes representing all four TaMYB sub-families selected for detailed characterization. There were 42 genes in TaMYB-1R sub-family, 332 genes in TaMYB-2R sub-family, 31 genes in TaMYB-3R sub-family, and 4 genes in TaMYB-4R sub-family, along with the number of domains, E-value, and domains present in each gene. Each of the 409 TaMYB genes were renamed with “Gene Name” from TaMYB-001 to TaMYB-409. Supplementary Table S4: Details of the conserved motifs discovered in the 409 TaMYB TF genes. Supplementary Table S5: Duplicated gene pairs with their percentage of similarity and identity, chromosomal location and position, and difference in chromosomal position. The 66 pairs of segmental duplicate (SD) genes with >90% similarity and identity with largest gene sequence were identified based on similarity and identity matrix generated from Sequence Identity and Similarity (SIAS) online tool. Supplementary Table S6: Details of genes in each cluster and transcript per million (tpm) values of 409 TaMYB TF genes in in silico expression analysis under abiotic stress using wheat expression database. There were nine clusters, with number of genes grouped in the clusters ranging from 2 genes in cluster 7 to 254 genes in cluster 9. Supplementary Table S7: The 48 pairs of gene-specific primers used in RT-qPCR for gene expression analysis. Supplementary Figures: Supplementary Figure S1: Phylogenetic grouping, distribution of conserved motifs, and gene structure of 409 TaMYB family TF genes. The phylogenetic tree was constructed using MEGAX with bootstrap values of 1000 repeats, motif composition was analyzed using online MEME tool, and gene structure was analyzed using GSDS online server. In the motif graph, the colored blocks represent the position of motifs in corresponding proteins coding TF genes, block fonts indicate the length of motifs, and grey lines connecting the colored bocks represent non-conserved sequences. In the gene structure graph, the yellow blocks are exons, green blocks are UTRs, and grey lines are introns. The relative position of each motif, exon, intron, and UTR can be determined with the help of scale displayed just below the corresponding graph. Supplementary Figure S2: The expression pattern profiling of 409 TaMYB TF genes under abiotic stress using publicly available transcriptome data. The intensity of color shows the level of gene expression, where blue color shows the least expressed genes and red color shows the highest expressed genes. The genes are clustered into nine clusters based on their expression values, forming a bigger cluster (cluster 9) of relatively lowest expressed genes (62.1%) and a smaller cluster (cluster 1) of highest expressed genes (4.4%) under abiotic stress. Supplementary Figure S3: Diagram illustrating the effect of heat stress on seedling root depth in all three genotypes: (a) Perenjori, (b) Brazil 32, and (c) Yitpi. Supplementary Figure S4: Glasshouse experiments showing the three different stages of wheat growth: (a) sowing, (b) vegetative growth, and (c) reproductive growth approaching physiological maturity.
Author Contributions
M.M.M. performed conceptualization, investigation, methodology, data curation, formal analysis, and writing—original draft. H.L. was involved in conceptualization, funding acquisition, methodology, formal analysis, resources, supervision, and writing—review and editing. G.Y. provided conceptualization, funding acquisition, methodology, formal analysis, resources, supervision, and writing—review and editing. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The University of Western Australia International Fee Scholarship and a University Postgraduate Award, as a sponsorship to the PhD of MM. The research is partly supported by the Global Innovation Linkages Project (GIL53853) from the Australian Department of Industry, Science and Resources.
Data Availability Statement
All data in this study are available in this article and its Supplementary Files.
Acknowledgments
The authors would like to acknowledge the technical support and valuable suggestions from Robert Creasy, Bill Piasini, Lu Lu, Sun Kumar Gurung, Mukesh Chaudhary, Sultan Mia, Candy Taylor, Kosala Ranathunge, Dipesh Maharjan, and Wei San Wong.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Poor, P.; Nawaz, K.; Gupta, R.; Ashfaque, F.; Khan, M.I.R. Ethylene Involvement in the Regulation of Heat Stress Tolerance in Plants. Plant Cell Rep. 2022, 41, 675–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lin, H.; Lin, H. Diverse Roles of MYB Transcription Factors in Plants. J. Integr. Plant Biol. 2025, 1–24. [Google Scholar] [CrossRef]
- Asseng, S.; Foster, I.A.N.; Turner, N.C. The Impact of Temperature Variability on Wheat Yields. Glob. Change Biol. 2011, 17, 997–1012. [Google Scholar] [CrossRef]
- Xie, C.; Mosjidis, J.A. Seedling-Selection Effects on Morphological Traits of Mature Plants in Red Clover. Theor. Appl. Genet. 1995, 91, 1032–1036. [Google Scholar] [CrossRef]
- Lu, L.; Liu, H.; Wu, Y.; Yan, G. Wheat Genotypes Tolerant to Heat at Seedling Stage Tend to Be Also Tolerant at Adult Stage: The Possibility of Early Selection for Heat Tolerance Breeding. Crop J. 2022, 10, 1006–1013. [Google Scholar] [CrossRef]
- EI-Daim, I.A.; Bejai, S.; Meijer, J. Improved Heat Stress Tolerance of Wheat Seedlings by Bacterial Seed Treatment. Plant Soil 2014, 379, 337–350. [Google Scholar] [CrossRef]
- Mishra, S.; Spaccarotella, K.; Gido, J.; Samanta, I.; Chowdhary, G. Effects of Heat Stress on Plant-Nutrient Relations: An Update on Nutrient Uptake, Transport, and Assimilation. Int. J. Mol. Sci. 2023, 24, 15670. [Google Scholar] [CrossRef]
- Samalova, M.; Gahurova, E.; Hejatko, J. Expansin-Mediated Developmental and Adaptive Responses: A Matter of Cell Wall Biomechanics? Quant. Plant Biol. 2022, 3, e11. [Google Scholar] [CrossRef]
- González-García, M.P.; Conesa, C.M.; Lozano-Enguita, A.; Baca-González, V.; Simancas, B.; Navarro-Neila, S.; Sánchez-Bermúdez, M.; Salas-González, I.; Caro, E.; Castrillo, G.; et al. Temperature Changes in the Root Ecosystem Affect Plant Functionality. Plant Commun. 2023, 4, 100514. [Google Scholar] [CrossRef]
- Li, S.; Chang, X.; Wang, C.; Jing, R. Mapping QTLs for Seedling Traits and Heat Tolerance Indices in Common Wheat. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1525–1533. [Google Scholar]
- Reynolds, M.; Manes, Y.; Izanloo, A.; Langridge, P. Phenotyping Approaches for Physiological Breeding and Gene Discovery in Wheat. Ann. Appl. Biol. 2009, 155, 309–320. [Google Scholar] [CrossRef]
- Magar, M.M.; Liu, H.; Yan, G. Genome-Wide Analysis of AP2/ERF Superfamily Genes in Contrasting Wheat Genotypes Reveals Heat Stress-Related Candidate Genes. Front. Plant Sci. 2022, 13, 853086. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Chattha, M.U.; Khan, I.; Chattha, M.B.; Barbanti, L.; Aamer, M.; Iqbal, M.M.; Nawaz, M.; Mahmood, A.; Ali, A.; et al. Heat Stress in Cultivated Plants: Nature, Impact, Mechanisms, and Mitigation Strategies—A Review. Plant Biosyst.-Int. J. Deal. All Asp. Plant Biol. 2020, 155, 211–234. [Google Scholar] [CrossRef]
- Pradhan, S.; Babar, M.A.; Bai, G.; Khan, J.; Shahi, D.; Avci, M.; Guo, J.; McBreen, J.; Asseng, S.; Gezan, S.; et al. Genetic Dissection of Heat-Responsive Physiological Traits to Improve Adaptation and Increase Yield Potential in Soft Winter Wheat. BMC Genom. 2020, 21, 315. [Google Scholar] [CrossRef]
- Saini, N.; Nikalje, G.C.; Zargar, S.M.; Suprasanna, P. Molecular Insights into Sensing, Regulation and Improving of Heat Tolerance in Plants. Plant Cell Rep. 2022, 41, 799–813. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjarvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef]
- Kumar, S.; Kumari, P.; Kumar, U.; Grover, M.; Singh, A.K.; Singh, R.; Sengar, R.S. Molecular Approaches for Designing Heat Tolerant Wheat. J. Plant Biochem. Biotechnol. 2013, 22, 359–371. [Google Scholar] [CrossRef]
- Lal, M.K.; Tiwari, R.K.; Gahlaut, V.; Mangal, V.; Kumar, A.; Singh, M.P.; Paul, V.; Kumar, S.; Singh, B.; Zinta, G. Physiological and Molecular Insights on Wheat Responses to Heat Stress. Plant Cell Rep. 2022, 41, 501–518. [Google Scholar] [CrossRef]
- Liao, C.; Zheng, Y.; Guo, Y. MYB30 Transcription Factor Regulates Oxidative and Heat Stress Responses through ANNEXIN-Mediated Cytosolic Calcium Signaling in Arabidopsis. New Phytolologist 2017, 216, 163–177. [Google Scholar] [CrossRef]
- Mariyam, S.; Kumar, V.; Roychoudhury, A.; Ghodake, G.S.; Muneer, S.; Duhan, J.S.; Ahmad, F.; Sharma, R.K.; Singh, J.; Seth, C.S. Functional Diversification and Mechanistic Insights of MYB Transcription Factors in Mediating Plant Growth and Development, Secondary Metabolism, and Stress Responses. J. Plant Growth Regul. 2025. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB Transcription Factor Genes as Regulators for Plant Responses: An Overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Martin, C. Multifunctionality and Diversity within the Plant MYB-Gene Family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef]
- Haider, S.; Iqbal, J.; Naseer, S.; Shaukat, M.; Abbasi, B.A.; Yaseen, T.; Zahra, S.A.; Mahmood, T. Unfolding Molecular Switches in Plant Heat Stress Resistance: A Comprehensive Review. Plant Cell Rep. 2022, 41, 775–798. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The Regulatory C1 Locus of Zea Mays Encodes a Protein with Homology to Myb Proto-oncogene Products and with Structural Similarities to Transcriptional Activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-Wide Classification and Expression Analysis of MYB Transcription Factor Families in Rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Azameti, M.K.; Ranjan, A.; Singh, P.K.; Gaikwad, K.; Singh, A.K.; Dalal, M.; Arora, A.; Rai, V.; Padaria, J.C. Transcriptome Profiling Reveals the Genes and Pathways Involved in Thermo-Tolerance in Wheat (Triticum aestivum L.) Genotype Raj 3765. Sci. Rep. 2022, 12, 14831. [Google Scholar] [CrossRef]
- Feng, C.; Andreasson, E.; Maslak, A.; Mock, H.P.; Mattsson, O.; Mundy, J. Arabidopsis MYB68 in Development and Responses to Environmental Cues. Plant Sci. 2004, 167, 1099–1107. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Liu, X.; Yuan, G.; Hou, H.; Teng, N. A Novel R2R3-MYB Transcription Factor LlMYB305 from Lilium Longiflorum Plays a Positive Role in Thermotolerance via Activating Heat-Protective Genes. Environ. Exp. Bot. 2021, 184, 104399. [Google Scholar] [CrossRef]
- Deeba, F.; Sultana, T.; Javaid, B.; Mahmood, T.; Naqvi, S.M.S. Molecular Characterization of a MYB Protein from Oryza Sativa for Its Role in Abiotic Stress Tolerance. Braz. Arch. Biol. Technol. 2017, 60, e17160352. [Google Scholar] [CrossRef]
- El-kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The Rice R2R3-MYB Transcription Factor OsMYB55 Is Involved in the Tolerance to High Temperature and Modulates Amino Acid Metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef]
- Meng, X.; Wang, J.R.; Wang, G.D.; Liang, X.Q.; Li, X.D.; Meng, Q.W. An R2R3-MYB Gene, LeAN2, Positively Regulated the Thermo-Tolerance in Transgenic Tomato. J. Plant Physiol. 2015, 175, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Yang, B.; Xian, B.; Chen, B.; Yan, J.; Chen, Q.; Gao, S.; Zhao, P.; Han, F.; Xu, J.; et al. The R2R3-MYB Transcription Factor BnaMYB111L from Rapeseed Modulates Reactive Oxygen Species Accumulation and Hypersensitive-like Cell Death. Plant Physiol. Biochem. 2020, 147, 280–288. [Google Scholar] [CrossRef]
- Chen, S.; Niu, X.; Guan, Y.; Li, H. Genome-Wide Analysis and Expression Profiles of the MYB Genes in Brachypodium distachyon. Plant Cell Physiol. 2017, 58, 1777–1788. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular Characterization of 60 Isolated Wheat MYB Genes and Analysis of Their Expression during Abiotic Stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature Increase Reduces Global Yields of Major Crops in Four Independent Estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Tyagi, A.K.; Khurana, J.P.; Khurana, P. Identification and Characterization of High Temperature Stress Responsive Genes in Bread Wheat (Triticum aestivum L.) and Their Regulation at Various Stages of Development. Plant Mol. Biol. 2011, 75, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-Wide Identification of R2R3-MYB Family in Wheat and Functional Characteristics of the Abiotic Stress Responsive Gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef]
- Sukumaran, S.; Lethin, J.; Liu, X.; Pelc, J.; Zeng, P.; Hassan, S.; Aronsson, H. Genome-Wide Analysis of MYB Transcription Factors in the Wheat Genome and Their Roles in Salt Stress Response. Cells 2023, 12, 1431. [Google Scholar] [CrossRef]
- Mia, M.S.; Liu, H.; Wang, X.; Zhang, C.; Yan, G. Root Transcriptome Profiling of Contrasting Wheat Genotypes Provides an Insight to Their Adaptive Strategies to Water Deficit. Sci. Rep. 2020, 10, 4854. [Google Scholar] [CrossRef]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Maruyama, S.; Eveleigh, R.J.M.; Archibald, J.M. Treetrimmer: A Method for Phylogenetic Dataset Size Reduction. BMC Res. Notes 2013, 6, 145. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheat. Science 2018, 361, 6403. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Yang, C.; Wang, J.; Yan, G.; Si, P.; Bai, Q.; Lu, Z.; Zhou, W.; Xu, L. Genome-Wide Identification of MYB Genes and Expression Analysis under Different Biotic and Abiotic Stresses in Helianthus annuus L. Ind. Crops Prod. 2020, 143, 111924. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Sallam, M.; Ghazy, A.; Ibrahim, A.; Alotaibi, M.; Ullah, N.; Al-Doss, A. Agro-Physiological Indices and Multidimensional Analyses for Detecting Heat Tolerance in Wheat Genotypes. Agronomy 2023, 13, 154. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research Advances of MYB Transcription Factors in Plant Stress Resistance and Breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Chanwala, J.; Khadanga, B.; Jha, D.K.; Sandeep, I.S.; Dey, N. MYB Transcription Factor Family in Pearl Millet: Genome-Wide Identification, Evolutionary Progression and Expression Analysis under Abiotic Stress and Phytohormone Treatments. Plants 2023, 12, 355. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-Wide Analysis and Expression Profiles of the StR2R3-MYB Transcription Factor Superfamily in Potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 Plays Dual Roles in Flavonoid Biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Seven, M.; Akdemir, H. DOF, MYB and TCP Transcription Factors: Their Possible Roles on Barley Germination and Seedling Establishment. Gene Expr. Patterns 2020, 37, 119116. [Google Scholar] [CrossRef]
- Firouzian, A.; Shafeinia, A.; Ghaffary, S.M.T.; Mohammadi, V.; Sadat, S. Terminal Heat Tolerance in Bread Wheat Determined by Agronomical Traits and SSR Markers. J. Plant Growth Regul. 2022, 42, 2041–2052. [Google Scholar] [CrossRef]
- Flores, P.C.; Yoon, J.S.; Kim, D.Y.; Seo, Y.W. Transcriptome Analysis of MYB Genes and Patterns of Anthocyanin Accumulation During Seed Development in Wheat. Evol. Bioinforma. 2022, 18, 11769343221093340. [Google Scholar] [CrossRef]
- Kooiker, M.; Drenth, J.; Glassop, D.; McIntyre, C.L.; Xue, G.P. TaMYB13-1, a R2R3 MYB Transcription Factor, Regulates the Fructan Synthetic Pathway and Contributes to Enhanced Fructan Accumulation in Bread Wheat. J. Exp. Bot. 2013, 64, 3681–3696. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, Y.; Du, J.; Li, H.; Wei, B.; Wang, Y.; Li, Y.; Yu, G.; Liu, H.; Zhang, J.; et al. ZmMYB14 Is an Important Transcription Factor Involved in the Regulation of the Activity of the ZmBT1 Promoter in Starch Biosynthesis in Maize. FEBS J. 2017, 284, 3079–3099. [Google Scholar] [CrossRef]
- Wang, L.; Gao, W.; Wu, X.; Zhao, M.; Qu, J.; Huang, C.; Zhang, J. Genome-Wide Characterization and Expression Analyses of Pleurotus Ostreatus MYB Transcription Factors during Developmental Stages and under Heat Stress Based on de Novo Sequenced Genome. Int. J. Mol. Sci. 2018, 19, 2052. [Google Scholar] [CrossRef]
- Zhang, X.; Xiong, H.; Liu, A.; Zhou, X.; Peng, Y.; Li, Z.; Luo, G.; Tian, X.; Chen, X. Microarray Data Uncover the Genome-Wide Gene Expression Patterns in Response to Heat Stress in Rice Post-Meiosis Panicle. J. Plant Biol. 2014, 57, 327–336. [Google Scholar] [CrossRef]
- Heyn, P.; Kalinka, A.T.; Tomancak, P.; Neugebauer, K.M. Introns and Gene Expression: Cellular Constraints, Transcriptional Regulation, and Evolutionary Consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.H.; Seong, H.J.; Yang, W.H.; Jung, W. Growth Responses and Differences in Gene Expression Depending on Cultivation Temperature between Alternative Type Wheat Varieties. J. Crop Sci. Biotechnol. 2019, 23, 47–55. [Google Scholar] [CrossRef]
- Chaudhary, C.; Sharma, N.; Khurana, P. Decoding the Wheat Awn Transcriptome and Overexpressing TaRca1beta in Rice for Heat Stress Tolerance. Plant Mol. Biol. 2021, 105, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Sihag, P.; Kumar, U.; Sagwal, V.; Kapoor, P.; Singh, Y.; Mehla, S.; Balyan, P.; Mir, R.R.; Varshney, R.K.; Singh, K.P.; et al. Effect of Terminal Heat Stress on Osmolyte Accumulation and Gene Expression during Grain Filling in Bread Wheat (Triticum aestivum L.). Plant Genome 2024, 17, e20307. [Google Scholar] [CrossRef]
- Liao, J.L.; Zhou, H.W.; Peng, Q.; Zhong, P.A.; Zhang, H.Y.; He, C.; Huang, Y.J. Transcriptome Changes in Rice (Oryza sativa L.) in Response to High Night Temperature Stress at the Early Milky Stage. BMC Genom. 2015, 16, 18. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, H.C.; Zhang, X.S.; Guo, Q.X.; Bian, S.M.; Wang, J.Y.; Zhai, L.L. VcMYB4a, an R2R3-MYB Transcription Factor from Vaccinium Corymbosum, Negatively Regulates Salt, Drought, and Temperature Stress. Gene 2020, 757, 144935. [Google Scholar] [CrossRef]
- Sakai, M.; Yamagishi, M.; Matsuyama, K. Repression of Anthocyanin Biosynthesis by R3-MYB Transcription Factors in Lily (Lilium spp.). Plant Cell Rep. 2019, 38, 609–622. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Yamada, E.; Saito, M.; Fujita, K.; Nishihara, M. Heterologous Expression of Gentian MYB1R Transcription Factors Suppresses Anthocyanin Pigmentation in Tobacco Flowers. Plant Cell Rep. 2013, 32, 1925–1937. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-Type MYB Transcription Factor of Arabidopsis, Negatively Regulates Salt Stress by Modulating ABA Signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).