Antibiotics Resistance and PGPR Traits of Endophytic Bacteria Isolated in Arid Region of Morocco
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Rhizospheric Soils and Plant Materials
2.2. Isolation of Endophytic Bacteria
2.3. Antibiotics Resistance
2.4. Production of Indole 3-Acetic Acid
2.5. Production of Siderophores
2.6. Solubilization of the Insoluble Inorganic Phosphate
3. Statistical Analysis
4. Results
4.1. Isolation and Identification of the Strains
4.2. Resistance to Antibiotics
4.3. Production of Indole-3-Acetic Acid (IAA)
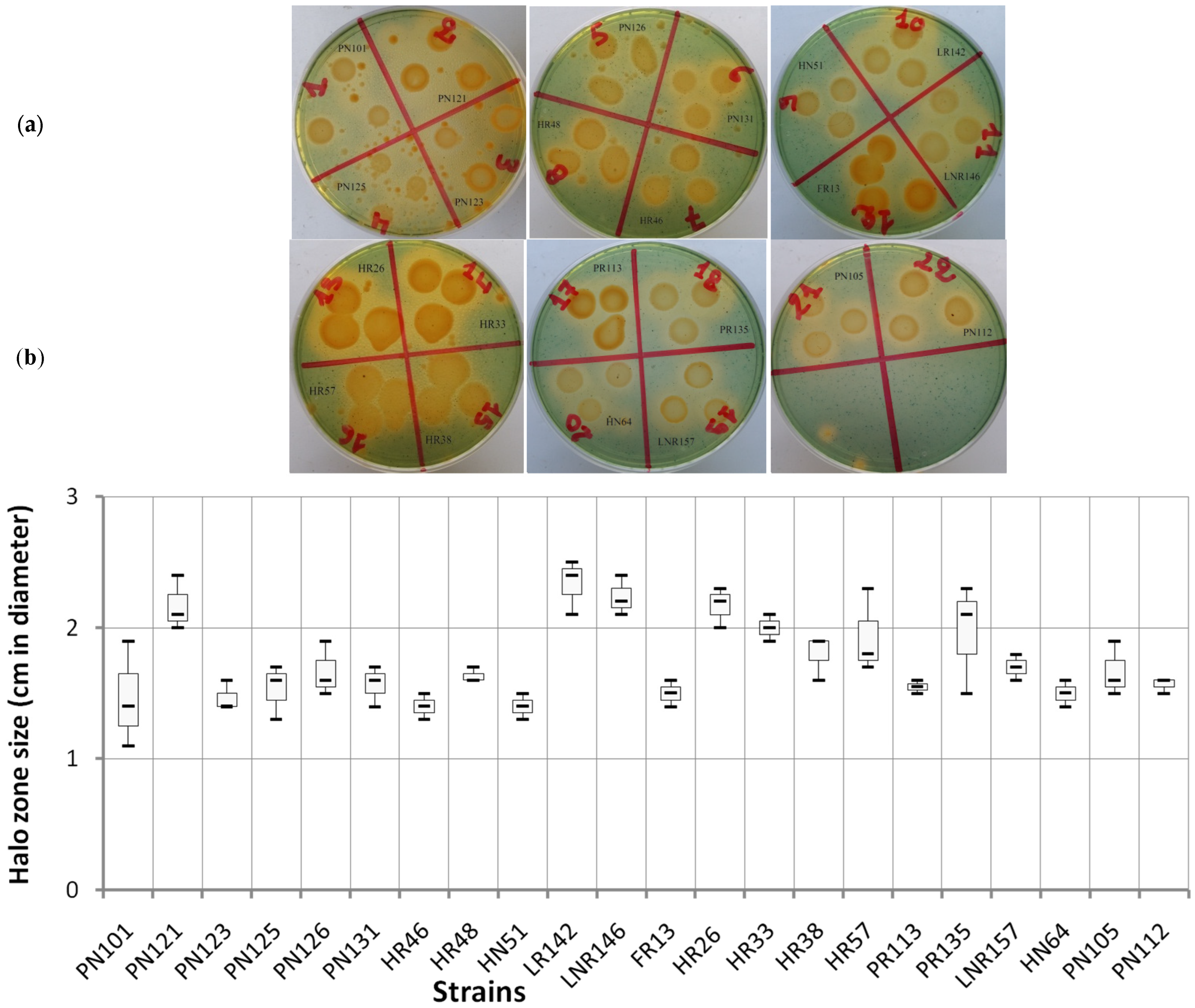
4.4. Production of Siderophores
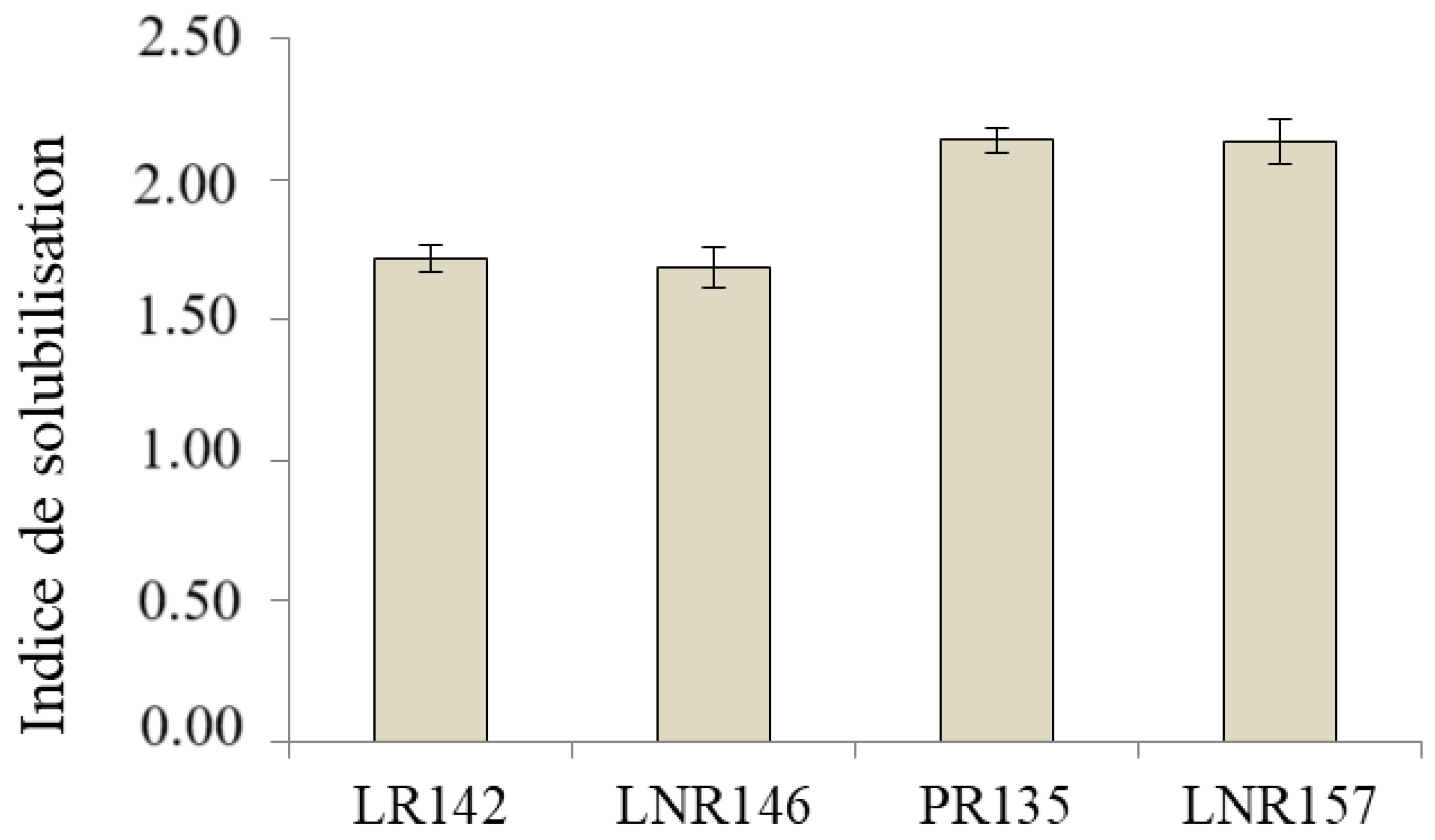
4.5. Solubilization of Phosphate
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Herman, R.P.; Provencio, K.R.; Herrera-Matos, J.; Torrez, R.J. Resource islands predict the distribution of heterotrophic bacteria in Chihuahuan Desert soils. Appl. Environ. Microbiol. 1995, 61, 1816–1821. [Google Scholar] [CrossRef]
- Grego, S.; Moscatelli, M.C.; Di Mattia, E.; Marinari, S.; Cacciari, I. Activité biochimique de la rhizosphère d’Acacia raddiana au nord et du sud du Sahara. In Un Arbre au Désert: Acacia raddiana; Grouzis, M., Le Floc’h, E., Eds.; IRD: Paris, France, 2003; pp. 231–247. ISBN 2-7099-1522-7. [Google Scholar]
- Lal, B.; Khanna, S. Selection of salt-tolerant Rhizobium isolates of Acacia nilotica. World J. Microbiol. Biotechnol. 1994, 10, 637–639. [Google Scholar] [CrossRef]
- Taoufiq, K.; Faghire, M.; Tahrouch, S.; Nejemeddine, M.; Oufdou, K.; Hatimi, A. Isolation and identification of endophytic bacteria isolated from legumes nodules and roots cultivated in acacia rhizosphere soils collected from Tata-Akka region of Morocco. Indian J. Nat. Sci. 2018. [Google Scholar]
- Zahran, H.H. Rhizobia from wild legumes: Diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J. Biotechnol. 2001, 91, 143–153. [Google Scholar] [CrossRef]
- Zahran, H.H. Enhancement of rhizobia–legumes symbioses and nitrogen fixation for crops productivity improvement. In Microbial Strategies for Crop Improvement; Khan, M.S., Zaidi, A., Musarrat, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 227–254. [Google Scholar]
- Essendoubi, M.; Brhada, F.; Eljamali, J.E.; Filali-Maltouf, A.; Bonnassie, S.; Georgeault, S.; Blanco, C.; Jebbar, M. Osmoadaptative responses in the rhizobia nodulating Acacia isolated from south-eastern Moroccan Sahara. Environ. Microbiol. 2007, 9, 603–611. [Google Scholar] [CrossRef]
- Naseem, H.; Bano, A. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J. Plant Interact. 2014, 9, 689–701. [Google Scholar] [CrossRef]
- Faghire, M.; Mandri, B.; Oufdou, K.; Bargaz, A.; Ghoulam, C.; Ramírez-Bahena, M.H.; Velázquez, E.; Peix, A. Identification at the species and symbiovar levels of strains nodulating Phaseolus vulgaris in saline soils of the Marrakech region (Morocco) and analysis of the otsA gene putatively involved in osmotolerance. Syst. Appl. Microbiol. 2012, 35, 156–164. [Google Scholar] [CrossRef]
- Faghire, M.; Farissi, M.; Taoufiq, K.; Fghire, R.; Bargaz, A.; Mandri, B.; Oufdou, K.; Laury, A.; Drevon, J.J.; Ghoulam, C. Genotypic variation of nodules’ enzymatic activities in symbiotic nitrogen fixation among common bean (Phaseolus vulgaris L.) genotypes grown under salinity constraint. Symbiosis 2013, 60, 115–122. [Google Scholar] [CrossRef]
- Bargaz, A.; Faghire, M.; Farissi, M.; Drevon, J.J.; Ghoulam, C. Oxidative stress in the root nodules of Phaseolus vulgaris is induced under conditions of phosphorus deficiency. Acta Physiol. Plant. 2013, 35, 1633–1644. [Google Scholar] [CrossRef]
- Faghire, M.; Bargaz, A.; Farissi, M.; Palma, F.; Mandri, B.; Lluch, C.; TejeraGarcía, N.A.; Herrera-Cervera, J.A.; Oufdou, K.; Ghoulam, C. Effect of salinity on nodulation, nitrogen fixation and growth of common bean (Phaseolus vulgaris) inoculated with rhizobial strains isolated from the Haouz region of Morocco. Symbiosis 2011, 55, 69–75. [Google Scholar] [CrossRef]
- Benidire, L.; Lahrouni, M.; El Khalloufi, F.; Gottfert, M.; Oufdou, K. Effects of Rhizobium leguminosarum inoculation on growth, nitrogen uptake and mineral assimilation in Viciafaba plants under salinity stress. J. Agric. Sci. Technol. 2017, 889–901. [Google Scholar]
- Benbrahim, K.F.; Berrada, H.; El Ghachtouli, N.; Ismaili, M. Les acacias: Des plantes fixatrices d’azote prometteuses pour le développement durable des zones arides et semi-arides [Acacia: PromisingNitrogen fixing trees for sustainabledevelopment in arid and semi-arid areas]. Int. J. Innov. Appl. Stud. 2014, 8, 46. [Google Scholar]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Scher, F.M.; Baker, R. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 1982, 72, 1567–1573. [Google Scholar] [CrossRef]
- Halder, A.K.; Mishra, A.K.; Bhattacharyya, P.; Chakrabartty, P.K. Solubilization of rock phosphate by Rhizobium and Bradyrhizobium. J. Gen. Appl. Microbiol. 1990, 36, 81–92. [Google Scholar] [CrossRef]
- Datta, C.; Basu, P.S. Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanuscajan. Microbiol. Res. 2000, 155, 123–127. [Google Scholar] [CrossRef]
- Peix, A.; Rivas-Boyero, A.A.; Mateos, P.F.; Rodriguez-Barrueco, C.; Martinez-Molina, E.; Velazquez, E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of Mesorhizobiummediterraneum under growth chamber conditions. Soil Biol. Biochem. 2001, 33, 103–110. [Google Scholar] [CrossRef]
- Idris, E.E.; Iglesias, D.J.; Talon, M.; Borriss, R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant-Microbe Interact. 2007, 20, 619–626. [Google Scholar] [CrossRef]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef]
- Kyungseok, P.; Kloepper, J.W.; Ryu, C.M. Rhizobacterial exopolysaccharides elicit induced resistance on cucumber. J. Microbiol. Biotechnol. 2008, 18, 1095–1100. [Google Scholar]
- Karadayi, M.; Gulluce, M.; Dogan, S.; Gulluce, E. Isolation and Molecular Characterization of Bacteria with Plant Growth Promoting Characteristics from Magnesite Mining Fields in Kütahya-Turkey. J. Appl. Biol. Sci. 2016, 10, 27–32. [Google Scholar]
- Adesemoye, A.O.; Torbert, H.A.; Kloepper, J.W. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb. Ecol. 2009, 58, 921–929. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 1–15. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Cleves, P.; Barka, E.A. Use of plant growth-promoting bacteria for sustainable agriculture. J. Plant Nutr. Soil Sci. 2019, 182, 563–579. [Google Scholar]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes are more than just endosymbionts. Funct. Plant Biol. 2006, 33, 701–712. [Google Scholar]
- de Carvalho, C.A.A.; Teixeira SM, R.; Figueira, A.V. Plant growth-promoting bacteria in agriculture: Benefits and applications. Microorganisms 2021, 9, 1478. [Google Scholar]
- Mitter, B.; Pfaffenbichler, N.; Riedel, T. Endophytes: The Plant Microbiome and Its Impact on Plant Health and Agriculture. J. Appl. Microbiol. 2017, 123, 938–949. [Google Scholar]
- Kumar, A.; Pandey, S.S.; Kumar, V. Endophytic bacteria: A novel approach for the enhancement of plant growth and stress tolerance. Environ. Exp. Bot. 2020, 176, 104041. [Google Scholar]
- Broughton, W.J.; Hernandez, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.) model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef]
- Manchanda, G.; Garg, N. Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant. 2008, 30, 595–618. [Google Scholar] [CrossRef]
- Naamala, J.; Jaiswal, S.K.; Dakora, F.D. Antibiotics resistance in Rhizobium: Type, process, mechanism and benefit for agriculture. Curr. Microbiol. 2016, 72, 804–816. [Google Scholar] [CrossRef]
- Howie, W.J.; Suslow, T.V. Role of antibiotic biosynthesis in the inhibition of Pythium ultimum in the cotton spermosphere and rhizosphere by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 1991, 4, 393–399. [Google Scholar] [CrossRef]
- Yokoyama, T.; Ando, S.; Tsuchiya, K. Serological properties and intrinsic antibiotic resistance of soybean bradyrhizobia isolated from Thailand. Soil Sci. Plant Nutr. 1999, 45, 505–515. [Google Scholar] [CrossRef]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. Int. Soc. Microb. Ecol. J. 2007, 1, 191. [Google Scholar] [CrossRef]
- Nwosu, V.C. Antibiotic resistance with particular reference to soil microorganisms. Res. Microbiol. 2001, 152, 421–430. [Google Scholar] [CrossRef]
- Vincent, J.M. The cultivation, isolation and maintenance of rhizobia. In A Manual for the Practical Study of Root-Nodule; Vincent, J.M., Ed.; Blackwell Scientific Publications: Oxford, UK, 1970; pp. 1–13. [Google Scholar]
- Wöhler, I. Auxin-indole derivatives in soils determined by a colorimetric method and by high performance liquid chromatography. Microbiol. Res. 1997, 152, 399–405. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Sultana, S.; Alam, S.; Karim, M.M. Screening of siderophore-producing salt-tolerant rhizobacteria suitable for supporting plant growth in saline soils with iron limitation. J. Agric. Food Res. 2021, 4, 100–150. [Google Scholar] [CrossRef]
- Fernandes Júnior, P.I.; Lima, A.A.D.; Passos, S.R.; Gava, C.A.T.; Oliveira, P.J.D.; Rumjanek, N.G.; Xavier, G.R. Phenotypic diversity and amylolytic activity of fast-growing rhizobia from pigeonpea [Cajanuscajan (L.) Millsp. Braz. J. Microbiol. 2012, 43, 1604–1612. [Google Scholar] [CrossRef]
- Singh, A.K.; Bhatt, R.P.; Pant, S.; Bedi, M.K.; Naglot, A. Characterization of Rhizobium isolated from root nodules of Trifolium alexandrinum. J. Agric. Technol. 2011, 7, 1705–1723. [Google Scholar]
- Kimenju, J.W.; Karanja, N.K.; Macharia, I. Plant parasitic nematodes associated with common bean in Kenya and the effect of Meloidogyne infection on bean nodulation. Afr. Crop Sci. J. 1999, 7, 503–510. [Google Scholar] [CrossRef]
- Melakeberhan, H.; Brooke, R.C.; Webster, J.M. Relationship between physiological response of French beans of different age to Meloidogyne incognita and subsequent yield loss. Plant Pathol. 1987, 35, 203–213. [Google Scholar] [CrossRef]
- Wang, E.T.; Tan, Z.Y.; Willems, A.; Fernández-López, M.; Reinhold-Hurek, B.; Martínez-Romero, E. Sinorhizobiummorelense sp. nov., a Leucaena leucocephala-associated bacterium that is highly resistant to multiple antibiotics. Int. J. Syst. Evol. Microbiol. 2002, 52, 1687–1693. [Google Scholar] [CrossRef]
- Amarger, N Rhizobia in the field. Adv. Agron. 2001, 73, 109–168.
- Ramesh, A.; Sharma, S.K.; Sharma, M.P.; Yadav, N.; Joshi, O.P. Plant growth-promoting traits in Enterobacter cloacae subsp. dissolvens MDSR9 isolated from soybean rhizosphere and its impact on growth and nutrition of soybean and wheat upon inoculation. Agric. Res. 2014, 3, 53–66. [Google Scholar] [CrossRef]
- Khalifa, A.Y.; Alsyeeh, A.M.; Almalki, M.A.; Saleh, F.A. Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef]
- Arredondo-Peter, R.; Escamilla, E. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein banding patterns among Rhizobium leguminosarumbiovarphaseoli strains isolated from the Mexican bean Phaseoluscoccineus. Appl. Environ. Microbioology 1993, 59, 3960–3962. [Google Scholar] [CrossRef]
- Ramirez, M.E.; Israel, D.W.; Wollum, A.G., II. Phenotypic and genotypic diversity of similar serotypes of soybean bradyrhizobia from two soil populations. Soil Biol. Biochem. 1997, 29, 1539–1545. [Google Scholar] [CrossRef]
- El-Akhal, M.R.; Rincon, A.; Mourabit, N.E.; Pueyo, J.J.; Barrijal, S. Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of peanut (Arachishypogaea L.) grown in Moroccan soils. J. Basic Microbiol. 2009, 49, 415–425. [Google Scholar] [CrossRef]
- Khamna, S.; Yokota, A.; Peberdy, J.F.; Lumyong, S. Indole-3-acetic acid production by Streptomyces sp. isolated from some Thai medicinal plant rhizosphere soils. Eur. Asian J. BioSci. 2010, 4. [Google Scholar] [CrossRef]
- Mohite, B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Haas, H. Molecular genetics of fungal siderophore biosynthesis and uptake: The role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 2003, 62, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Azmat, R. The impact of siderophore secretion by Pseudomonas stutzeri to chelating cu metal in solution culture. Pak. J. Bot. 2014, 46, 383–387. [Google Scholar]
- Kesaulya, H.; Hasinu, J.V.; Tuhumury, G.N. Potential of Bacillus spp. produces siderophoresinsuppressing the wilt disease of banana plants. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, England, 2018; Volume 102, p. 012016. [Google Scholar]
- Arora, N.K.; Kang, S.C.; Maheshwari, D.K. Isolation of siderophore-producing strains of Rhizobium meliloti and their biocontrol potential against Macrophominaphaseolina that causes charcoal rot of groundnut. Curr. Sci. 2001, 81, 673–677. [Google Scholar]
- Kesaulya, H.; Zakaria, B.; Syaiful, S.A. Isolation and physiological characterization of PGPR from potato plant rhizosphere in medium land of Buru Island. Procedia Food Sci. 2015, 3, 190–199. [Google Scholar] [CrossRef]
- Halder, A.K.; Chakrabartty, P.K. Solubilization of inorganic phosphate by Rhizobium. Folia Microbiol. 1993, 38, 325–330. [Google Scholar] [CrossRef]
- Rodíguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Peix, A.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; Rodríguez-Barrueco, C.; Velazquez, E. Pseudomonasrhizosphaerae sp. nov., a novel speciesthatactively solubilizes phosphate in vitro. Int. J. Syst. Evol. Microbiol. 2003, 53, 2067–2072. [Google Scholar] [CrossRef]
- Rosas, S.B.; Andrés, J.A.; Rovera, M.; Correa, N.S. Phosphate-solubilizing Pseudomonasputida can influence the rhizobia–legume symbiosis. Soil Biol. Biochem. 2006, 38, 3502–3505. [Google Scholar] [CrossRef]
- Rfaki, A.; Nassiri, L.; Ibijbijen, J. Phosphate-solubilizing bacteria in the rhizosphere of some cultivated legumes from Meknes region, Morocco. Br. Biotechnol. J. 2014, 4, 946–956. [Google Scholar] [CrossRef]
- Rfaki, A.; Nassiri, L.; Ibijbijen, J. Isolation and Characterization of phosphate solubilizing bacteria from the rhizosphere of faba bean (Viciafaba L.) in Meknes Region, Morocco. Br. Microbiol. Res. J. 2015, 6, 247–254. [Google Scholar] [CrossRef]
- Marra, L.M.; Oliveira, S.M.D.; Soares, C.R.F.S.; Moreira, F.M.D.S. Solubilisation of inorganic phosphates by inoculant strains from tropical legumes. Sci. Agric. 2011, 68, 603–609. [Google Scholar] [CrossRef]
- Sridevi, M.; Mallaiah, K.V.; Yadav, N.C.S. Phosphate solubilization by Rhizobium isolates from Crotalaria species. J. Plant Sci. 2007, 2, 635–639. [Google Scholar] [CrossRef]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Richardson, A.E.; Barea, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Zou, N.; Dart, P.J.; Marcar, N.E. Interaction of salinity and rhizobial strain on growth and N2-fixation by Acacia ampliceps. Soil Biol. Biochem. 1995, 27, 409–413. [Google Scholar] [CrossRef]
- Sadki, I.; Taoufiq, K.; Aberchane, L.; El Biari, K.; Tahrouch, S.; Oufdou, K.; Faghire, M.; Hatimi, A. Effect of inoculation by indigenous endophytic bacteria from the arid region in Morocco (Tata-Akka) on the antioxidative responses of Phaseolus vulgaris L. J. Mater. Environ. Sci. 2021, 12, 205–216. [Google Scholar]


| Antibiotic | Quantity of Antibiotic/Disc µg | |
|---|---|---|
| E15 | Erythromycin | 15 |
| CIP5 | Ciprofloxacin | 5 |
| SAM20 | Ampicillin–sulbactam | 20 |
| TET30 | Tetracycline | 30 |
| E15 | CIP5 | SAM20 | TET30 | |||||
|---|---|---|---|---|---|---|---|---|
| Growth | I.Z.D. | Growth | I.Z.D. | Growth | I.Z.D. | Growth | I.Z.D. | |
| PN101 | + | 0 o | − | 3 b | − | 2.3 e | − | 3 b |
| PN121 | − | 1.4 l | + | 0 o | − | 2.1 fg | − | 1.9 hi |
| PN123 | + | 0 o | − | 2.1 fg | − | 1.8 ij | − | 3 b |
| PN125 | + | 0 o | − | 1.7 jk | − | 1.6 k | − | 1.4 l |
| PN126 | + | 0 | + | 0 | − | 2.2 | − | 1.9 |
| PN131 | + | 0 o | − | 3.2 a | − | 1.7 jk | − | 3.3 a |
| HR46 | − | 1.3 lm | + | 0 o | − | 1.2 m | − | 2.1 fg |
| HR48 | − | 1.2 m | − | 1.4 l | − | 1.8 ij | − | 1.7 jk |
| HN51 | + | 0 o | + | 0 o | − | 1.8 ij | − | 2.6 d |
| LR142 | + | 0 o | + | 0 o | + | 0 o | − | 1.9 hi |
| LNR146 | + | 0 o | − | 1.3 lm | + | 0 o | − | 2.9 bc |
| FR13 | + | 0 o | − | 2.8 c | − | 1.8 ij | − | 2.9 bc |
| HR26 | + | 0 o | + | 0 o | − | 2.1fg | − | 3 b |
| HR33 | + | 0 o | + | 0 o | + | 0 o | + | 0 o |
| HR38 | + | 0 o | + | 0 o | − | 1.8 ij | − | 2.9 bc |
| HR57 | + | 0 o | + | 0 o | − | 2 gh | + | 0 o |
| PR113 | + | 0 o | + | 0 o | − | 1.6 k | − | 1.6 k |
| PR135 | + | 0 o | − | 3 b | − | 1.8 ij | − | 1.9 hi |
| LNR157 | + | 0 o | − | 1.4 l | + | 0 o | − | 2.1 fg |
| HN64 | + | 0 o | − | 2.5 d | − | 2.2 ef | − | 2.6 d |
| PN105 | + | 0 o | − | 2.1 fg | − | 1.4 l | − | 1.8 ij |
| PN112 | + | 0 o | − | 3 b | + | 0 o | − | 1.9 hi |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taoufiq, K.; Aberchane, L.; Amri, O.; Oufdou, K.; El Biari, K.; Talbi, A.; Fghire, R.; Ouachtak, H.; Faghire, M. Antibiotics Resistance and PGPR Traits of Endophytic Bacteria Isolated in Arid Region of Morocco. Int. J. Plant Biol. 2024, 15, 1063-1076. https://doi.org/10.3390/ijpb15040075
Taoufiq K, Aberchane L, Amri O, Oufdou K, El Biari K, Talbi A, Fghire R, Ouachtak H, Faghire M. Antibiotics Resistance and PGPR Traits of Endophytic Bacteria Isolated in Arid Region of Morocco. International Journal of Plant Biology. 2024; 15(4):1063-1076. https://doi.org/10.3390/ijpb15040075
Chicago/Turabian StyleTaoufiq, Khadijattou, Laila Aberchane, Oukacha Amri, Khalid Oufdou, Khouzaima El Biari, Abdeslem Talbi, Rachid Fghire, Hassan Ouachtak, and Mustapha Faghire. 2024. "Antibiotics Resistance and PGPR Traits of Endophytic Bacteria Isolated in Arid Region of Morocco" International Journal of Plant Biology 15, no. 4: 1063-1076. https://doi.org/10.3390/ijpb15040075
APA StyleTaoufiq, K., Aberchane, L., Amri, O., Oufdou, K., El Biari, K., Talbi, A., Fghire, R., Ouachtak, H., & Faghire, M. (2024). Antibiotics Resistance and PGPR Traits of Endophytic Bacteria Isolated in Arid Region of Morocco. International Journal of Plant Biology, 15(4), 1063-1076. https://doi.org/10.3390/ijpb15040075






