Genomic Prediction of Root Traits via Aerial Traits in Soybean Using Canonical Variables
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotypic Data Analysis
2.3. Phenotypic Data Analysis
2.4. Genomic Prediction Model
2.5. Evaluation of the Methodology
3. Results
3.1. Canonical Correlation Analysis
3.2. Heritability and Prediction Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borém, A.; Miranda, G.V.; Fritsche-Neto, R. Melhoramento de Plantas, 7th ed.; Editora UFV: Viçosa, Brazil, 2017. [Google Scholar]
- Silva, A.F.; Sediyama, T.; Silva, F.C.S.; Bezerra, A.R.G.; Ferreira, L.V. Correlation and Path Analysis of Yield Components in Soybean Varieties. Turk. J. Field Crops 2015, 10, 177–179. [Google Scholar] [CrossRef]
- Shea, Z.; Singer, W.M.; Zhang, B. Soybean Production, Versatility, and Improvement. In Legume Crops; Hasanuzzaman, M., Ed.; Intechopen: London, UK, 2020; Volume 1, pp. 1–22. [Google Scholar] [CrossRef]
- Ponnusha, B.S.; Subramaniyam, S.; Pasupathi, P.; Subramaniyam, B.; Virumandy, R. Antioxidant and Antimicrobial Properties of Glycine Max-A Review. Int. J. Curr. Biol. Med. Sci. 2011, 1, 49–62. [Google Scholar]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global Synthesis of Drought Effects on Food Legume Production. PLoS ONE 2015, 10, e0127401. [Google Scholar] [CrossRef] [PubMed]
- Polania, J.A.; Poschenrieder, C.; Beebe, S.; Rao, I.M. Effective Use of Water and Increased Dry Matter Partitioned to Grain Contribute to Yield of Common Bean Improved for Drought Resistance. Front. Plant Sci. 2016, 7, 660. [Google Scholar] [CrossRef] [PubMed]
- Waraich, E.A.; Ahmad, R.; Ashraf, M.Y. Role of Mineral Nutrition in Alleviation of Drought Stress in Plants. Aust. J. Crop Sci. 2011, 5, 764–777. [Google Scholar]
- Fenta, B.A.; Beebe, S.E.; Kunert, K.J.; Burridge, J.D.; Barlow, K.M.; Lynch, J.P.; Foyer, C.H. Field Phenotyping of Soybean Roots for Drought Stress Tolerance. Agronomy 2014, 4, 418–435. [Google Scholar] [CrossRef]
- Bucksch, A.; Burridge, J.; York, L.M.; Das, A.; Nord, E.; Weitz, J.S.; Lynch, J.P. Image-Based High-Throughput Field Phenotyping of Crop Roots. Plant Physiol. 2014, 166, 470–486. [Google Scholar] [CrossRef]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer Vision and Machine Learning Enabled Soybean Root Phenotyping Pipeline. Plant Methods 2020, 16, 5. [Google Scholar] [CrossRef]
- Andrade, L.R.B.d.; Sousa, M.B.; Oliveira, E.J.; Resende, M.D.V.; Azevedo, C.F. Cassava Yield Traits Predicted by Genomic Selection Methods. PLoS ONE 2019, 14, e0224920. [Google Scholar] [CrossRef]
- Heffner, E.L.; Jannink, J.L.; Iwata, H.; Souza, E.; Sorrells, M.E. Genomic Selection Accuracy for Grain Quality Traits in Biparental Wheat Populations. Crop Sci. 2011, 51, 2597–2606. [Google Scholar] [CrossRef]
- Hotelling, H. Relations Between Two Sets of Variates. Biometrika 1936, 28, 321–377. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating Crop Genetic Gains with Genomic Selection. Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Schnebly, S.R.; Rajcan, I. Accuracy of genomic prediction for seed oil concentration in high-oleic soybean populations using a low-density marker panel. Crop Sci. 2021, 61, 4012–4021. [Google Scholar] [CrossRef]
- Jia, Y.; Jannink, J.L. Multiple-Trait Genomic Selection Methods Increase Genetic Value Prediction Accuracy. Genetics 2012, 192, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Persa, R.; Bernardeli, A.; Jarquin, D. Prediction Strategies for Leveraging Information of Associated Traits under Single- and Multi-Trait Approaches in Soybeans. Agriculture 2020, 10, 308. [Google Scholar] [CrossRef]
- Hayashi, T.; Iwata, H. A Bayesian method and its variational approximation for prediction of genomic breeding values in multiple traits. BMC Bioinform. 2013, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Kizilkaya, K.; Zeng, J.; Garrick, D.; Fernando, R. Genomic Prediction from Multiple-Trait Bayesian Regression Methods Using Mixture Priors. Genetics 2018, 209, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Gianola, D.; Hernández-Suárez, C.M.; Martín-Vallejo, J. Multi-Trait, Multi-Environment Deep Learning Modeling for Genomic-Enabled Prediction of Plant Traits. G3 Genes Genomes Genet. 2018, 8, 3829–3840. [Google Scholar] [CrossRef]
- Saba, M.; Aaron, K.; Hu, G.; Wang, L.; Patrick, S.S. Multi-trait Genomic Selection Methods for Crop Improvement. Genetics 2020, 215, 931–945. [Google Scholar] [CrossRef]
- Apresentação do Município de Viçosa. Available online: https://www.vicosa.mg.gov.br/abrir_arquivo.aspx/Anexo_I__Apresentacao_Vicosa?cdLocal=2&arquivo=%7BC1D6CDDA-DDE4-5D26-DEA7-CE57C00D1CB7%7D.pdf (accessed on 2 January 2024).
- Nascimento, H.R.d.; Oliveira, L.M.; Duarte, A.B.; Dantas, S.A.G.; Ferreira, D.d.O.; Rosmaninho, L.B.d.C.; Cavallin, I.C.; da Cunha, F.F.; da Silva, F.L. A New Methodological Approach for Simulating Water Deficit in Soybean Genotypes. J. Agron. Crop Sci. 2021, 207, 946–955. [Google Scholar] [CrossRef]
- Bernardo, S.; Mantovani, E.C.; Silva, D.D.; Soares, A.A. Manual de Irrigação, 9th ed.; Editora UFV: Viçosa, Brazil, 2014. [Google Scholar]
- Fehr, W.R.; Caviness, C.E. Stages of Soybean Development; Iowa State University: Ames, IA, USA, 1977. [Google Scholar]
- Silva, F.; Borém, A.; Sediyama, T.; Câmara, G. Soja: Do Plantio à Colheita, 2nd ed.; Oficina de Textos: São Paulo, Brazil, 2022. [Google Scholar]
- WinRHIZO 2021. Available online: https://regentinstruments.com/assets/images_winrhizo/WinRHIZO_2021.pdf (accessed on 2 January 2024).
- Resende, M.D.V. Genética Biométrica e Estatística No Melhoramento de Plantas Perenes; Embrapa, Informação Tecnológica: Brasília, Brazil, 2002; 975 p. [Google Scholar]
- Hotelling, H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Hotelling, H. Simplified Calculation of Principal Components. Psychometrika 1936, 1, 27–35. [Google Scholar] [CrossRef]
- Ossani, C.; Cirillo, M.A.; Paulo, M.; Ossani, C. Package ‘Mvar.Pt’. 2023. Available online: https://cran.r-project.org/web/packages/MVar.pt/index.html (accessed on 2 January 2024).
- R Core Team. R: A Language and Environment for Statistical Computing, R Version 4.3.1; R Foundation for Statistical Computing: Vienna, Austria, 2023. Available online: https://www.R-project.org/ (accessed on 2 January 2024).
- Xu, S. Mapping quantitative trait loci by controlling polygenic background effects. Genetics 2013, 195, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the r Package Sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Leite WD, S.; Unêda-Trevisoli, S.H.; Silva FM, D.; Silva AJ, D.; Mauro, A.O.D. Identification of superior genotypes and soybean traits by multivariate analysis and selection index. Rev. Ciência Agronômica 2018, 49, 491–500. [Google Scholar] [CrossRef]
- Woyann, L.G.; Meira, D.; Matei, G.; Zdziarski, A.D.; Dallacorte, L.V.; Madella, L.A.; Benin, G. Selection indexes based on linear-bilinear models applied to soybean breeding. Agron. J. 2020, 112, 175–182. [Google Scholar] [CrossRef]
- Beche, E.; Gillman, J.D.; Song, Q.; Nelson, R.; Beissinger, T.; Decker, J.; Shannon, G.; Scaboo, A.M. Genomic prediction using training population design in interspecific soybean populations. Mol. Breed. 2021, 41, 15. [Google Scholar] [CrossRef]
- Khan, M.A.; Rai, A.; Mishra, D.C.; Budhlakoti, N.; Satpathy, S.; Majumdar, S.G. Comparative study of multi-trait genomic and phenotypic selection indexes for selection of superior genotypes. Indian J. Genet. Plant Breed. 2023, 83, 88–94. [Google Scholar]
- Dayoub, E.; Lamichhane, J.R.; Schoving, C.; Debaeke, P.; Maury, P. Early-Stage Phenotyping of Root Traits Provides Insights into the Drought Tolerance Level of Soybean Cultivars. Agronomy 2021, 11, 188. [Google Scholar] [CrossRef]
- Ferreira, D.F. Estatística Multivariada, 1st ed.; Editora UFLA: Lavras, Brazil, 2008. [Google Scholar]
- Paixão, P.T.M.; Nascimento, A.C.C.; Nascimento, M.; Azevedo, C.F.; Oliveira, G.F.; da Silva, F.L.; Caixeta, E.T. Factor Analysis Applied in Genomic Selection Studies in the Breeding of Coffea Canephora. Euphytica 2022, 218, 42. [Google Scholar] [CrossRef] [PubMed]
- De Ron, A.M.; Rodiño, A.P. Analysis of the Genetic Diversity of Crops and Associated Microbiota. Agronomy 2023, 13, 2132. [Google Scholar] [CrossRef]
- Karim, K.M.R.; Rafii, M.Y.; Misran, A.; Ismail, M.F.; Harun, A.R.; Ridzuan, R.; Chowdhury, M.F.N.; Hosen, M.; Yusuff, O.; Haque, M.A. Genetic Diversity Analysis among Capsicum annuum Mutants Based on Morpho-Physiological and Yield Traits. Agronomy 2022, 12, 2436. [Google Scholar] [CrossRef]
- Getnet, B.A. Genetic variability, heritability and expected genetic advance in soybean [Glycine max (L.) Merrill] genotypes. Agric. For. Fish. J. 2018, 7, 108–112. [Google Scholar] [CrossRef]
- Xavier, A.; Muir, W.M.; Rainey, K.M. Assessing Predictive Properties of Genome-Wide Selection in Soybeans. G3 Genes Genomes Genet. 2016, 6, 2611–2616. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Song, S.; Wang, W.; Wang, C.; Li, H.; Wang, F.; Li, S.; Sun, X. Screening diverse soybean genotypes for drought tolerance by membership function value based on multiple traits and drought-tolerant coefficient of yield. BMC Plant Biol. 2020, 20, 321. [Google Scholar] [CrossRef]
- Conte, M.V.D.; Carneiro, P.C.S.; Resende, M.D.V.; Silva, F.L.; Peternelli, L.A. Overcoming collinearity in path analysis of soybean [Glycine max (L.) Merr.] grain oil content. PLoS ONE 2020, 15, e0233290. [Google Scholar] [CrossRef]
- Bandillo, N.B.; Jarquin, D.; Posadas, L.G.; Lorenz, A.J.; Graef, G.L. Genomic Selection Performs as Effectively as Phenotypic Selection for Increasing Seed Yield in Soybean. Plant Genome 2023, 16, e20285. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez, P.; Hickey, J.; Burgueño, J.; Ornella, L.; Cerón-Rojas, J.; Zhang, X.; Dreisigacker, S.; Babu, R.; Li, L.; et al. Genomic Prediction in CIMMYT Maize and Wheat Breeding Programs. Heredity 2014, 112, 48–60. [Google Scholar] [CrossRef] [PubMed]
| Groups | Traits | Canonical Pairs | |
|---|---|---|---|
| 1° | 2° | ||
| Aerial (A1 and A2) | HD | −13.79 | 1.89 |
| PH | 0.39 | −0.10 | |
| Root (R1 and R2) | RDM | −49.58 | 12.20 |
| RV | −10.28 | 77.64 | |
| PSA | 1.05 | −8.04 | |
| TRL | −0.03 | 0.29 | |
| r | 0.59 * | 0.26 * | |
| p-value | 4.80 × 10−10 | 1.07 × 10−2 | |
| Groups | Traits | |
|---|---|---|
| Aerial | HD | 0.14 |
| PH | 0.37 | |
| Root | RDM | 0.66 |
| RV | 0.09 | |
| PSA | 0.19 | |
| TRL | 0.21 | |
| Latent Variable | A1 | 0.32 |
| Groups | Traits | ||||
|---|---|---|---|---|---|
| Aerial | HD | 0.61 | 0.0219 | 0.29 | 0.0401 |
| PH | 0.73 | 0.0153 | 0.52 | 0.0291 | |
| Root | RDM | 0.59 | 0.0208 | 0.44 | 0.0297 |
| RV | 0.55 | 0.0271 | 0.18 | 0.0431 | |
| PSA | 0.47 | 0.0272 | 0.25 | 0.0415 | |
| TRL | 0.51 | 0.0231 | 0.24 | 0.0397 | |
| Latent variable | A1 | 0.57 | 0.0246 | 0.35 | 0.0305 |
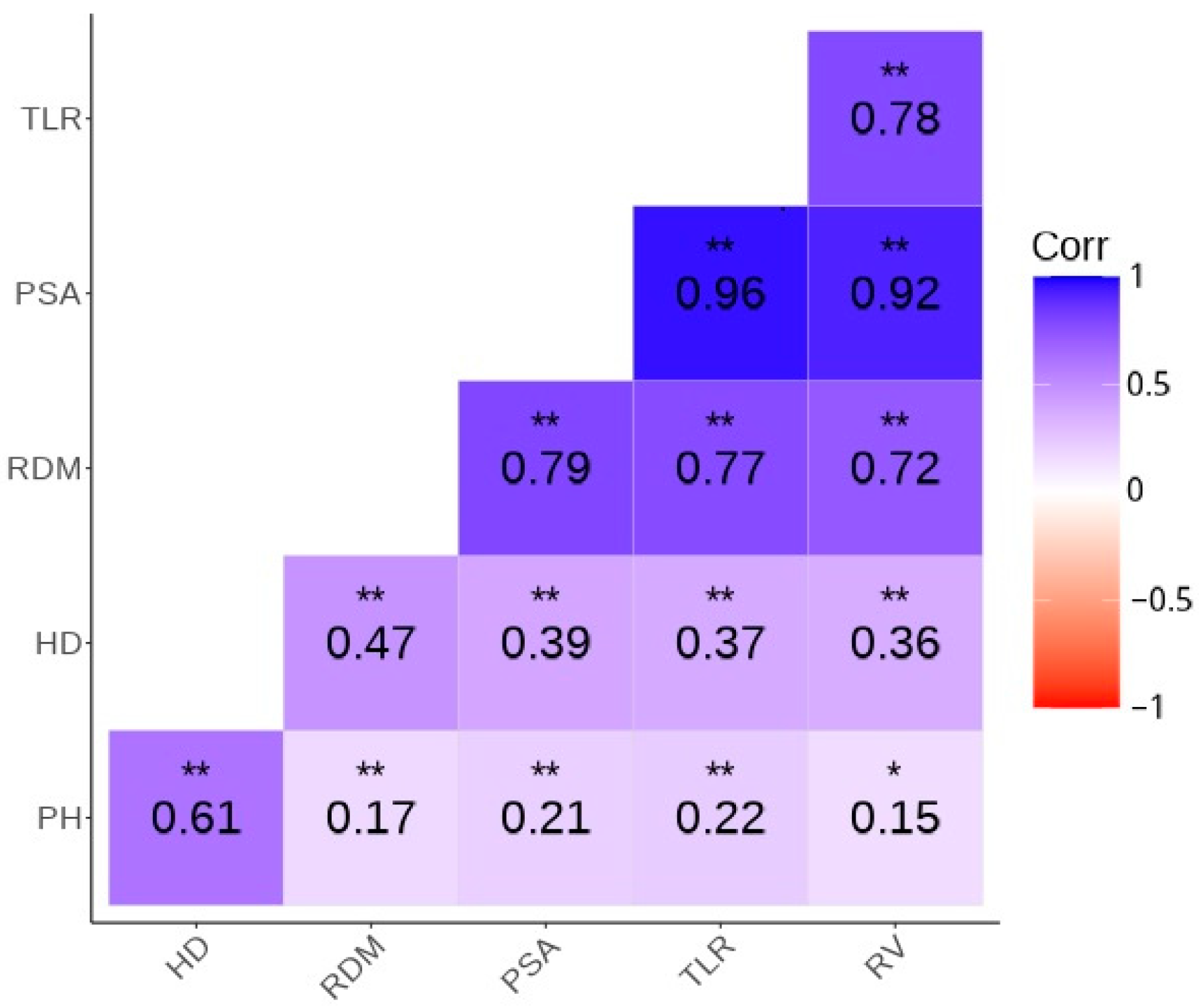
| Trait | PSA | HD | TLR | PH | RDM | RV | A1 |
|---|---|---|---|---|---|---|---|
| PSA | 1 | - | - | - | - | - | - |
| HD | 0.54 (0.03) | 1 | - | - | - | - | - |
| TLR | 0.88 (0.03) | 0.54 (0.03) | 1 | - | - | - | - |
| PH | 0.40 (0.02) | 0.51 (0.02) | 0.52 (0.02) | 1 | - | - | - |
| RDM | 0.63 (0.03) | 0.60 (0.02) | 0.60 (0.03) | 0.41 (0.02) | 1 | - | - |
| RV | 0.79 (0.03) | 0.51 (0.02) | 0.72 (0.04) | 0.46 (0.02) | 0.56 (0.024) | 1 | - |
| A1 | 0.50 (0.02) | 0.70 (0.03) | 0.49 (0.02) | 0.32 (0.02) | 0.69 (0.01) | 0.46 (0.02) | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagae, V.S.; Costa, N.M.E.P.d.L.d.; Suela, M.M.; Ferreira, D.d.O.; Nascimento, A.C.C.; Azevedo, C.F.; Silva, F.L.d.; Nascimento, M. Genomic Prediction of Root Traits via Aerial Traits in Soybean Using Canonical Variables. Int. J. Plant Biol. 2024, 15, 242-252. https://doi.org/10.3390/ijpb15020020
Sagae VS, Costa NMEPdLd, Suela MM, Ferreira DdO, Nascimento ACC, Azevedo CF, Silva FLd, Nascimento M. Genomic Prediction of Root Traits via Aerial Traits in Soybean Using Canonical Variables. International Journal of Plant Biology. 2024; 15(2):242-252. https://doi.org/10.3390/ijpb15020020
Chicago/Turabian StyleSagae, Vitor Seiti, Noé Mitterhofer Eiterer Ponce de Leon da Costa, Matheus Massariol Suela, Dalton de Oliveira Ferreira, Ana Carolina Campana Nascimento, Camila Ferreira Azevedo, Felipe Lopes da Silva, and Moysés Nascimento. 2024. "Genomic Prediction of Root Traits via Aerial Traits in Soybean Using Canonical Variables" International Journal of Plant Biology 15, no. 2: 242-252. https://doi.org/10.3390/ijpb15020020
APA StyleSagae, V. S., Costa, N. M. E. P. d. L. d., Suela, M. M., Ferreira, D. d. O., Nascimento, A. C. C., Azevedo, C. F., Silva, F. L. d., & Nascimento, M. (2024). Genomic Prediction of Root Traits via Aerial Traits in Soybean Using Canonical Variables. International Journal of Plant Biology, 15(2), 242-252. https://doi.org/10.3390/ijpb15020020









