Abstract
Farmers hoping to manage cropping systems sustainably are turning to cover crops to help mitigate plant pathogens. Plants with biofumigant properties are used to control soil-borne pathogens in agricultural settings, especially in till systems, where the brassicas are incorporated into the soil as green manure or seed meal. The effect of these crops is not well studied in no-till systems; thus, it is hard to know if they are as effective as green manure. Whether or not these cover crops can effect changes during a single growth season has not yet been studied. This study compared the response of the soil microbial community to four different brassica cover crops, two of which are commonly used in vineyards (Sinapis alba L. (white mustard) and Raphanus sativus (L.) Domin (tillage radish)) as well as two brassicas that are native or naturalized to the Okanagan (Capsella bursa-pastoris (L.) Medik. (Shepherd’s purse) and Boechera holboelli (Hornem.) Á. Löve and D. Löve (Holbøll’s rockcress)). Cover crops did not affect fungal species richness, but B. holboelli recover crops were associated with increased evenness among fungal taxa. Both C. bursa-pastoris and S. alba had lower levels of plant parasitic nematodes compared to non-brassica controls. These results were apparent only after a single growing season, which indicates growers could use this approach as needed, minimizing long-term exposure to biofumigants for beneficial soil microbes.
Keywords:
biofumigant; cover crop; diversity; fungi; glucosinolate; mycorrhizal fungi; nematode; ring nematode 1. Introduction
Vineyards around the world suffer significant economic loss from the destruction of crop due to soil-borne root diseases [1]. These include diseases caused by fungal pathogens such as Black-foot disease (caused by several fungal species in the genera Campylocarpon, Cylindrocarpon, Cylindrocladiella and Ilyonectria) and Petri disease (Phaeomoniella chlamydospore) as well as plant parasitic nematodes (PPNs) such as root-knot nematode (Meloidogyne spp.) and ring nematode (Mesocriconema xenoplax).
One way of controlling soil-borne fungal disease is through the use of fungicidal cover crops, particularly those in the Brassicaceae. Chemicals produced by these plants, glucosinolates (GSLs), have been found to be effective in the suppression of bacteria [2], fungi [3], nematodes [4], weeds [5] and insect pests [6]. Activation typically occurs with the maceration of plant tissues [7], therefore, treatment of soils with brassicas is commonly carried out in the form of green manure or seed meal applications.
Not all biofumigant cover crops are created equal. There is variation in GSLs among brassica taxa, which results in differential fungal [8,9,10] and nematode toxicity [11,12,13,14,15]. However, there is little guidance for farmers in choosing which brassica would be best for their particular pathogen challenge.
The broad-spectrum nature of GSLs is both good and bad—good in that they should theoretically inhibit all pathogens but bad in that they also affect beneficials. The use of biofumigants can lead to shifts in soil microbial communities [11,16,17,18,19,20,21]. Thus, growers need to be judicious in their use of biofumigant crops—restricting their use over time as to not impinge upon soil microbial functioning [22]. For example, biofumigant crops can have deleterious effects on essential beneficial microbes such as arbuscular mycorrhizal fungi (AMF) and free-living nematodes (FLNs) [23,24]). There is evidence that these crops may negatively affect AMF by inhibiting spore germination [25,26], suppressing the AM symbioses [27,28]. The effects of brassicas on FLNs have mixed results, with some studies reporting an increase in FLNs [29,30], while others reporting no change [13,31]. Understanding how beneficial microbes are affected by brassica crops is largely unexplored, yet is an important aspect of crop management.
If growers can use biofumigant crops strategically, deleterious effects on beneficials may be greatly reduced. By choosing the appropriate brassica taxa and applying it for the minimum amount of time, undesired effects to non-target organisms may be avoided. The aim of this study was to investigate how different species of intact brassica cover crops influence the soil fungal and nematode communities, including both pathogenic and beneficial organisms, in a vineyard setting over the course of a single growing season.
2. Materials and Methods
2.1. Vineyard Location and Site Conditions
The field study took place in Tantalus Vineyards, Kelowna, BC, Canada (latitude 49°50′5″ N, longitude −119°27′33″ W) from May 2020 to September 2020. This region lies within the Okanagan Valley geographical indication for viticulture (http://www.bcvqa.ca/wine-regions-of-bc/ (accessed on 23 June 2021). The field in question had been used as a horse pasture until 2012 and was fallow from 2012 to 2020. Vitis vinifera cv. Pinot noir (ungrafted) were planted with one shovel scoop of compost per vine (Glengrow: https://www.kelowna.ca/city-services/okanagan-compost/compost-we-produce/glengrow (accessed on 20 June 2021) at the beginning of the experiment in early June 2020.
2.2. Cover Crops
Two cultivars commonly used as cover crops were chosen: tillage radish (Raphanus sativus L.) and white mustard (Sinapis alba L.) as well as two cover crops native or naturalized to the Okanagan Valley (Shepherd’s purse (Capsella bursa-pastoris (L.) Medik.) and rockcress (Boechera holboelli (Hornem.) Á. Löve and D. Löve)). S. alba seeds were obtained from Richter’s, Ontario, Canada. R. sativus seeds were obtained from William Dam Seeds, Dundas, Ontario, Canada. C. bursa-pastoris seeds were obtained from Xeriscape Endemic Nursery, West Kelowna, BC, Canada. B. holboelli seeds were obtained from SeedsCo Community Conservation, Kelowna, BC, Canada.
2.3. Establishment of the Experiment
Field plots were prepared in May 2020 by removing all growth in the plots by a finger weeder (approximately 2–4 cm deep) and manual removal of all vegetation apart from control plots, where vegetation was not removed. Vegetation in the interrow remained undisturbed for all treatments. Seeds for each treatment were weighed ahead of time to achieve their respective seeding rates (Table 1). B. holboelli and C. bursa-pastoris were planted by sprinkling the seeds onto the soil while S. alba and R. sativus seeds were planted 2–3 cm deep individually and covered. Treatments were planted in a 5 × 5 randomized block design, with each row representing a block and plots within blocks representing experimental units for a total of 25 experimental units. One cover crop species was planted per plot (n = 5). Each plot contained five evenly spaced grapevines, which were planted 2 weeks after cover crop seeding.

Table 1.
Seeding rates for each brassica species used in the field experiment. Calculations based on recommendations from seed suppliers.
2.4. Plot Maintenance
Plants were watered and fertilized as per vineyard management protocols (4.5 metric tonnes/acre of compost (Glengrow) in spring—no additional fertilizer). Vines were irrigated by drip irrigation every other day for 2 h at 1.89 L per hour. Interrow cover crops were irrigated by microsprinklers for 4 h every 2 weeks. Water came on in May and continued through the season. Plots were hand-weeded as needed, to ensure the treatment species was the dominant undervine cover crop.
2.5. Estimated Biomass of Brassica Cover Crops
To estimate the amount of biomass produced by each of our brassica cover crops, the dry biomass of a randomly chosen single plant from each of the plots (roots included) was measured at the end of the growing season (September 2020). The number of individuals in each of the plots was estimated using a 50 × 50 cm quadrat, then multiplied by the mean dry biomass of the representative individuals by the total number of individuals to obtain a measure of dry biomass/m2. Vegetation biomass was not measured for control plots.
2.6. Soil Sampling
Three soil cores (3 cm × 20 cm deep) were taken from each plot at the end of the growing season (September 2020), combined in one plastic zipper storage bag and homogenized by shaking the bag before subsequent analyses. A subsample was stored in a paper bag and dried at 40 °C for 48 h for deoxyribose nucleic acid (DNA) extraction, soil chemistry and spore extraction. The soil was dried to ensure an equal amount of soil was used for each extraction. The dried soil was stored at −20 °C. The remaining wet soil was stored at 4 °C for nematode extraction. Soil pH was determined by mixing a 1:1 ratio of dried soil (10 g) with reverse osmosis water and measuring using an Orion Star A111 benchtop pH meter (Thermo Fisher Scientific, Waltham, MA, USA). Soil samples from each plot were examined separately.
2.7. Analysis of Effect of Cover Crops on AMF Abundance
To determine the influence of brassica species on AMF abundance in the soil, spore extractions were performed using 20 mg of dried soil from each plot using the protocol described in Gerdemann and Nicolson [32].
2.8. Fungal Community Analysis
DNA was extracted from each dried soil sample (500 mg) using FastDNA Spin Kit for Soil (MP Biomedicals, Irvine, CA, USA) and eluted with 100 µL DNase-free water. DNA quantification was carried out using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Extracted DNA was then sent to the Centre for Comparative Genomics and Evolutionary Bioinformatics (Dalhousie University, Halifax, NS, USA) for NextGen sequencing with the MiSeq (Illumina, Inc., San Diego, CA, USA). Amplicon sequencing of the ITS2 (Internal Transcribed Spacer) sub-region was performed for each plot using primers ITS86F 5′-GTGAATCATCGAATCTTTGAA-3′ and ITS4R 5′-TCCTCCGCTTATTGATATGC-3′ to target all fungi [33]. Raw reads were uploaded on NCBI (http://www.ncbi.nlm.nih.gov/bioproject/753154 (accessed on 9 August 2021)).
2.9. Fungal Community Sequencing
Raw sequence data were processed in QIIME2 (version 2020.2, https://qiime2.org). Due to the poor quality of the reverse reads, only forward reads were used for analyses. Of the five control plots, only three plots were successfully sequenced. Sequences were denoised using Divisive Amplicon Denoising Algorithm (DADA2 pipeline) [34] with the specifications: fragment length 6 to 279. Truncation length was determined by where the quality score starts to drop below 25. Features were identified for taxa comparison by using the QIIME2 q2-feature-classifier plugin and the naïve Bayes classifier trained on the UNITE database for QIIME reference sequences for fungi (version 8.2, dynamic) [35]. Alpha and beta diversity analyses were performed in QIIME2 using the core-metrics-phylogenetics plugin with a sampling depth of 2355 that retained 42.60% of all features in 100% of samples. Alpha diversity metrics used included Shannon diversity index (SDI), Faith’s phylogenetic diversity index (FPD), Pielou’s evenness and Observed Operational Taxonomic Units (OTU) (species richness). Beta diversity compared differences in OTUs in each plot based on cover crop treatment and metrics included Weighted UniFrac [36], Unweighted UniFrac [37] and Bray Curtis [38] dissimilarities.
2.10. Nematode Extraction and Counting
All soil nematodes were extracted from 100 cm3 of wet soil within 1 month of sampling using a modified wet sieving and sucrose flotation technique [39]. Nematodes extracted from each sample were counted using a gridded counting dish under an Olympus CK 2 inverted light microscope at 40× magnification (Olympus, Tokyo, Japan). To determine the individual abundances of PPNs and FLNs as well as the ratio of PPNs to FLNs, PPNs and FLNs were separately counted in one transect of the counting plate representing 1/12 of the total sample. PPNs were distinguished by the presence of a distinct stylet while all other nematodes were considered FLNs.
2.11. Statistical Analysis
2.11.1. Fungal Community
Correlation testing to relate mean diversity per cover crop (SDI, FPD, evenness and richness) with estimated mean brassica biomass per cover crop was carried out in QIIME2 using the alpha diversity metrics generated with core-metrics–phylogenetics and the qiime diversity alpha-correlation command [40].
Differences in alpha diversity among cover crop treatments were tested using one-way analysis of variance (ANOVA) with cover crop treatment as a fixed factor and block as a random factor (stats package version 4.0.5) [41] (R statistical software (version 1.4.1106)). Post hoc comparisons of means were performed using Tukey–Kramer post hoc test with p set at 0.05 (stats package version 2.0.5) [41]. Planned contrasts were conducted using the contrasts function (stats package version 4.0.5) [41] and the aov function from the car package (version 3.0-10) [42] to compare the control to all brassica treatments and to compare native brassica treatments to non-native brassica treatments. Planned contrasts were conducted for fungal species evenness, SDI, FPD and observed OTUs. Differences in beta diversity of OTUs among cover crop treatments was analyzed in QIIME2 using beta-group-significance (permutational analysis of variance (PERMANOVA), permutations = 999) [43].
Differential abundance of features among cover crop treatments was carried out using analysis of composition of microbiomes (ANCOM) in QIIME2. Before analysis, features were filtered to those that had a minimum frequency of 10 and appeared in a minimum of two samples. ANCOM was run on all features based on differences between treatments.
Taxonomic data for the Phylum, Class and Species levels (levels 2, 3 and 7) were exported from QIIME2 for further analysis in R. PERMANOVA using adonis2 (vegan package version 2.5-7) [44] was run at each of these taxonomic levels to determine differences in composition among cover crop treatments. A square root transformation was performed on the data before running the analysis to minimize the influence of the most abundant groups. A PERMANOVA was also run on all the phylotypes within each of the classes. If any significance was found, it was followed by a pairwise analysis using the vegan package add-on pairwise.adonis (Martinez Arbizu). All PERMANOVA tests used 999 permutations and Bray for calculating pairwise distances.
2.11.2. Nematode Community
A one-way ANOVA was run in R studio (stats package version 4.0.5) [41] with cover crop treatment as a fixed factor and block as a random factor to determine if brassica treatment had an effect on the nematode population. Brassica species were compared with total nematode abundance, estimated abundance of PPN and estimated abundance of FLNs in the soil. To meet the assumption of normality, FLN values were log transformed before final analysis. To determine if brassicas had an effect on the nematode population when compared to the control, or if native brassica treatments differed compared to non-native brassica treatments, a planned contrast was conducted using the contrasts function (stats package version 4.0.5) [41] and the aov function from the car package (version 3.0-10) [42]. These planned contrasts were conducted on total nematode abundance, PPN abundance and FLN abundance.
To determine how the treatment species influenced the nematode community composition in terms of PPN to FLN ratio (a binary outcome), a mixed effect logistic regression model was carried out in R (package lme4 version 1.1-26) [45] using block as a random factor and cover crop species as a fixed effect. This model was then run through ANOVA from the car package (version 3.0-10) [42]. This model was selected to show which treatments were more or less likely to alter the relative abundance of PPNs to FLNs compared to the control.
Correlation testing to relate each nematode measurement (total, PPNs, FLNs and PPN/FLN ratio) with estimated brassica biomass was carried out in R studio using the cor_test function from the rstatix package (version 0.7.0) [46]. Estimated biomass did not meet the normality assumption, so a non-parametric Spearman correlation was conducted with a confidence level of 0.95 [40].
3. Results
3.1. Cover Crop Establishment
At the time of harvest in September 2020, tillage radish had the highest estimated biomass per individual (54.9 g ± 10.4 g), followed by white mustard (19.5 g ± 6.9 g), then rockcress (1.1 g ± 0.2 g) and Shepherd’s purse (0.8 g ± 0.2 g) (Table S1). At the time of harvest, white mustard and Shepherd’s purse were at senescence, but Shepherd’s purse had started senescing in August 2020. Tillage radish was green and seeding, and rockcress was green and at maturity.
3.2. Effect of Brassica Cover Crops on Soil Fungal Diversity and Community Composition
Of the 127,143 sequences examined, 21.03% could not be identified past the phylum/class level. There was no significant difference in soil fungal community composition among cover crop treatments (Figure 1). The most abundant class in our communities was Sordariomycetes (44.50% of total reads), which contains a range of fungi including some plant pathogens [47]. Sordariomycetes was primarily represented by the family Nectriaceae (43.54% of Sordariomycetes), which is known to contain some plant pathogens as well as several species used as biocontrol agents [47]. These reads were not identified past the family level. Next most common was in the family Lasiosphaeriaceae (29.97% of Sordariomycetes), which are saprobes [48]. The genus Coniochaeta was also found in abundance (13.37% of Sordariomycetes). Species of this genus are known to be pathogens of woody hosts, including grapevines [49]. There are also some species in this genus that are known to be beneficial in that they are inhibitory to plant pathogenic fungi [49].
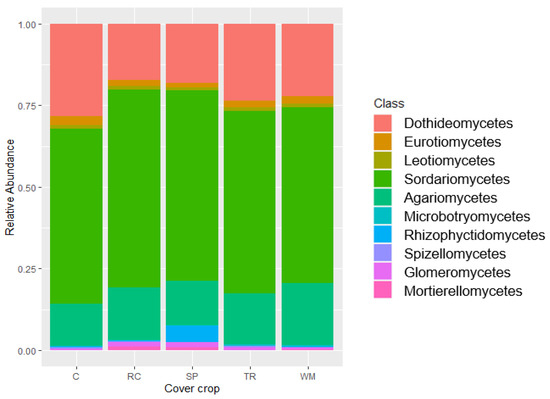
Figure 1.
Relative abundance of soil fungi (10 most abundant classes) grown with different cover crop treatments. C, undisturbed control; RC, rockcress; SP, Shepherd’s purse; TR, tillage radish; WM, white mustard. Values were obtained in QIIME2. PERMANOVA; F = 1.320; p = 0.237; R2 = 0.227. Dispersion; p = 0.918.
The next most abundant class found in our soil was Dothideomycetes (16.85% of total reads), which is mainly composed of saprobes but contains some plant pathogens [50]. Dothideomycetes was primarily represented by the genus Curvularia (89.58% of Dothideomycetes) (94% of which was species Curvularia ryleyi, Y.P. Tan and R.G. Shivas). C. ryleyi is known to be a pathogen of the genus Saproblus.
The class Agaricomycetes (12.26% of total reads) was the next most abundant and was primarily represented by the order Agaricales (30.65% of Agaricomycetes) and the family Cantharellaceae (8.47% of Agaricomycetes). The order Agaricales mainly contains saprobes but also ectomycorrhizal fungi and plant pathogens [51]. The reads found in our study were not identified further than the order Agaricales so it cannot be conferred what the main role in the community is. Other classes detected compromised less than 2% of total reads. There were no taxa whose abundance differed significantly among treatments (ANCOM).
Among the brassica plants, rockcress treatments had higher evenness than control plots evenness (Tukey–Kramer; p = 0.054) (Figure 2). We detected a trend for differences in fungal community evenness and Shannon Diversity Index (SDI) between brassica cover crops and the control (Table S2), but not between native and non-native brassica treatments (Table S3), or among different cover crop treatments (Table S4).
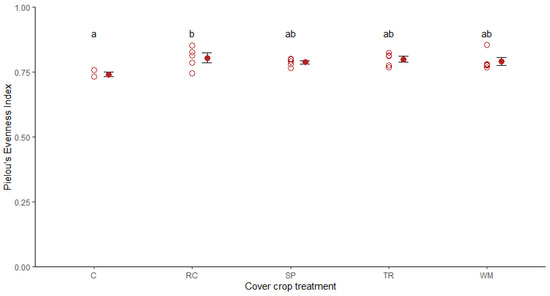
Figure 2.
Pielou’s evenness index for soil fungal communities associated with different cover crop treatments. C, undisturbed control; RC, rockcress; SP, Shepherd’s purse; TR, tillage radish; WM, white mustard (n = 3 for control, n = 5 for all other treatments). Hollow red circles represent individual replicates, solid red dots and black lines represent mean and standard error. Group means sharing the same letter are not significantly different.
One-way analysis of variance (ANOVA) indicated that there was no statistical difference among cover crop treatments in terms of SDI, species richness, or Faith’s Phylogenetic Diversity (FPD) (Table S5). Across all treatments, average evenness was 0.79 ± 0.03 (Figure 2), average SDI was 5.84 ± 0.29 (Figure S1), average species richness was 173 ± 32 (Figure S2) and average FPD was 46.95 ± 8.01 (Figure S3). There was no correlation between estimated brassica biomass and any of the fungal diversity metrics (Table S5).
ANOVA comparing all four brassica treatments did not indicate differences in AMF spore count in the soil among treatments. Planned contrasts also did not indicate any differences between brassica treatments and the control (p = 0.955) with respect to AMF spore abundance. Similarly, there were no differences in AMF spore abundance between native and cultivated brassica species (p = 0.958).
3.3. Do Intact Brassica Cover Crops Affect Soil Nematode Abundance and Community Composition?
Taken as a whole, we could not detect significant differences in abundance of nematodes among brassica cover crop treatments (p = 0.392). Abundance ranged from 780 nematodes per 100 mL soil to 1932 nematodes per 100 mL soil, with a mean of 1542 ± 625 nematodes per 100 mL soil. Similarly, there were no differences in total nematode abundance between the cultivated and native brassica species.
Cover crops did significantly affect the ratio of PPN to FLN (p = 0.008) (Figure 3). Shepherd’s purse (p = 0.036) and white mustard (p = 0.009) had a significantly lower PPN:FLN ratio to when compared to the control (Figure 3). In comparison to the control, Shepherd’s purse lowered the odds of finding a PPN by 46% and the white mustard treatment lowered the odds by 52%.
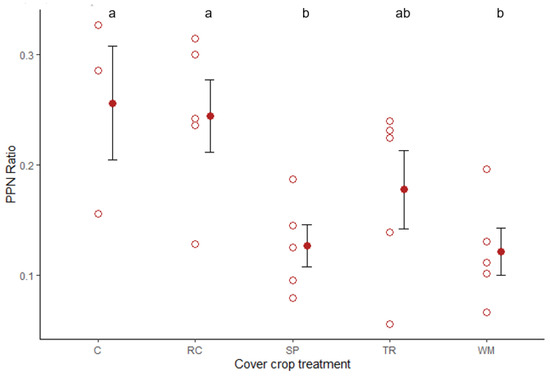
Figure 3.
Effect of cover crop treatment on parasitic (PPN) to free-living (FLN) nematode ratio. C, undisturbed control; RC, rockcress; SP, Shepherd’s purse; TR, tillage radish; WM, white mustard (n = 5). Data obtained using a mixed effect logistic regression model with plot number as a random factor. Hollow red circles represent individual replicates, solid red dots and black lines represent mean and standard error. ANOVA; χ2 = 13.87; df = 4; p = 0.008. Group means sharing the same letter are not significantly different.
4. Discussion
4.1. Brassica Cover Crops Effect on Soil Fungal Diversity
Our cover crop treatments significantly affected the evenness of soil fungal communities. In particular, the rockcress treatment had higher evenness compared to control. This suggests that rockcress was able to mediate competition among microbes by reducing dominance of strong competitors. In soils with high pathogen load, this may have a dramatic effect on soil suppressiveness. This is supported by the fact that Shannon Diversity Index (SDI) was lowest in controls, suggesting that brassicas reduce dominance among soil fungi. The low diversity of control soils was surprising, as they had much higher plant diversity [52,53]. This report is the first to document that brassica green manure or seed meal changes to soil fungal communities may result from changes to evenness, rather than richness. In this way, brassicas may engender increased microbial activity, in the absence of compositional changes [54,55,56].
4.2. Brassica Effects on AMF
Glomeromycota read abundance was not significantly different between treatments (Figure 3) and there was no significant difference in AMF spore count in the soil between treatments. This was unexpected as the hypothesis was that AMF spores would be less abundant in brassica treatments as Brassica sp. are considered non-mycorrhizal and sometimes known to inhibit germination of AMF spores [25,26,57]. In a field study by Pellerin et al. (2007), they found no evidence of a negative effect of the incorporation of brassica residues on the colonization of crop roots by AMF [58]. Another study by Tong et al. (2014) looked at the interactions between a brassica crop and a mycorrhizal crop when grown together [59]. They found the presence of a mycorrhizal crop with the brassica stimulated the GSL production/activation through constant invasion/contact with AMF. It is possible that growing the brassica crops in the presence of the mycorrhizal grapevine negated any negative biofumigant effects the brassica would have on AMF if grown alone. The AMF spore count alone does not reflect changes to the relationship between the grapevines and AMF. Future studies should look at AMF colonization in the grapevine roots.
4.3. Brassica Effects on Nematodes
When comparing the relative abundance of PPNs and FLNs, both Shepherd’s purse and white mustard had significantly lower PPN:FLN ratio than the control. Multiple studies have shown nematode communities react differently depending on GSL profiles produced by different crops [11,12,60,61,62]. The PPNs present in this field may be particularly susceptible to the GSLs that are secreted by Shepherd’s purse and white mustard, while some of the FLNs present may be stimulated by it. A reason that rockcress may be less efficient in reducing PPN populations relative to FLNs is that the rockcress plant is relatively small compared to the other brassicas and it has a shallow root system. Many PPNs of interest live deep in the soil profile, which may be unaffected by rockcress root presence and exudates [63]. The host status of these brassica species for the plant parasitic nematode species in the soil needs additional study.
Total nematode abundance as well as FLN abundance were found to be positively correlated with estimated brassica biomass while PPN abundance was not. This suggests the brassica biomass was affecting the FLN population more than the PPN population. A study by Lu et al. (2016) found a positive correlation between plant biomass (mixed brassica and non-brassica) with nematode abundance [64], but many other studies comparing total nematode population to non-brassica plant biomass have not found a correlation [65,66,67].
5. Conclusions
In our study, brassica cover treatments had measurable effects on both soil fungal and nematode communities after only one field growing season. Such changes were not at the expense of either beneficial fungus (AMF) nor free living nematodes (FLNs). Our study found that even naturally occurring, common vineyard crops (Shepherd’s purse) can provide additional benefits than they are currently used for. Further studies should include challenging crops with pathogens to see if observed changes result in lower disease incidence. Additionally, further studies that include looking at any functional changes that may be occurring in soil community would be beneficial as there may be changes that occur in the community on a functional level that are not reflected in the community analysis. For now, these results suggest a promising and nature-based alternative to tillage, allowing for short-term manipulation of soil microbial communities.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijpb14040081/s1, Figure S1. Shannon diversity index of fungal communities in soil samples from plots with different cover crop treatments; Figure S2. Species richness as measured by number of observed OTUs in fungal communities in soil samples from plots with different cover crop treatments; Figure S3. Faith’s Phylogenetic Diversity index of fungal communities in soil samples from plots with different cover crop treatments; Table S1. Mean biomass (grams) for individual plants from each brassica treatment; Table S2. Planned comparison results for indices of diversity of the soil fungal community between all brassica cover crop treatments (tillage radish, white mustard, shepherd’s purse, rockcress) and the undisturbed control; Table S3. Planned comparison results for indices of diversity of the soil fungal community between native brassica cover crop treatments (shepherd’s purse and rockcress) and non-native brassica cover crop treatments (tillage radish and white mustard); Table S4. One-way ANOVA results for indices of diversity of the soil fungal community among different cover crop treatments (tillage radish, white mustard, shepherd’s purse, rockcress); Table S5. Spearman correlation coefficients relating estimated brassica cover crop biomass to various diversity measurements (Pielou’s evenness Index, Shannon Diversity Index (SDI), Species richness, and Faith Phylogenetic Diversity (FPD)). Includes all brassica treatments (tillage radish, white mustard, shepherd’s purse, and rockcress); Table S6. PERMANOVA results for fungal community composition of different cover crop treatments (tillage radish, white mustard, shepherd’s purse, rockcress, and control) at different taxonomic levels; Table S7. PERMANOVA results for beta diversity of detected fungal OTUs of different cover crop treatments.
Author Contributions
Conceptualization, C.O. and M.M.H.; methodology, C.O., T.F. and M.M.H.; software, C.O.; validation, C.O., T.F. and M.M.H.; formal analysis, C.O.; investigation, C.O.; resources, C.O., T.F. and M.M.H.; data curation, C.O.; writing—original draft preparation, C.O.; writing—review and editing C.O., T.F. and M.M.H.; visualization, C.O.; supervision, T.F. and M.M.H.; project administration, M.M.H.; Funding acquisition, M.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agriculture and Agrifood Canada Going Forward Grape Cluster Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw reads can be found at http://www.ncbi.nlm.nih.gov/bioproject/753154.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Úrbez-Torres, J.R.; Haag, P.; Bowen, P.; O’Gorman, D.T. Grapevine Trunk Diseases in British Columbia: Incidence and Characterization of the Fungal Pathogens Associated with Black Foot Disease of Grapevine. Plant Dis. 2014, 98, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Barton, M.; Pendleton, P. Controlled Release of Allyl Isothiocyanate for Bacteria Growth Management. Food Control 2012, 23, 478–484. [Google Scholar] [CrossRef]
- Tiznado-Hernandez, M.-E.; Troncoso-Rojas, R. Control of Fungal Diseases with Isothiocyanates. Stewart Postharvest Rev. 2008, 2, 1–14. [Google Scholar] [CrossRef]
- Eugui, D.; Escobar, C.; Velasco, P.; Poveda, J. Glucosinolates as an effective tool in plant-parasitic nematodes control: Exploiting natural plant defenses. Appl. Soil Ecol. 2022, 176, 104497. [Google Scholar] [CrossRef]
- Norsworthy, J.K.; Brandenberger, L.; Burgos, N.R.; Riley, M. Weed Suppression in Vigna unguiculata with a Spring-Seeded Brassicaceae Green Manure. Crop Prot. 2005, 24, 441–447. [Google Scholar] [CrossRef]
- Hopkins, R.J.; Van Dam, N.M.; Van Loon, J.J.A. Role of Glucosinolates in Insect-Plant Relationships and Multitrophic Interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Nakamura, K.; Asai, Y.; Wada, T.; Tanaka, K.; Matsuo, T.; Okamoto, S.; Meijer, J.; Kitamura, Y.; Nishikawa, A.; et al. Comparison of the Glucosinolate-Myrosinase Systems among Daikon (Raphanus sativus, Japanese White Radish) Varieties. J. Agric. Food Chem. 2008, 56, 2702–2707. [Google Scholar] [CrossRef]
- Drobnica, L.; Zemanova, M.; Nemec, P.; Antos, K.; Kristian, P.; Stullerova, A.; Knoppova, V. Antifungal Activity of Isothiocyanates and Related Compounds I. Naturally Occurring Isothiocyanates and Their Analogues. Appl. Microbiol. 1967, 15, 701–709. [Google Scholar] [CrossRef]
- Manici, L.M.; Lazzeri, L.; Palmieri, S. In Vitro Fungitoxic Activity of Some Glucosinolates and Their Enzyme-Derived Products toward Plant Pathogenic Fungi. J. Agric. Food Chem. 1997, 45, 2768–2773. [Google Scholar] [CrossRef]
- Sarwar, M.; Kirkegaard, J.A.; Wong, P.T.W.; Desmarchelier, J.M. Biofumigation Potential of Brassicas III. In Vitro Toxicity of Isothiocyanates to Soil-Borne Fungal Pathogens. Plant Soil 1998, 201, 103–112. [Google Scholar] [CrossRef]
- Mocali, S.; Landi, S.; Curto, G.; Dallavalle, E.; Infantino, A.; Colzi, C.; D’Errico, G.; Roversi, P.F.; D’Avino, L.; Lazzeri, L. Resilience of Soil Microbial and Nematode Communities after Biofumigant Treatment with Defatted Seed Meals. Ind. Crops Prod. 2015, 75, 79–90. [Google Scholar] [CrossRef]
- Reardon, C.L.; Strauss, S.L.; Mazzola, M. Changes in Available Nitrogen and Nematode Abundance in Response to Brassica Seed Meal Amendment of Orchard Soil. Soil. Biol. Biochem. 2013, 57, 22–29. [Google Scholar] [CrossRef]
- Gruver, L.S.; Weil, R.R.; Zasada, I.A.; Sardanelli, S.; Momen, B. Brassicaceous and Rye Cover Crops Altered Free-Living Soil Nematode Community Composition. Appl. Soil Ecol. 2010, 45, 1–12. [Google Scholar] [CrossRef]
- Zasada, I.A.; Meyer, S.L.F.; Morra, M.J. Brassicaceous Seed Meals as Soil Amendments to Suppress the Plant-Parasitic Nematodes Pratylenchus penetrans and Meloidogyne incognita. J. Nematol. 2009, 41, 221–227. [Google Scholar] [PubMed]
- Dahlin, P.; Hallmann, J. New Insights on the Role of Allyl Isothiocyanate in Controlling the Root Knot Nematode Meloidogyne hapla. Plants 2020, 9, 603. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.; Yang, H.; Chang, Z. Effect of Biofumigation and Chemical Fumigation on Soil Microbial Community Structure and Control of Pepper Phytophthora Blight. World J. Microbiol. Biotechnol. 2014, 30, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.S.; Hu, P.; Hollister, E.B.; Rothlisberger, K.L.; Somenahally, A.; Provin, T.L.; Hons, F.M.; Gentry, T.J. Impact of Indian Mustard (Brassica juncea) and Flax (Linum usitatissimum) Seed Meal Applications on Soil Carbon, Nitrogen, and Microbial Dynamics. Appl. Environ. Soil Sci. 2012, 2012, 351609. [Google Scholar] [CrossRef]
- Omirou, M.; Rousidou, C.; Bekris, F.; Papadopoulou, K.K.; Menkissoglou-Spiroudi, U.; Ehaliotis, C.; Karpouzas, D.G. The Impact of Biofumigation and Chemical Fumigation Methods on the Structure and Function of the Soil Microbial Community. Microb. Ecol. 2011, 61, 201–213. [Google Scholar] [CrossRef]
- Larkin, R.P.; Griffin, T.S.; Honeycutt, C.W. Rotation and Cover Crop Effects on Soilborne Potato Diseases, Tuber Yield, and Soil Microbial Communities. Plant Dis. 2010, 94, 1491–1502. [Google Scholar] [CrossRef]
- Cohen, M.F.; Yamasaki, H.; Mazzola, M. Brassica napus Seed Meal Soil Amendment Modifies Microbial Community Structure, Nitric Oxide Production and Incidence of Rhizoctonia Root Rot. Soil Biol. Biochem. 2005, 37, 1215–1227. [Google Scholar] [CrossRef]
- Richards, A.; Estaki, M.; Úrbez-Torres, J.R.; Bowen, P.; Lowery, T.; Hart, M. Cover Crop Diversity as a Tool to Mitigate Vine Decline and Reduce Pathogens in Vineyard Soils. Diversity 2020, 12, 128. [Google Scholar] [CrossRef]
- Waisen, P.; Cheng, Z.; Sipes, B.S.; Wang, K.H. Biofumigation Effects of Brassicaceous Cover Crops on Soil Health in Cucurbit Agroecosystems in Hawaii, USA. Pedosphere 2022, 32, 521–531. [Google Scholar] [CrossRef]
- Ingham, R.E.; Trofymow, J.A.; Ingham, E.R.; Coleman, D.C. Interactions of Bacteria, Fungi, and Their Nematode Grazers: Effects on Nutrient Cycling. Ecol. Monogr. 1985, 55, 119–140. [Google Scholar] [CrossRef]
- Freckman, D.W. Bacterivorous Nematodes and Organic-Matter Decomposition. Agric. Ecosyst. Environ. 1988, 24, 195–217. [Google Scholar] [CrossRef]
- Vierheilig, H.; Ocampo, J.A. Effect of Isothiocyanates on Germination of Spores of G. mosseae. Soil Biol. Biochem. 1990, 22, 1161–1162. [Google Scholar] [CrossRef]
- Schreiner, R.P.; Koide, R.T. Antifungal Compounds from the Roots of Mycotrophic and Non-Mycotrophic Plant. New Phytol. 1993, 123, 99–105. [Google Scholar] [CrossRef]
- Cipollini, D.; Cipollini, K. A Review of Garlic Mustard (Alliaria petiolata, Brassicaceae) as an Allelopathic Plant. J. Torrey Bot. Soc. 2016, 143, 339–348. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Rodgers, V.L.; Stinson, K.A.; Pringle, A. The Invasive Plant Alliaria petiolata (Garlic Mustard) Inhibits Ectomycorrhizal Fungi in Its Introduced Range. J. Ecol. 2008, 96, 777–783. [Google Scholar] [CrossRef]
- Stirling, B.G.; Wilson, E.; Stirling, A.; Pankhurst, C.; Moody, P.; Bell, M. Organic Amendments Enhance Biological Suppression of Plant-Parasitic Nematodes in Sugarcane Soils. In Proceedings of the 2003 Conference of the Australian Society of Sugar Cane Technologists, Townsville, Australia, 6–9 May 2003; Volume 25. [Google Scholar]
- Valdes, Y.; Viaene, N.; Moens, M. Effects of Yellow Mustard Amendments on the Soil Nematode Community in a Potato Field with Focus on Globodera rostochiensis. Appl. Soil Ecol. 2012, 59, 39–47. [Google Scholar] [CrossRef]
- Vervoort, M.T.W.; Vonk, J.A.; Brolsma, K.M.; Schütze, W.; Quist, C.W.; de Goede, R.G.M.; Hoffland, E.; Bakker, J.; Mulder, C.; Hallmann, J.; et al. Release of Isothiocyanates Does Not Explain the Effects of Biofumigation with Indian Mustard Cultivars on Nematode Assemblages. Soil Biol. Biochem. 2014, 68, 200–207. [Google Scholar] [CrossRef]
- Gerdemann, J.W.; Nicolson, T.H. Spores of Mycorrhizal Endogone Species Extracted from Soil by Wet Sieving and Decanting. Trans. Br. Mycol. Soc. 1963, 46, 235–244. [Google Scholar] [CrossRef]
- Vancov, T.; Keen, B. Amplification of Soil Fungal Community DNA Using the ITS86F and ITS4 Primers. FEMS Microbiol. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Fungi; UNITE Community, 2020. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, T. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content. K. Dan. Vidensk. Selsk. 1948, 5, 1–5. [Google Scholar]
- Jenkins, W.R. A Rapid Centrifugal-Flotation Technique for Separating Nematodes from Soil. Plant Dis. Rep. 1964, 48, 692. [Google Scholar]
- Spearman, C. The Proof and Measurement of Association between Two Things. Am. J. Psychol. 1904, 15, 72–101. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.; Guillaume, B.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://github.com/vegandevs/vegan (accessed on 10 October 2023).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statstical Tests. 2021. Available online: https://rpkgs.datanovia.com/rstatix/ (accessed on 10 October 2023).
- Bao, D.F.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Perera, R.H.; Thiyagaraja, V.; Hongsanan, S.; Wanasinghe, D.N.; Shen, H.W.; Tian, X.G.; Yang, L.Q.; et al. Taxonomy, phylogeny and evolution of freshwater Hypocreomycetidae (Sordariomycetes). Fungal Divers. 2023, 121, 1–94. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Purahong, W.; Zhang, W.; Wubet, T.; Li, X.; Liu, M.; Zhao, W.; Hyde, K.D.; Liu, J.H.; Yan, J. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 2018, 90, 1–84. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Nityagovsky, N.N.; Kiselev, K.V. Biodiversity of endophytic bacteria and fungi of wild grapes Vitis amurensis Rupr. In BIO Web of Conferences; EDP Sciences: Les Ulis, France, 2021; Volume 39. [Google Scholar]
- Pem, D.; Jeewon, R.; Chethana, K.W.T.; Hongsanan, S.; Doilom, M.; Suwannarach, N.; Hyde, K.D. Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 2021, 109, 283–319. [Google Scholar] [CrossRef]
- Kalichman, J.; Kirk, P.M.; Matheny, P.B. A compendium of generic names of agarics and Agaricales. Taxon 2020, 69, 425–447. [Google Scholar] [CrossRef]
- Thapa, V.R.; Ghimire, R.; Acosta-Martínez, V.; Marsalis, M.A.; Schipanski, M.E. Cover Crop Biomass and Species Composition Affect Soil Microbial Community Structure and Enzyme Activities in Semiarid Cropping Systems. Appl. Soil Ecol. 2021, 157, 103735. [Google Scholar] [CrossRef]
- Steinauer, K.; Chatzinotas, A.; Eisenhauer, N. Root Exudate Cocktails: The Link between Plant Diversity and Soil Microorganisms? Ecol. Evol. 2016, 6, 7387–7396. [Google Scholar] [CrossRef]
- Hu, P.; Hollister, E.B.; Somenahally, A.C.; Hons, F.M.; Gentry, T.J. Soil Bacterial and Fungal Communities Respond Differently to Various Isothiocyanates Added for Biofumigation. Front. Microbiol. 2015, 5, 729. [Google Scholar] [CrossRef]
- Hollister, E.B.; Hu, P.; Wang, A.S.; Hons, F.M.; Gentry, T.J. Differential Impacts of Brassicaceous and Nonbrassicaceous Oilseed Meals on Soil Bacterial and Fungal Communities. Microbiol. Ecol. 2012, 83, 632–641. [Google Scholar] [CrossRef]
- Siebers, M.; Rohr, T.; Ventura, M.; Schü Tz, V.; Thies, S.; Kovacic, F.; Jaeger, K.-E.; Berg, M.; Dö Rmann, P.; Schulz, M. Disruption of Microbial Community Composition and Identification of Plant Growth Promoting Microorganisms after Exposure of Soil to Rapeseed-Derived Glucosinolates. PLoS ONE 2018, 13, e0200160. [Google Scholar] [CrossRef]
- Vierheilig, H.; Bennett, R.; Kiddle, G.; Kaldorf, M.; Ludwig-Müller, J. Differences in Glucosinolate Patterns and Arbuscular Mycorrhizal Status of Glucosinolate-Containing Plant Species. New Phytol. 2000, 146, 343–352. [Google Scholar] [CrossRef]
- Pellerin, S.; Mollier, A.; Morel, C.; Plenchette, C. Effect of Incorporation of Brassica napus L. Residues in Soils on Mycorrhizal Fungus Colonisation of Roots and Phosphorus Uptake by Maize (Zea mays L.). Europ. J. Agron. 2007, 26, 113–120. [Google Scholar] [CrossRef]
- Tong, Y.; Gabriel-Neumann, E.; Krumbein, A.; Ngwene, B.; George, E.; Schreiner, M. Interactive Effects of Arbuscular Mycorrhizal Fungi and Intercropping with Sesame (Sesamum indicum) on the Glucosinolate Profile in Broccoli (Brassica oleracea Var. Italica). Environ. Exp. Bot. 2014, 109, 288–295. [Google Scholar] [CrossRef]
- Potter, M.J.; Vanstone, V.A.; Davies, K.A.; Kirkegaard, J.A.; Rathjen, A.J. Reduced Susceptibility of Brassica napus to Pratylenchus neglectus in Plants with Elevated Root Levels of 2-Phenylethyl Glucosinolate. J. Nematol. 1999, 31, 291–298. [Google Scholar] [PubMed]
- Zasada, I.A.; Ferris, H. Sensitivity of Meloidogyne javanica and Tylenchulus semipenetrans to Isothiocyanates in Laboratory Assays. Phytopathology 2003, 93, 747–750. [Google Scholar] [CrossRef]
- Zasada, I.A.; Ferris, H. Nematode Suppression with Brassicaceous Amendments: Application Based upon Glucosinolate Profiles. Soil Biol. Biochem. 2004, 36, 1017–1024. [Google Scholar] [CrossRef]
- Goodell, P.; Ferris, H. Plant-Parasitic Nematode Distributions in an Alfalfa Field. J. Nematol. 1980, 12, 136–141. [Google Scholar]
- Lu, Z.-B.; Dong, D.-F.; Yang, B.; Li, L.-L.; Yu, Y.; Ouyang, F.; Ge, F.; Verma, V.-C.; Men, X.-Y. Effects of Crop Species Richness on the Community of Soil Nematodes in an Experimental Agro-Ecosystem. Eur. J. Soil Biol. 2016, 73, 26–33. [Google Scholar] [CrossRef]
- Viketoft, M. Effects of Six Grassland Plant Species on Soil Nematodes: A Glasshouse Experiment. Soil Biol. Biochem. 2008, 40, 906–915. [Google Scholar] [CrossRef]
- Cortois, R.; Veen, G.F.C.; Duyts, H.; Abbas, M.; Strecker, T.; Kostenko, O.; Eisenhauer, N.; Scheu, S.; Gleixner, G.; De Deyn, G.B.; et al. Possible Mechanisms Underlying Abundance and Diversity Responses of Nematode Communities to Plant Diversity. Ecosphere 2017, 8, e01719. [Google Scholar] [CrossRef]
- De Deyn, G.B.; Raaijmakers, C.E.; Van Ruijven, J.; Berendse, F.; Van Der Putten, W.H. Plant Species Identity and Diversity Effects on Different Trophic Levels of Nematodes in the Soil Food Web. Okios 2004, 106, 576–586. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).