Novel Sources of Resistance to Stagonospora nodorum and Role of Effector-Susceptibility Gene Interactions in Wheat of Russian Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Objects
2.2. Experimental Design
2.3. Plant Resistance/Susceptibility Assay to NEs SnToxA, SnTox1 and SnTox3
2.4. Isolation of DNA in Plants and Fungi and Performing the Polymerase Chain Reaction (PCR)
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results
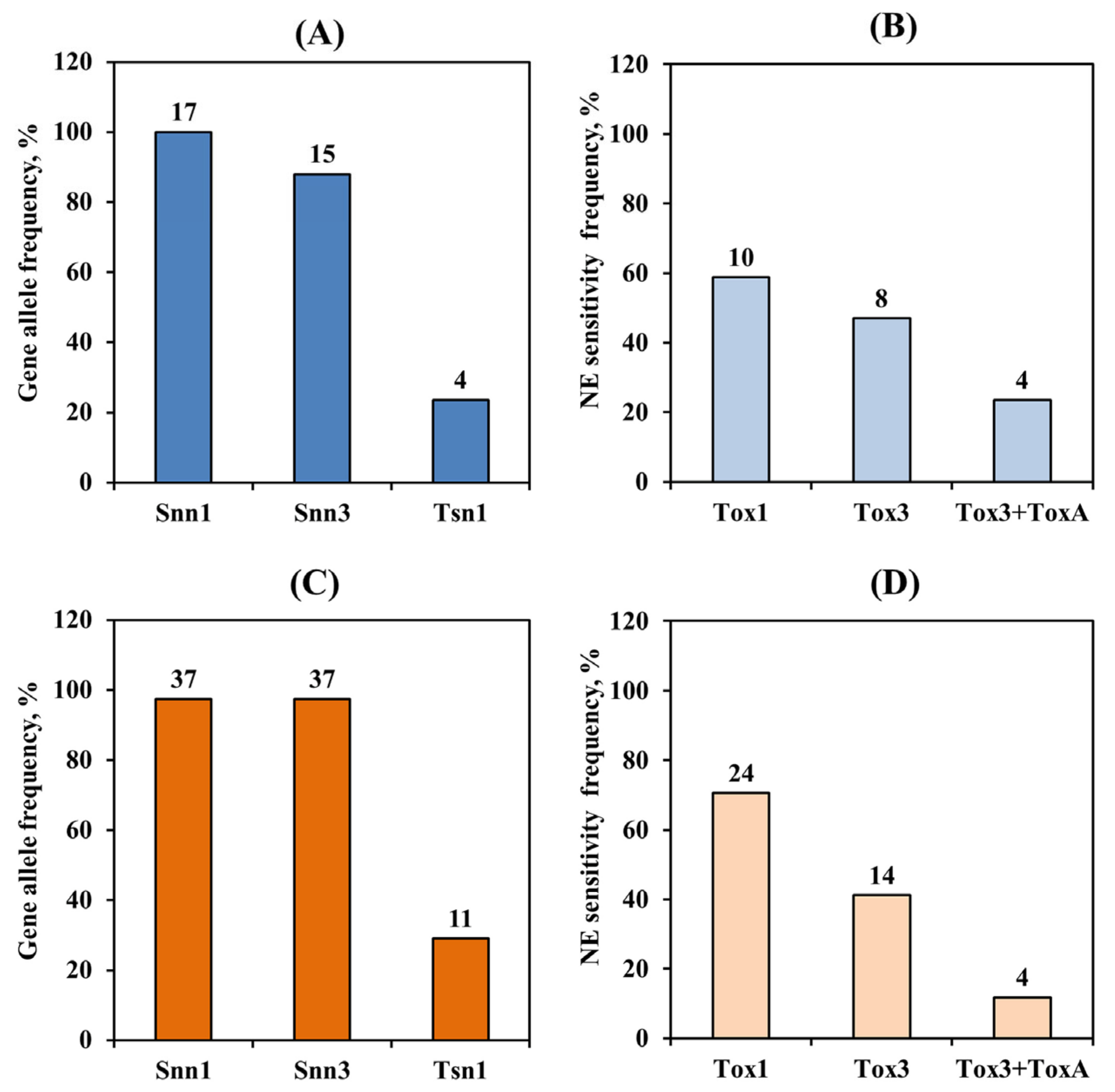
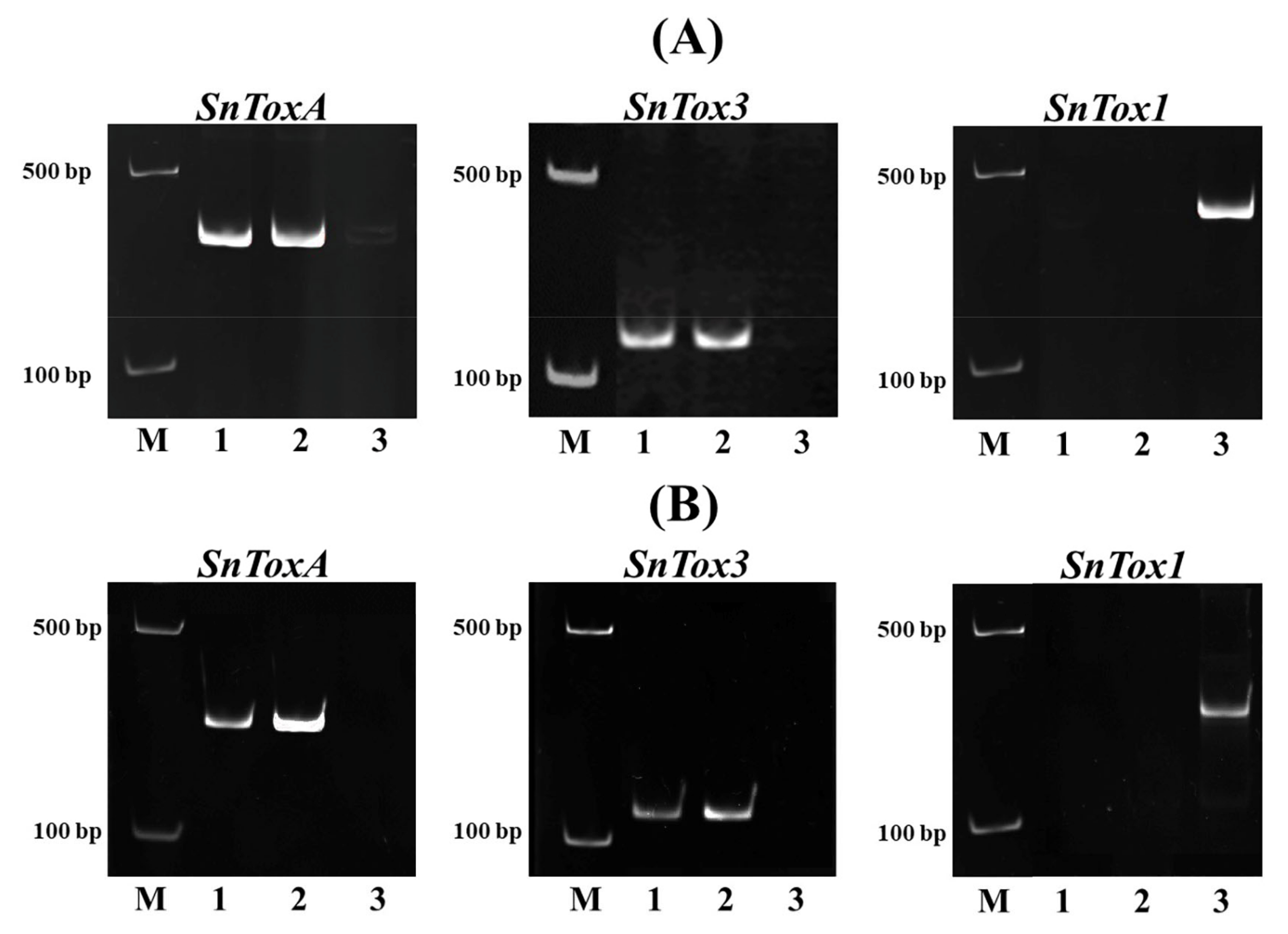
3.1. Analysis of the Allelic State of the Tsn1, Snn1 Genes and the Snn3-B1 Locus
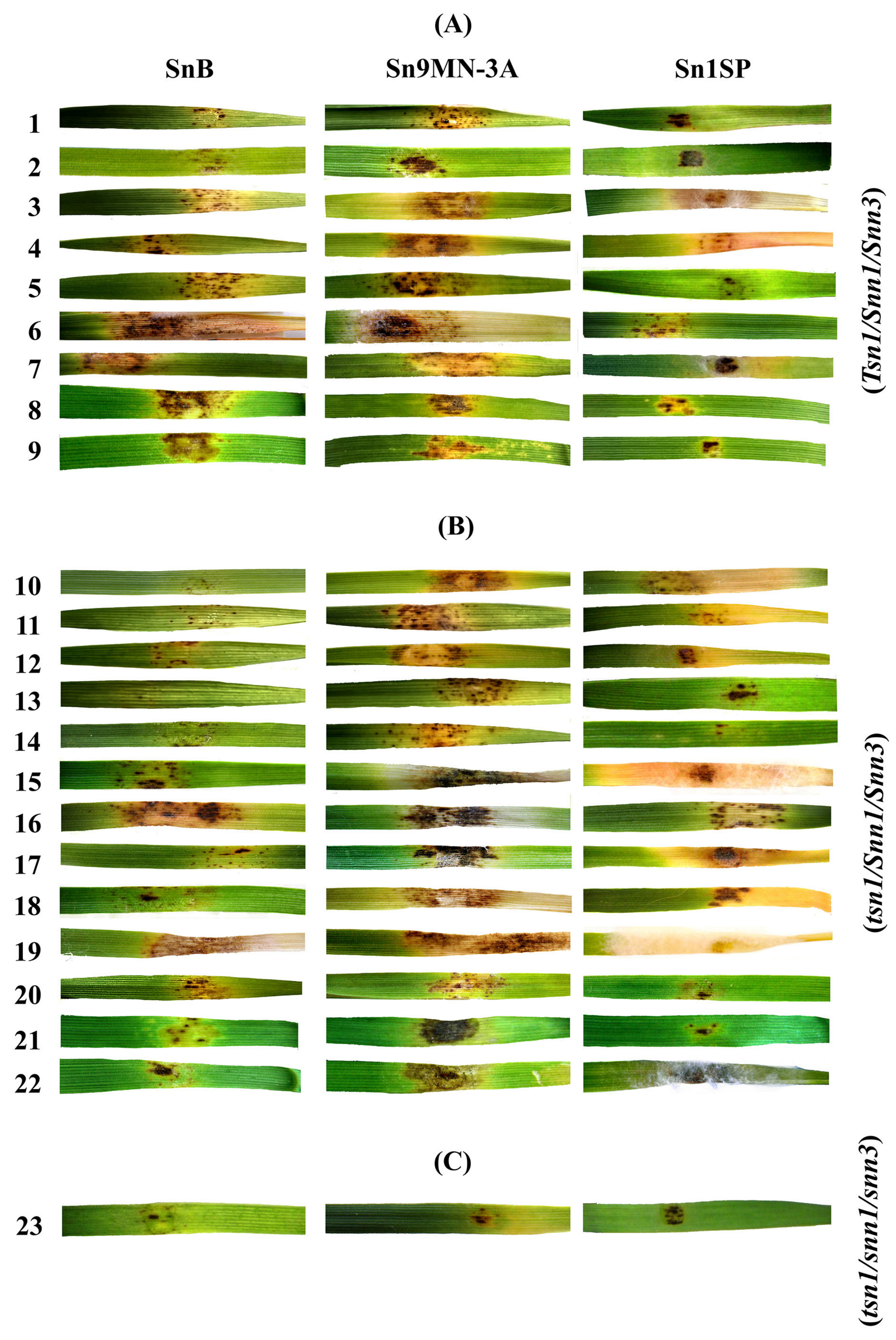
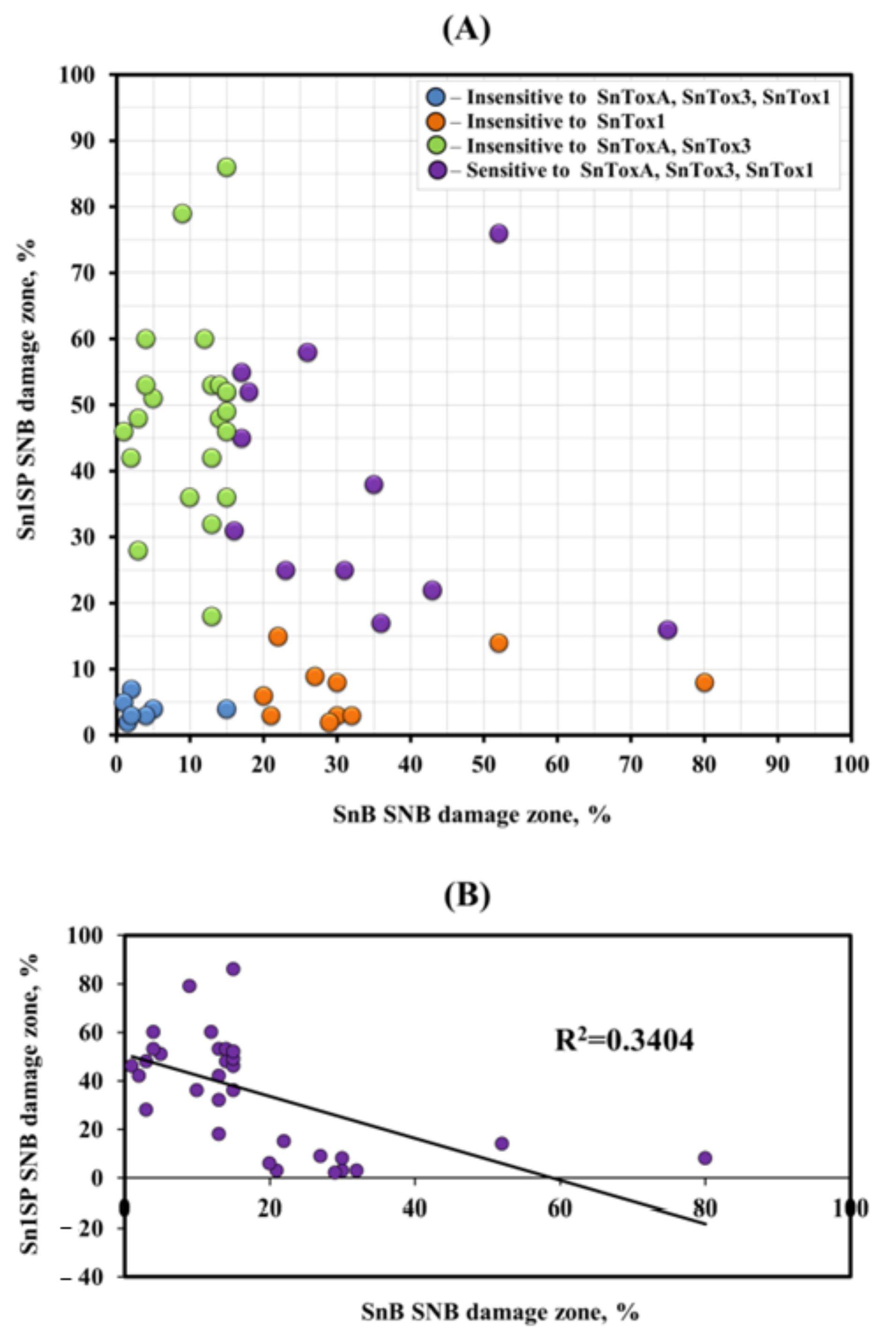
3.2. Analysis of Resistance/Susceptibility of Wheat Accessions to NEs SnToxA, SnTox1 and SnTox3
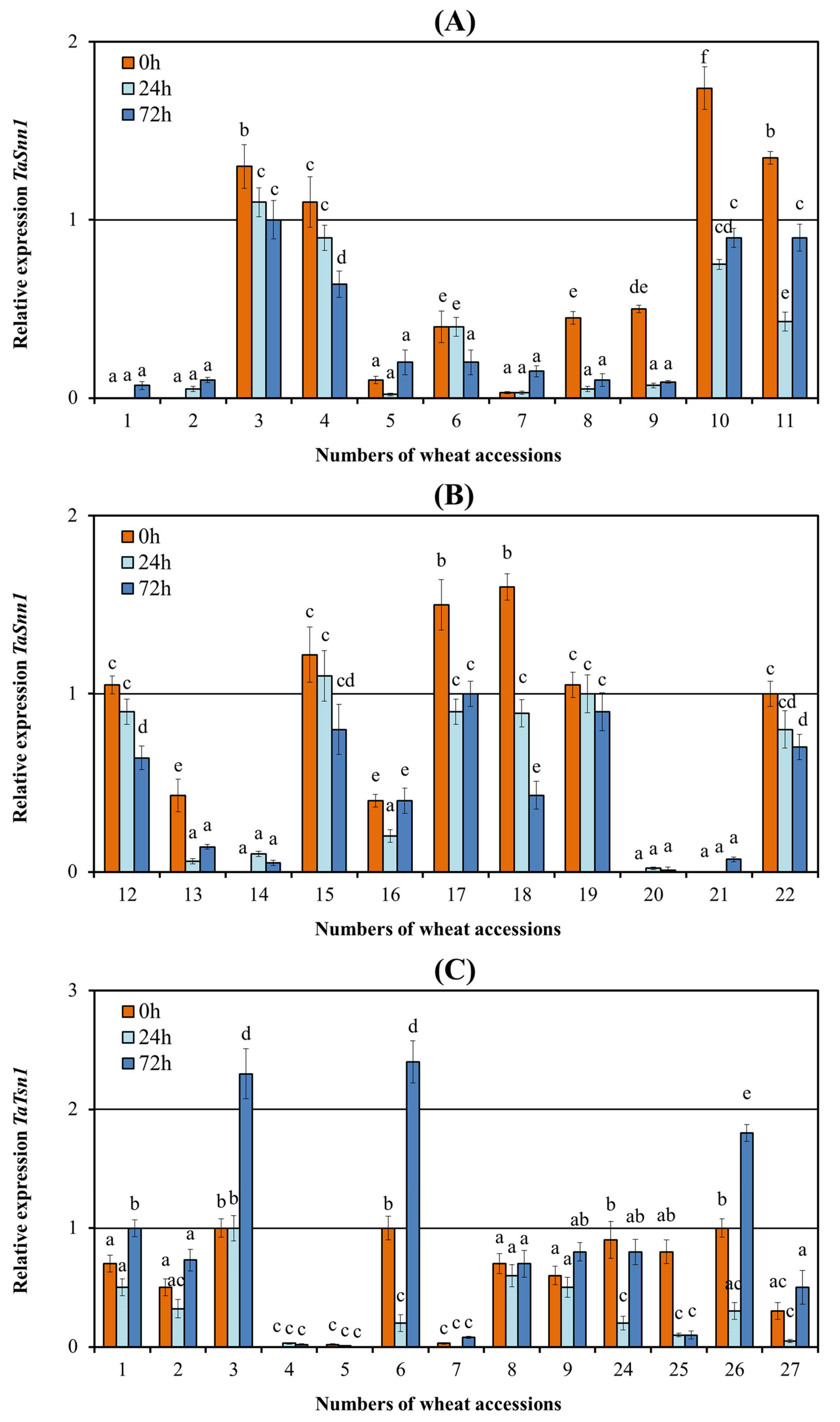
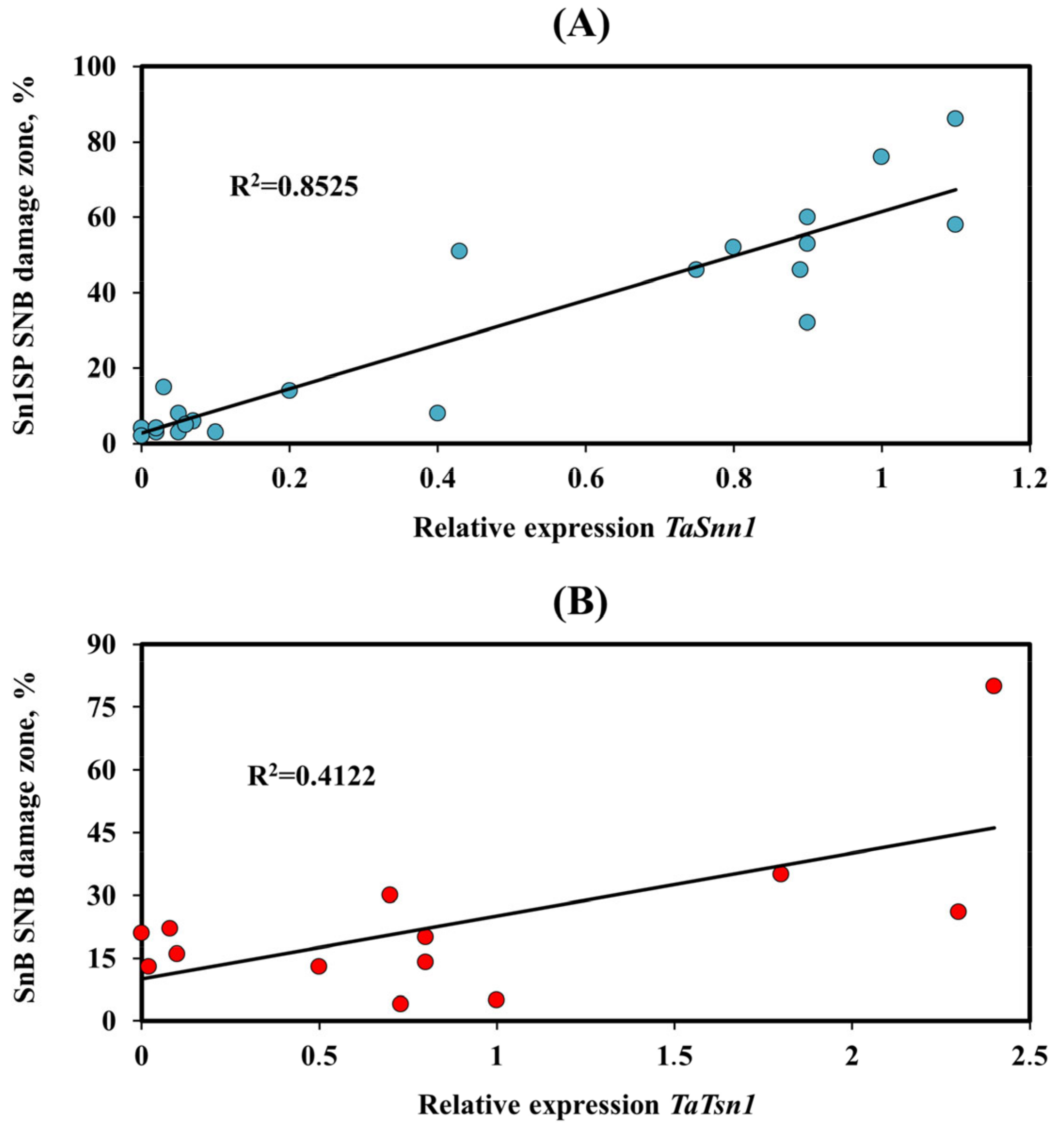
3.3. Analysis of the Expression of the Tsn1 and Snn1 Susceptibility Genes
4. Discussion
4.1. Effector Sensitivity in Russian Wheat Panel and Its Relevance to SNB
4.2. The Role of Various SnTox-Snn Interactions in the Development of SNB
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Solomon, P.S.; Lowe, R.G.T.; Tan, K.-C.; Waters, O.D.C.; Oliver, R.P. Stagonospora nodorum: Cause of Stagonospora nodorum blotch of wheat. Mol. Plant Pathol. 2006, 7, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ficke, A.; Cowger, C.; Bergstrom, G.; Brodal, G. Understanding yield loss and pathogen biology to improve disease management: Septoria nodorum blotch-a case study in wheat. Plant Dis. 2018, 102, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Cowger, C.; Ward, B.; Brown-Guedira, G.; Brown, J.K.M. Role of effector-sensitivity gene interactions and durability of quantitative resistance to Septoria nodorum Blotch in Eastern U.S. Wheat. Front. Plant Sci. 2020, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Haugrud, A.R.P.; Zhang, Z.; Friesen, T.L.; Faris, J.D. Genetics of resistance to Septoria nodorum blotch in wheat. Theor. Appl. Genet. 2022, 135, 3685–3707. [Google Scholar] [CrossRef]
- Downie, R.C.; Lin, M.; Corci, B.; Ficke, A.; Lillemo, M.; Oliver, R.P.; Phan, H.T.T.; Tan, K.-C.; Cockram, J. Septoria nodorum blotch of wheat: Disease management and resistance breeding in the face of shifting disease dynamics and a changing environment. Phytopathology 2021, 111, 906–920. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.C.; Solomon, P.S. Just the surface: Advances in the discovery and characterization of necrotrophic wheat effectors. Curr. Opin. Microbiol. 2018, 46, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.C.; Oliver, R.P.; Solomon, P.S.; Moffat, C.S. Proteinaceous necrotrophic effectors in fungal virulence. Funct. Plant Biol. 2010, 37, 907–912. [Google Scholar] [CrossRef]
- Faris, J.D.; Friesen, T.L. Plant genes hijacked by necrotrophic fungal pathogens. Curr. Opin. Plant Biol. 2020, 56, 74–80. [Google Scholar] [CrossRef]
- Navathe, S.; Yadav, P.S.; Chand, R.; Mishra, V.K.; Vasistha, N.K.; Meher, P.K.; Joshi, A.K.; Gupta, P.K. ToxA-Tsn1 interaction for spot blotch susceptibility in Indian wheat: An example of inverse gene-for-gene relationship. Plant Dis. 2020, 104, 71. [Google Scholar] [CrossRef]
- Friesen, T.L.; Stukenbrock, E.H.; Liu, Z.H.; Meinhardt, S.; Ling, H.; Faris, J.D.; Rasmussen, J.B.; Solomon, P.S.; McDonald, B.A.; Oliver, R.P. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 2006, 38, 953–956. [Google Scholar] [CrossRef]
- Friesen, T.L.; Zhang, Z.; Solomon, P.S.; Oliver, R.P.; Faris, J.D. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 2008, 146, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, Z.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Friesen, T.L.; Xu, S.S.; Shi, G.; Liu, Z.; Rasmussen, J.B.; Faris, J.D. Two putatively homoeologous wheat genes mediate recognition of SnTox3 to confer effector-triggered susceptibility to Stagnonospora nodorum. Plant J. 2011, 65, 27–38. [Google Scholar] [CrossRef]
- Abeysekara, N.S.; Faris, J.D.; Chao, S.; McClean, P.E.; Friesen, T.L. Whole-genome analysis of Stagonospora nodorum blotch resistance and validation of the SnTox4-Snn4 interaction in hexaploidy wheat. Phytopathology 2012, 102, 94–104. [Google Scholar] [CrossRef]
- Gao, Y.; Faris, J.D.; Liu, Z.; Kim, Y.M.; Syme, R.A.; Oliver, R.P.; Xu, S.S.; Friesen, T.L. Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum-wheat pathosystem. Mol. Plant Microbe Interact 2015, 28, 615–625. [Google Scholar] [CrossRef]
- Shi, G.; Zhang, Z.; Friesen, T.L.; Bansal, U.; Cloutier, S.; Wicker, T.; Rasmussen, J.B.; Faris, J.D. Marker development, saturation mapping, and high-resolution mapping of the Septoria nodorum blotch susceptibility gene Snn3-B1 in wheat. Mol. Genet. Genom. 2016, 291, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Zhang, Z.; Friesen, T.L.; Raats, D.; Fahima, T.; Brueggeman, R.S.; Lu, S.; Trick, H.N.; Liu, Z.; Chao, W.; et al. The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2016, 2, e1600822. [Google Scholar] [CrossRef]
- Kariyawasam, G.K.; Richards, J.K.; Wyatt, N.A.; Running, K.; Xu, S.S.; Liu, Z.; Borowicz, P.; Faris, J.D.; Friesen, T.L. The Parastagonospora nodorum necrotrophic effector SnTox5 targets the wheat gene Snn5 and facilitates entry into the leaf mesophyll. New Phytol. 2022, 233, 409–426. [Google Scholar] [CrossRef]
- Zhang, Z.; Running, K.L.D.; Seneviratne, S.; Peters-Haugrud, A.R.; Szabo-Hever, A.; Shi, G.; Luo, M.-C.; Brueggeman, R.; Xu, S.S.; Friesen, T.L.; et al. A protein kinase-major sperm protein gene hijacked by a necrotrophic fungal pathogen triggers disease susceptibility in wheat. Plant J. 2021, 106, 720–732. [Google Scholar] [CrossRef]
- Richards, J.K.; Kariyawasam, G.K.; Seneviratne, S.; Wyatt, N.A.; Xu, S.S.; Liu, Z.; Faris, J.D.; Friesen, T.L. A triple threat: The Parastagonospora nodorum SnTox267 effector exploits three distinct host genetic factors to cause disease in wheat. New Phytol. 2022, 233, 427–442. [Google Scholar] [CrossRef]
- Haugrud, A.R.P.; Zhang, Z.; Richards, J.; Friesen, T.L.; Faris, J.D. Genetics of vaiable disease expression conferred by inverse gene-for-gene interactions in the wheat-Parastagonospora nodorum Pathosystem. Plant Physiol. 2019, 180, 420–434. [Google Scholar] [CrossRef]
- Ruud, A.K.; Dieseth, J.A.; Lillemo, M. Effects of three Parastagonospora nodorum necrotrophic effectors on spring wheat under Norwegian field conditions. Crop Sci. 2018, 58, 159–168. [Google Scholar] [CrossRef]
- John, E.; Jacques, S.; Phan, H.T.T.; Liu, L.; Pereira, D.; Croll, D.; Singh, K.B.; Oliver, R.P.; Tan, K.-C. Variability in an effector gene promoter of a necrotrophic fungal pathogen dictates epistasis and effector-triggered susceptibility in wheat. PLoS Pathog. 2022, 18, e1010149. [Google Scholar] [CrossRef] [PubMed]
- Veselova, S.; Nuzhnaya, T.; Burkhanova, G.; Rumyantsev, S.; Maksimov, I. Reactive oxygen species in host plant are required for an early defense response against attack of Stagonospora nodorum Berk. necrotrophic effectors SnTox. Plants 2021, 10, 1586. [Google Scholar] [CrossRef]
- Fraaije, B.; Lovell, D.; Baldwin, S. Septoria epidemics on wheat: Combined use of visual assessment and PCR-based diagnostics to identify mechanisms of disease escape. Plant Prot. Sci. 2017, 38, 421–424. [Google Scholar] [CrossRef]
- Bertucci, M.; Brown-Guedira, G.; Murphy, J.P.; Cowger, C. Genes conferring sensitivity to Stagonospora nodorum necrotrophic effectors in Stagonospora nodorum blotch-susceptible U.S. wheat cultivars. Plant Dis. 2014, 98, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, M.J.; Piston, F.; Atienza, S.G. Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Veselova, S.V.; Sorokan, A.V.; Burkhanova, G.F.; Rumyantsev, S.D.; Cherepanova, E.A.; Alekseev, V.Y.; Sarvarova, E.R.; Kasimova, A.R.; Maksimov, I.V. By modulating the hormonal balance and ribonuclease activity of tomato plants Bacillus subtilis induces defense response against Potato Virus X and Potato Virus, Y. Biomolecules 2022, 12, 288. [Google Scholar] [CrossRef]
- Rumyantsev, S.D.; Alekseev, V.Y.; Sorokan, A.V.; Burkhanova, G.F.; Cherepanova, E.A.; Garafutdinov, R.R.; Maksimov, I.V.; Veselova, S.V. Additive effect of the composition of endophytic bacteria Bacillus subtilis on systemic resistance of wheat against greenbug aphid Schizaphis graminum due to lipopeptides. Life 2023, 13, 214. [Google Scholar] [CrossRef]
- Tan, K.C.; Waters, O.D.C.; Rybak, K.; Antoni, E.; Furuki, E.; Oliver, R.P. Sensitivity to three Parastagonospora nodorum necrotrophic effects in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci. 2014, 65, 150–158. [Google Scholar] [CrossRef]
- Downie, R.C.; Bouvet, L.; Furuki, E.; Gosman, N.; Gardner, K.A.; Mackay, I.J.; Campos Mantello, C.; Mellers, G.; Phan, H.T.T.; Rose, G.A.; et al. Assessing european wheat sensitivities to Parastagonospora nodorum necrotrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Front. Plant Sci. 2018, 9, 881. [Google Scholar] [CrossRef] [PubMed]
- Waters, O.D.C.; Lichtenzveig, J.; Rybak, K.; Friesen, T.L.; Oliver, R.P. Prevalence and importance of sensitivity to the Stagonospora nodorum necrotrophic effector SnTox3 in current Western Australian wheat cultivars. Crop Pasture Sci. 2011, 62, 556–562. [Google Scholar] [CrossRef]
- Phan, H.T.T.; Rybak, K.; Bertazzoni, S.; Furuki, E.; Dinglasan, E.; Hickey, L.T.; Oliver, R.P.; Tan, K.C. Novel sources of resistance to Septoria nodorum blotch in the Vavilov wheat collection identified by genome-wide association studies. Theor. Appl. Genet. 2018, 131, 1223–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Z.; Faris, J.D.; Oliver, R.P.; Syme, R.; McDonald, M.C.; McDonald, B.A.; Solomon, P.S.; Lu, S.; Shelver, W.L.; et al. The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 2012, 8, e1002467. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.; Rybak, K.; Furuki, E.; Breen, S.; Solomon, P.S.; Oliver, R.P.; Tan, K.C. Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 2016, 87, 343–354. [Google Scholar] [CrossRef]
- Friesen, T.L.; Chu, C.G.; Liu, Z.H.; Xu, S.S.; Halley, S.; Faris, J.D. Hostselective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor. Appl. Genet. 2009, 118, 1489–1497. [Google Scholar] [CrossRef]
- Oliver, R.P.; Solomon, P.S. Recent fungal diseases of crop plants: Is lateral gene transfer a common theme? Mol. Plant Microbe Interact 2008, 21, 287–293. [Google Scholar] [CrossRef]
- McDonald, M.C.; Oliver, R.P.; Friesen, T.L.; Brunner, P.C.; McDonald, B.A. Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 2013, 199, 241–251. [Google Scholar] [CrossRef]
- McDonald, M.C.; Ahren, D.; Simpfendorfer, S.; Milgate, A.; Solomon, P.S. The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. 2018, 19, 432–439. [Google Scholar] [CrossRef]
- Oliver, R.; Lichtenzveig, J.; Tan, K.C.; Waters, O.; Rybak, K.; Lawrence, J.; Friesen, T.; Burgess, P. Absence of detectable yield penalty associated with insensitivity to Pleosporales necrotrophic effectors in wheat grown in the West Australian wheat belt. Plant Pathol. 2014, 63, 1027–1032. [Google Scholar] [CrossRef]
- Ruud, A.K.; Windju, S.; Belova, T.; Friesen, T.L.; Lillemo, M. Mapping of SnTox3–Snn3 as a major determinant of field susceptibility to Septoria nodorum leaf blotch in the SHA3/CBRD × Naxos population. Appl. Genet. 2017, 130, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuzhnaya, T.; Veselova, S.; Burkhanova, G.; Rumyantsev, S.; Shoeva, O.; Shein, M.; Maksimov, I. Novel Sources of Resistance to Stagonospora nodorum and Role of Effector-Susceptibility Gene Interactions in Wheat of Russian Breeding. Int. J. Plant Biol. 2023, 14, 377-396. https://doi.org/10.3390/ijpb14020031
Nuzhnaya T, Veselova S, Burkhanova G, Rumyantsev S, Shoeva O, Shein M, Maksimov I. Novel Sources of Resistance to Stagonospora nodorum and Role of Effector-Susceptibility Gene Interactions in Wheat of Russian Breeding. International Journal of Plant Biology. 2023; 14(2):377-396. https://doi.org/10.3390/ijpb14020031
Chicago/Turabian StyleNuzhnaya, Tatyana, Svetlana Veselova, Guzel Burkhanova, Sergey Rumyantsev, Olesya Shoeva, Mikhail Shein, and Igor Maksimov. 2023. "Novel Sources of Resistance to Stagonospora nodorum and Role of Effector-Susceptibility Gene Interactions in Wheat of Russian Breeding" International Journal of Plant Biology 14, no. 2: 377-396. https://doi.org/10.3390/ijpb14020031
APA StyleNuzhnaya, T., Veselova, S., Burkhanova, G., Rumyantsev, S., Shoeva, O., Shein, M., & Maksimov, I. (2023). Novel Sources of Resistance to Stagonospora nodorum and Role of Effector-Susceptibility Gene Interactions in Wheat of Russian Breeding. International Journal of Plant Biology, 14(2), 377-396. https://doi.org/10.3390/ijpb14020031







