Emergence, Spread of Antimicrobial-Resistant Bacteria and Phylogenetic Relationships in Coastal Ecosystems—Gastropod Phorcus lineatus as a Bioindicator
Abstract
1. Introduction
2. Materials and Methods
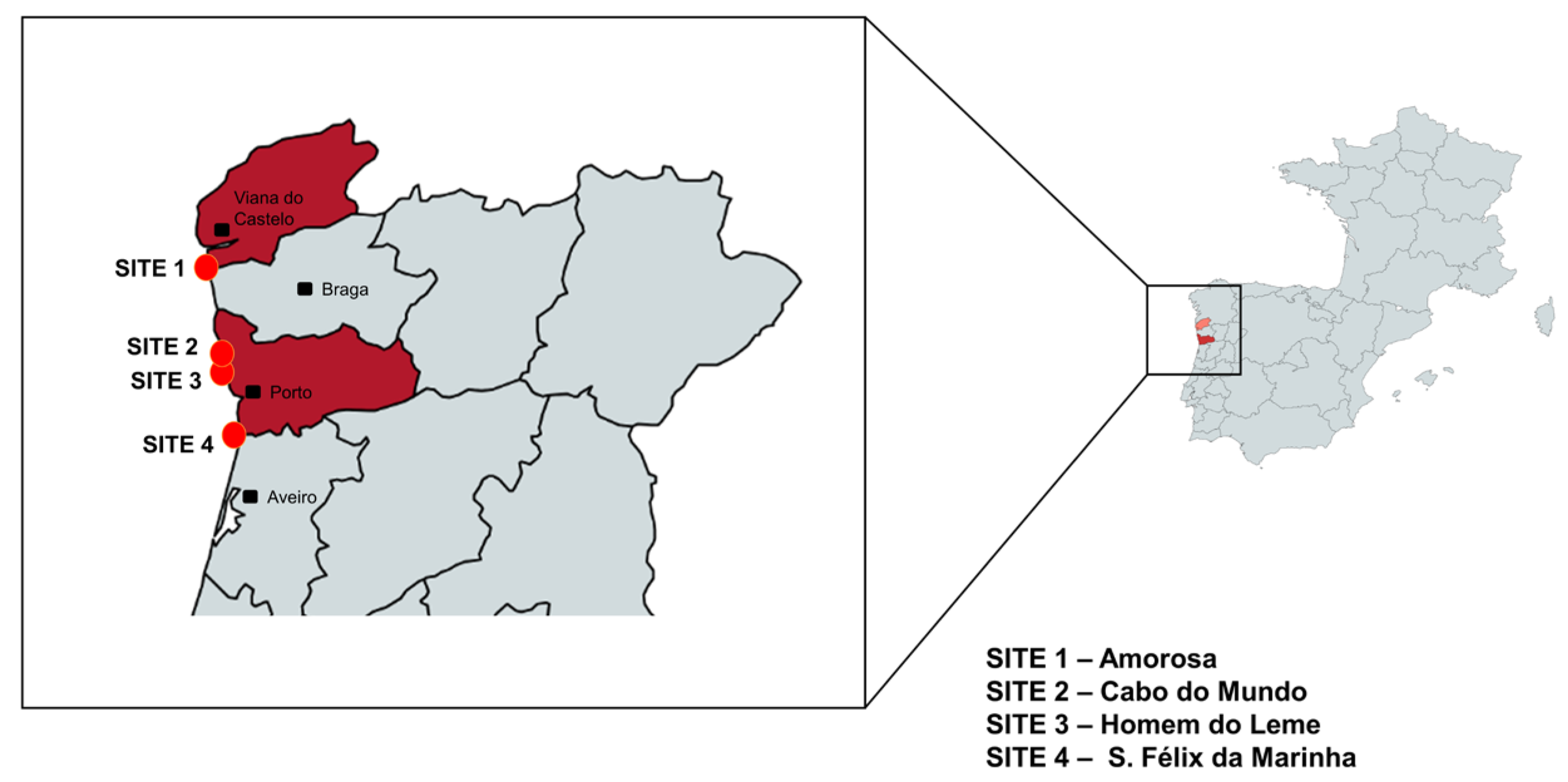
2.1. Study Area
2.2. Sampling
2.3. Preparation of Samples and Bacterial Culture
2.4. Identification of Pure Bacterial Isolates
2.5. Antimicrobial Susceptibility Testing
2.6. Determination of E. coli Pathotypes and Phylogenetic Grouping
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Okoye, C.O.; Nyaruaba, R.; Ita, R.E.; Okon, S.U.; Addey, C.I.; Ebido, C.C.; Opabunmi, A.O.; Okeke, E.S.; Chukwudozie, K.I. Antibiotic Resistance in the Aquatic Environment: Analytical Techniques and Interactive Impact of Emerging Contaminants. Environ. Toxicol. Pharmacol. 2022, 96, 103995. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding Human Health Risks Caused by Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARG) in Water Environments: Current Knowledge and Questions to Be Answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Du, L.; Liu, W. Occurrence, Fate, and Ecotoxicity of Antibiotics in Agro-Ecosystems. A Review. Agron. Sustain. Dev. 2012, 32, 309–327. [Google Scholar] [CrossRef]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.-H.; Xu, H. Stepwise Impact of Urban Wastewater Treatment on the Bacterial Community Structure, Antibiotic Contents, and Prevalence of Antimicrobial Resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef]
- Dasí, D.; Camaró-Sala, M.L.; González, A.; García-Ferrús, M.; Jiménez-Belenguer, A.I.; Castillo, M.Á. Antibiotic Resistance in Seawater Samples from East Coast of Spain. Appl. Sci. 2024, 14, 1965. [Google Scholar] [CrossRef]
- Devarajan, N.; Laffite, A.; Graham, N.D.; Meijer, M.; Prabakar, K.; Mubedi, J.I.; Elongo, V.; Mpiana, P.T.; Ibelings, B.W.; Wildi, W.; et al. Accumulation of Clinically Relevant Antibiotic-Resistance Genes, Bacterial Load, and Metals in Freshwater Lake Sediments in Central Europe. Environ. Sci. Technol. 2015, 49, 6528–6537. [Google Scholar] [CrossRef]
- Gambino, D.; Savoca, D.; Sucato, A.; Gargano, V.; Gentile, A.; Pantano, L.; Vicari, D.; Alduina, R. Occurrence of Antibiotic Resistance in the Mediterranean Sea. Antibiotics 2022, 11, 332. [Google Scholar] [CrossRef] [PubMed]
- Hladicz, A.; Kittinger, C.; Zarfel, G. Tigecycline Resistant Klebsiella Pneumoniae Isolated from Austrian River Water. Int. J. Environ. Res. Public Health 2017, 14, 1169. [Google Scholar] [CrossRef]
- Blackburn, J.K.; Mitchell, M.A.; Blackburn, M.-C.H.; Curtis, A.; Thompson, B.A. Evidence of Antibiotic Resistance in Free-Swimming, Top-Level Marine Predatory Fishes. J. Zoo Wildl. Med. 2010, 41, 7–16. [Google Scholar] [CrossRef]
- Chiesa, L.M.; Nobile, M.; Malandra, R.; Panseri, S.; Arioli, F. Occurrence of Antibiotics in Mussels and Clams from Various FAO Areas. Food Chem. 2018, 240, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Müller, A.; Seinige, D.; Wohlsein, P.; Oliveira, M.; Steinhagen, D.; Kehrenberg, C.; Siebert, U. Occurrence of Antimicrobial-Resistant Escherichia coli in Marine Mammals of the North and Baltic Seas: Sentinels for Human Health. Antibiotics 2022, 11, 1248. [Google Scholar] [CrossRef]
- Ozbey, G.; Tanriverdi, E.S.; Basusta, A.; Lakshmanappa, Y.S.; Otlu, B.; Zigo, F. Investigation for the Presence of Bacteria and Antimicrobial Resistance Genes in Sea Snails (Rapana venosa). Ann. Agric. Environ. Med. 2023, 30, 235–243. [Google Scholar] [CrossRef]
- Saavedra, M.J.; Fernandes, C.; Teixeira, A.; Álvarez, X.; Varandas, S. Multiresistant Bacteria: Invisible Enemies of Freshwater Mussels. Environ. Pollut. 2022, 295, 118671. [Google Scholar] [CrossRef] [PubMed]
- Unger, N.R.; Ritter, E.; Borrego, R.; Goodman, J.; Osiyemi, O.O. Antibiotic Susceptibilities of Bacteria Isolated within the Oral Flora of Florida Blacktip Sharks: Guidance for Empiric Antibiotic Therapy. PLoS ONE 2014, 9, e104577. [Google Scholar] [CrossRef] [PubMed]
- Coutellec, M.-A.; Caquet, T. Gastropod Ecophysiological Response to Stress. In Physiology of Molluscs; Apple Academic Press: Palm Bay, FL, USA, 2017; ISBN 978-1-315-20712-4. [Google Scholar]
- Santos, D.; Varandas, S.; Carrola, J.S.; Saavedra, M.J.; Luzio, A.; Monteiro, S.M.; Cabecinha, E. Environmental Health Assessment of the Northwest Portuguese Coast—Biochemical Biomarker Responses in the Marine Gastropod Phorcus Lineatus. Water 2024, 16, 5. [Google Scholar] [CrossRef]
- Serratore, P.; Zavatta, E.; Bignami, G.; Lorito, L. Preliminary Investigation on the Microbiological Quality of Edible Marine Gastropods of the Adriatic Sea, Italy. Ital. J. Food Saf. 2019, 8, 7691. [Google Scholar] [CrossRef]
- Sousa, R.; Delgado, J.; González, J.A.; Freitas, M.; Henriques, P. Marine Snails of the Genus Phorcus: Biology and Ecology of Sentinel Species for Human Impacts on the Rocky Shores; IntechOpen: London, UK, 2017; ISBN 978-1-78923-081-9. [Google Scholar]
- Nowicki, S.; deLaurent, Z.R.; de Villiers, E.P.; Githinji, G.; Charles, K.J. The Utility of Escherichia coli as a Contamination Indicator for Rural Drinking Water: Evidence from Whole Genome Sequencing. PLoS ONE 2021, 16, e0245910. [Google Scholar] [CrossRef]
- Tanyitiku, M.N.; Nicholas, G.; Sullivan, J.J.; Petcheu, I.C.N.; On, S.L.W. Survival of Escherichia coli in Edible Land Snails: Implications for Heliciculture and Public Health. Pathogens 2024, 13, 204. [Google Scholar] [CrossRef]
- Bankers, L.; Dahan, D.; Neiman, M.; Adrian-Tucci, C.; Frost, C.; Hurst, G.D.D.; King, K.C. Invasive Freshwater Snails Form Novel Microbial Relationships. Evol. Appl. 2021, 14, 770–780. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Clermont, O.; Olier, M.; Hoede, C.; Diancourt, L.; Brisse, S.; Keroudean, M.; Glodt, J.; Picard, B.; Oswald, E.; Denamur, E. Animal and Human Pathogenic Escherichia coli Strains Share Common Genetic Backgrounds. Infect. Genet. Evol. 2011, 11, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Geurtsen, J.; de Been, M.; Weerdenburg, E.; Zomer, A.; McNally, A.; Poolman, J. Genomics and Pathotypes of the Many Faces of Escherichia coli. FEMS Microbiol. Rev. 2022, 46, fuac031. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST)—Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13. 2023. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 12 June 2025).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; ISBN 978-1-68440-067-6. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Contamination of Foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Pais, S.; Costa, M.; Barata, A.R.; Rodrigues, L.; Afonso, I.M.; Almeida, G. Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens. Antibiotics 2022, 12, 20. [Google Scholar] [CrossRef]
- Aranda, K.R.S.; Fabbricotti, S.H.; Fagundes-Neto, U.; Scaletsky, I.C.A. Single Multiplex Assay to Identify Simultaneously Enteropathogenic, Enteroaggregative, Enterotoxigenic, Enteroinvasive and Shiga Toxin-Producing Escherichia coli Strains in Brazilian Children. FEMS Microbiol. Lett. 2007, 267, 145–150. [Google Scholar] [CrossRef]
- Schmidt, H.; Knop, C.; Franke, S.; Aleksic, S.; Heesemann, J.; Karch, H. Development of PCR for Screening of Enteroaggregative Escherichia coli. J. Clin. Microbiol. 1995, 33, 701–705. [Google Scholar] [CrossRef]
- ISO/TS 13136:2012. Available online: https://www.iso.org/standard/53328.html (accessed on 2 September 2024).
- Bighiu, M.A.; Norman Haldén, A.; Goedkoop, W.; Ottoson, J. Assessing Microbial Contamination and Antibiotic Resistant Bacteria Using Zebra Mussels (Dreissena polymorpha). Sci. Total Environ. 2019, 650, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, F.; Pezzi, A.; Galletti, G.; Tamba, M.; Merialdi, G.; Piva, S.; Serraino, A.; Rubini, S. Antimicrobial Resistance Patterns in Salmonella Enterica Subsp. Enterica and Escherichia coli Isolated from Bivalve Molluscs and Marine Environment. Food Control 2021, 121, 107590. [Google Scholar] [CrossRef]
- Tabanelli, G.; Montanari, C.; Gardini, A.; Maffei, M.; Prioli, C.; Gardini, F. Environmental Factors Affecting Escherichia coli Concentrations in Striped Venus Clam (Chamelea gallina L.) Harvested in the North Adriatic Sea. J. Food Prot. 2017, 80, 1429–1435. [Google Scholar] [CrossRef]
- Varandas, S.; Fernandes, C.; Cabecinha, E.; Gomes, S.; da Silva, G.J.; Saavedra, M.J. Escherichia coli Phylogenetic and Antimicrobial Pattern as an Indicator of Anthropogenic Impact on Threatened Freshwater Mussels. Antibiotics 2023, 12, 1401. [Google Scholar] [CrossRef]
- Tahri, L.; Hafiane, F.Z.; Fekhaoui, M. Prevalence and Antibiotic Resistance of the Escherichia coli in the Groundwater (Tadla-Morocco). Groundw. Sustain. Dev. 2021, 13, 100572. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef]
- Guedes, B.; Godinho, O.; Lage, O.M.; Quinteira, S. Microbiological Quality, Antibiotic Resistant Bacteria and Relevant Resistance Genes in Ready-to-Eat Pacific Oysters (Magallana gigas). FEMS Microbiol. Lett. 2023, 370, fnad053. [Google Scholar] [CrossRef] [PubMed]
- Krahulcová, M.; Cverenkárová, K.; Koreneková, J.; Oravcová, A.; Koščová, J.; Bírošová, L. Occurrence of Antibiotic-Resistant Bacteria in Fish and Seafood from Slovak Market. Foods 2023, 12, 3912. [Google Scholar] [CrossRef]
- Scarano, C.; Piras, F.; Virdis, S.; Ziino, G.; Nuvoloni, R.; Dalmasso, A.; De Santis, E.P.L.; Spanu, C. Antibiotic Resistance of Aeromonas ssp. Strains Isolated from Sparus aurata Reared in Italian Mariculture Farms. Int. J. Food Microbiol. 2018, 284, 91–97. [Google Scholar] [CrossRef]
- Vignaroli, C.; Di Sante, L.; Leoni, F.; Chierichetti, S.; Ottaviani, D.; Citterio, B.; Biavasco, F. Multidrug-Resistant and Epidemic Clones of Escherichia coli from Natural Beds of Venus Clam. Food Microbiol. 2016, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahian, S.; Graham, J.P.; Halaji, M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli. Front Cell Infect Microbiol 2024, 14, 1387497. [Google Scholar] [CrossRef]
- Enne, V.I.; Delsol, A.A.; Roe, J.M.; Bennett, P.M. Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrob Agents Chemother 2006, 50, 3003–3010. [Google Scholar] [CrossRef]
- Bilozor, A.; Balode, A.; Chakhunashvili, G.; Chumachenko, T.; Egorova, S.; Ivanova, M.; Kaftyreva, L.; Kõljalg, S.; Kõressaar, T.; Lysenko, O.; et al. Application of Molecular Methods for Carbapenemase Detection. Front. Microbiol. 2019, 10, 1755. [Google Scholar] [CrossRef]
- Bodendoerfer, E.; Marchesi, M.; Imkamp, F.; Courvalin, P.; Böttger, E.C.; Mancini, S. Co-occurrence of aminoglycoside and β-lactam resistance mechanisms in aminoglycoside- non-susceptible Escherichia coli isolated in the Zurich area, Switzerland. Int J Antimicrob Agents. 2020, 56, 106019. [Google Scholar] [CrossRef]
- Anastasi, E.M.; Matthews, B.; Stratton, H.M.; Katouli, M. Pathogenic Escherichia coli Found in Sewage Treatment Plants and Environmental Waters. Appl. Environ. Microbiol. 2012, 78, 5536–5541. [Google Scholar] [CrossRef] [PubMed]
- Bong, C.W.; Low, K.Y.; Chai, L.C.; Lee, C.W. Prevalence and Diversity of Antibiotic Resistant Escherichia coli from Anthropogenic-Impacted Larut River. Front. Public Health 2022, 10, 794513. [Google Scholar] [CrossRef] [PubMed]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Faecal Contamination of Water. Environ. Sci. Pollut. Res. 2015, 22, 10969–10980. [Google Scholar] [CrossRef]
- Pokharel, P.; Dhakal, S.; Dozois, C.M. The Diversity of Escherichia coli Pathotypes and Vaccination Strategies against This Versatile Bacterial Pathogen. Microorganisms 2023, 11, 344. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The Population Genetics of Commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The Link between Phylogeny and Virulence in Escherichia coli Extraintestinal Infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli Phylogenetic Group Determination and Its Application in the Identification of the Major Animal Source of Fecal Contamination. BMC Microbiol. 2010, 10, 161. [Google Scholar] [CrossRef]
- Chakraborty, A.; Saralaya, V.; Adhikari, P.; Shenoy, S.; Baliga, S.; Hegde, A. Characterization of Escherichia coli Phylogenetic Groups Associated with Extraintestinal Infections in South Indian Population. Ann. Med. Health Sci. Res. 2015, 5, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Girardeau, J.P.; Dalmasso, A.; Bertin, Y.; Ducrot, C.; Bord, S.; Livrelli, V.; Vernozy-Rozand, C.; Martin, C. Association of Virulence Genotype with Phylogenetic Background in Comparison to Different Seropathotypes of Shiga Toxin-Producing Escherichia coli Isolates. J. Clin. Microbiol. 2005, 43, 6098–6107. [Google Scholar] [CrossRef] [PubMed]
- Nowrouzian, F.L.; Adlerberth, I.; Wold, A.E. Enhanced Persistence in the Colonic Microbiota of Escherichia coli Strains Belonging to Phylogenetic Group B2: Role of Virulence Factors and Adherence to Colonic Cells. Microbes Infect. 2006, 8, 834–840. [Google Scholar] [CrossRef]
- Pereira, A.; Santos, A.; Tacão, M.; Alves, A.; Henriques, I.; Correia, A. Genetic Diversity and Antimicrobial Resistance of Escherichia coli from Tagus Estuary (Portugal). Sci. Total Environ. 2013, 461–462, 65–71. [Google Scholar] [CrossRef]
- EU Reference Laboratory for E. coli, Identification and Characterization of SHIGA Toxin (Verocytotoxin)-Producing Escherichia coli (STEC/VTEC) by Real Time PCR Amplification of the Main Virulence Genes and the Genes Associated with the Serogroups Mainly Associated with Severe Human Infection. EURL-VTEC_Method 02_Rev1. Available online: https://www.iss.it/documents/20126/0/EURL-VTEC_Method_02_Rev_1.pdf/b5173cbd-5789-c729-b0c3-81da039e88c7?t=1644309049719 (accessed on 12 June 2025).
- EU Reference Laboratory for E. coli, Detection of Enteroaggregative Escherichia coli in Food by Real Time PCR Amplification of the aggR and aaiC Genes. EURL-VTEC_Method_05_Rev 2. Available online: https://www.iss.it/documents/20126/0/EURL-VTEC_Method_05_Rev+2.pdf/c0e054b4-0fbc-1189-8f3d-063d000af712?t=1644309220376 (accessed on 12 June 2025).
- EU Reference Laboratory for E. coli, Detection of Enteroinvasive Escherichia coli in Food by Real Time PCR Amplification of the ipaH Gene. EURL-VTEC_Method_07_Rev 0. Available online: https://www.iss.it/documents/20126/0/EURL-VTEC_Method_07_Rev+0.pdf/c4da4642-00ab-d350-d942-8d4469225ede?t=1644309249941 (accessed on 12 June 2025).
- EU Reference Laboratory for E. coli, Detection of Enterotoxigenic Escherichia coli in Food by Real Time PCR Amplification of the lt, sth, and stp Genes, Encoding the Heat-Labile and Heat-Stable Enterotoxins. EURL-VTEC_Method_08_Rev 0. Available online: https://www.iss.it/documents/20126/0/EURL-VTEC_Method_08_Rev+0.pdf/e82174f2-ea21-a67d-10e9-70df683940a8?t=1644309265000 (accessed on 12 June 2025).
- Fricker, M.; Messelhäußer, U.; Busch, U.; Scherer, S.; Ehling-Schulz, M. Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 2007, 73, 1892–1898. [Google Scholar] [CrossRef]

| Phylogroup | Target Gene | |||
|---|---|---|---|---|
| arpA | chuA | yjaA | TspE4.C2 | |
| A | + | - | - | - |
| A or C | + | - | + | - |
| B1 | + | - | - | + |
| B2 | - | + | + | - |
| B2 | - | + | - | + |
| B2 | - | + | + | + |
| E or D | + | + | - | - |
| E or D | + | + | - | + |
| E or Clade I | + | + | + | - |
| F | - | + | - | - |
| Unknown (a) | + | - | + | + |
| Antimicrobial Agent | SITE 1—Amorosa | SITE 2—Cabo do Mundo | SITE 3—Homem do Leme | |||
|---|---|---|---|---|---|---|
| Resistant | Susceptible | Resistant | Susceptible | Resistant | Susceptible | |
| % (No. Isolates/Total) | % (No. Isolates/Total) | % (No. Isolates/Total) | ||||
| AMP (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| AML (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| AMC (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| TIC (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| TIM (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| PRL (1) | -- | 100 (2/2) | 25 (3/12) | 75 (9/12) | -- | 100 (4/4) |
| TZP (1) | -- | 100 (2/2) | 25 (3/12) | 75 (9/12) | -- | 100 (4/4) |
| FOX (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| CAZ (1) | -- | 100 (2/2) | 8 (1/12) | 92 (11/12) | -- | 100 (4/4) |
| CTX (1) | -- | 100 (2/2) | 8 (1/12) | 92 (11/12) | -- | 100 (4/4) |
| CRO (1) | -- | 100 (2/2) | 8 (1/12) | 92 (11/12) | -- | 100 (4/4) |
| IMP (1) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| MEM (1) | -- | 100 (2/2) | 17 (2/12) | 83 (10/12) | -- | 100 (4/4) |
| ETP (1) | -- | 100 (2/2) | 17 (2/12) | 83 (10/12) | -- | 100 (4/4) |
| ATM (1) | -- | 100 (2/2) | 8 (1/12) | 92 (11/12) | -- | 100 (4/4) |
| LEV (2) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| CIP (2) | -- | 100 (2/2) | -- | 100 (12/12) | -- | 100 (4/4) |
| CN (3) | -- | 100 (2/2) | 25 (3/12) | 75 (9/12) | -- | 100 (4/4) |
| AK (3) | -- | 100 (2/2) | 33 (4/12) | 67 (8/12) | -- | 100 (4/4) |
| STX (4) | -- | 100 (2/2) | 8 (1/12) | 92 (11/12) | -- | 100 (4/4) |
| TE (5) | -- | 100 (2/2) | 25 (3/12) | 75 (9/12)) | -- | 100 (4/4) |
| FOS (6) | -- | 100 (2/2) | 17 (2/12) | 83 (10/12) | -- | 100 (4/4) |
| Multidrug-resistant (MDR) | 0 | 25% (3/12) | 0 | |||
| Multiple antibiotic resistance (MAR) index | <0.2 | 16.7% > 0.2 | <0.2 | |||
| Phylogroup | |||||
|---|---|---|---|---|---|
| A | B1 | B2 | A or C | E or D | |
| All Sites/Total | 5 (27.7%) | 4 (22.2%) | 5 (27.7%) | 2 (11.1%) | 2 (11.1%) |
| Site 1—Amorosa | 0 (0%) | 2 (11.1%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Site 2—Cabo do Mundo | 5 (27.7%) | 2 (11.1%) | 1 (5.5%) | 2 (11.1%) | 2 (11.1%) |
| Site 3—Homem do Leme | 0 (0%) | 0 (0%) | 4 (22.2%) | 0 (0%) | 0 (0%) |
| Antibiotic resistance pattern | PRL/TZP/CAZ/ CTX/CRO/MEM/ETP/ ATM/CN/AK/STX/TE/FOS | AK | AK | AK | CN/AK/TE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, D.; Pinto, A.R.; Barata, R.; Fernandes, C.; Guedes, H.; Almeida, G.; Cabecinha, E.; Monteiro, S.M.; Varandas, S.; Saavedra, M.J. Emergence, Spread of Antimicrobial-Resistant Bacteria and Phylogenetic Relationships in Coastal Ecosystems—Gastropod Phorcus lineatus as a Bioindicator. Microbiol. Res. 2025, 16, 133. https://doi.org/10.3390/microbiolres16060133
Santos D, Pinto AR, Barata R, Fernandes C, Guedes H, Almeida G, Cabecinha E, Monteiro SM, Varandas S, Saavedra MJ. Emergence, Spread of Antimicrobial-Resistant Bacteria and Phylogenetic Relationships in Coastal Ecosystems—Gastropod Phorcus lineatus as a Bioindicator. Microbiology Research. 2025; 16(6):133. https://doi.org/10.3390/microbiolres16060133
Chicago/Turabian StyleSantos, Dércia, Ana Rita Pinto, Rita Barata, Conceição Fernandes, Hugo Guedes, Gonçalo Almeida, Edna Cabecinha, Sandra M. Monteiro, Simone Varandas, and Maria José Saavedra. 2025. "Emergence, Spread of Antimicrobial-Resistant Bacteria and Phylogenetic Relationships in Coastal Ecosystems—Gastropod Phorcus lineatus as a Bioindicator" Microbiology Research 16, no. 6: 133. https://doi.org/10.3390/microbiolres16060133
APA StyleSantos, D., Pinto, A. R., Barata, R., Fernandes, C., Guedes, H., Almeida, G., Cabecinha, E., Monteiro, S. M., Varandas, S., & Saavedra, M. J. (2025). Emergence, Spread of Antimicrobial-Resistant Bacteria and Phylogenetic Relationships in Coastal Ecosystems—Gastropod Phorcus lineatus as a Bioindicator. Microbiology Research, 16(6), 133. https://doi.org/10.3390/microbiolres16060133













