Genotypic Characterisation and Risk Assessment of Virulent ESBL-Producing E. coli in Chicken Meat in Tunisia: Insights from Multi-Omics Machine Learning Perspective
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. Isolation and Identification of E. coli
2.3. Antibiotic Susceptibility Testing
2.4. Screening and Confirmation of ESBLs and Colistin Resistance
2.5. DNA Extraction PCR Condition
2.6. Molecular Study of Antimicrobial β-Lactamase and Carbapenemase Genes
2.7. Molecular Study of Non-β-Lactam Antibiotic Resistance Genes
2.8. Virulence Genotyping of E. coli Strains
2.9. Phylogenetic Analysis
2.10. Data Mining and Modelling Analysis
3. Results
3.1. Phenotype Evaluations
3.1.1. Prevalence of AMR Between Suppliers
3.1.2. Prevalence of AMR Across Antimicrobial Molecules
3.2. Genetic Assessment
3.2.1. Detection of Beta-Lactamase Coding Genes
3.2.2. Detection of Non-β-Lactams Resistance Coding Genes
3.2.3. Detection of Virulence Coding Genes
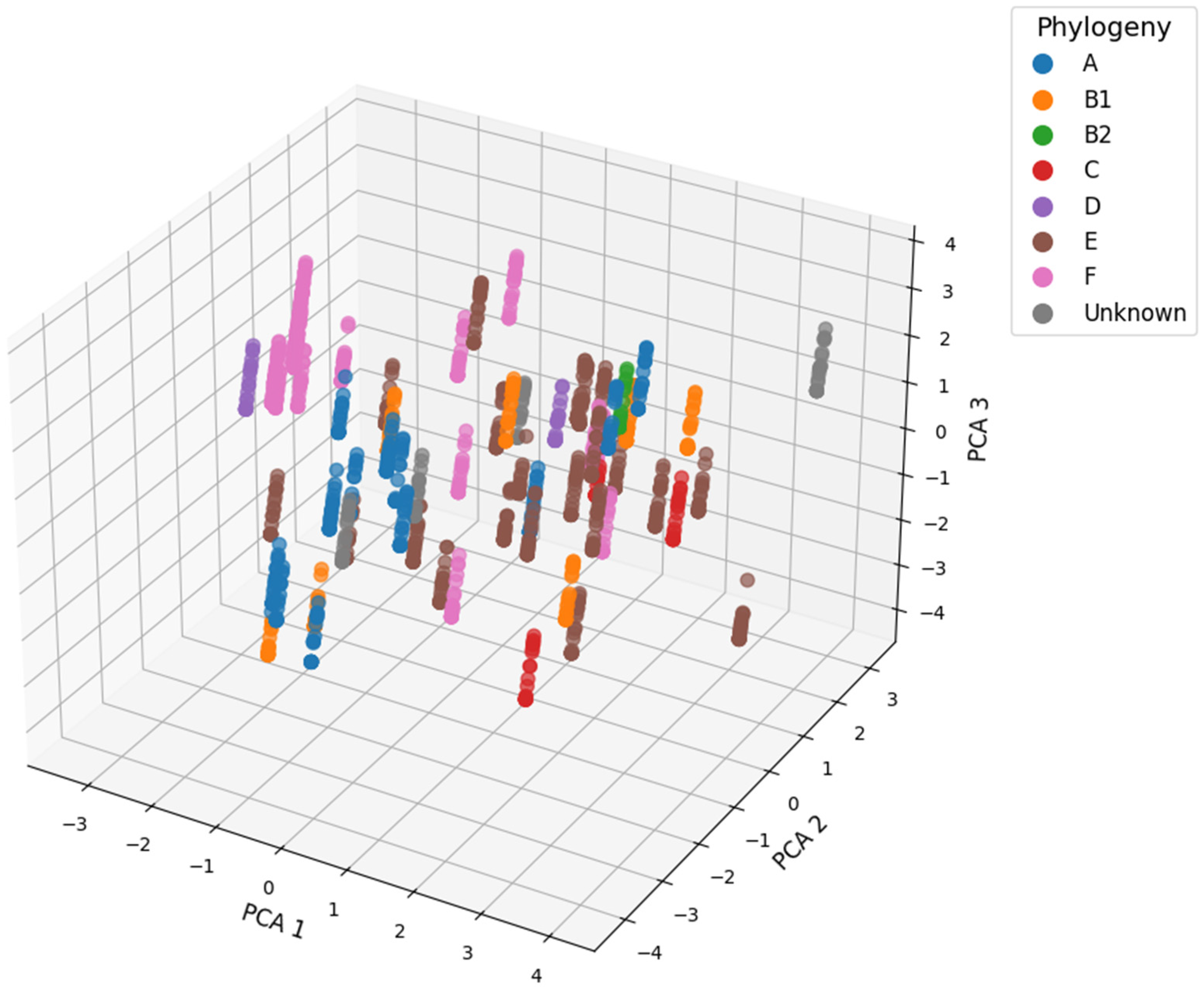
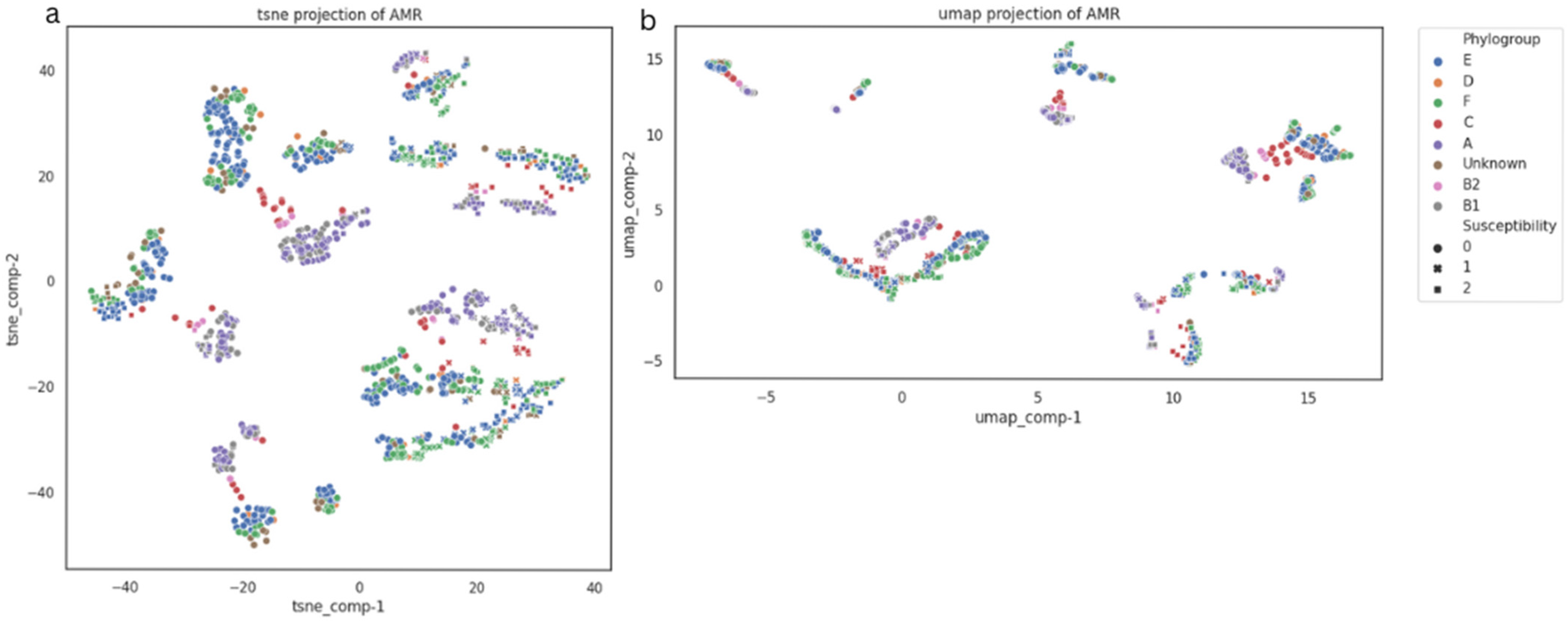
3.2.4. Phylogroups Assessment of E. coli Strains
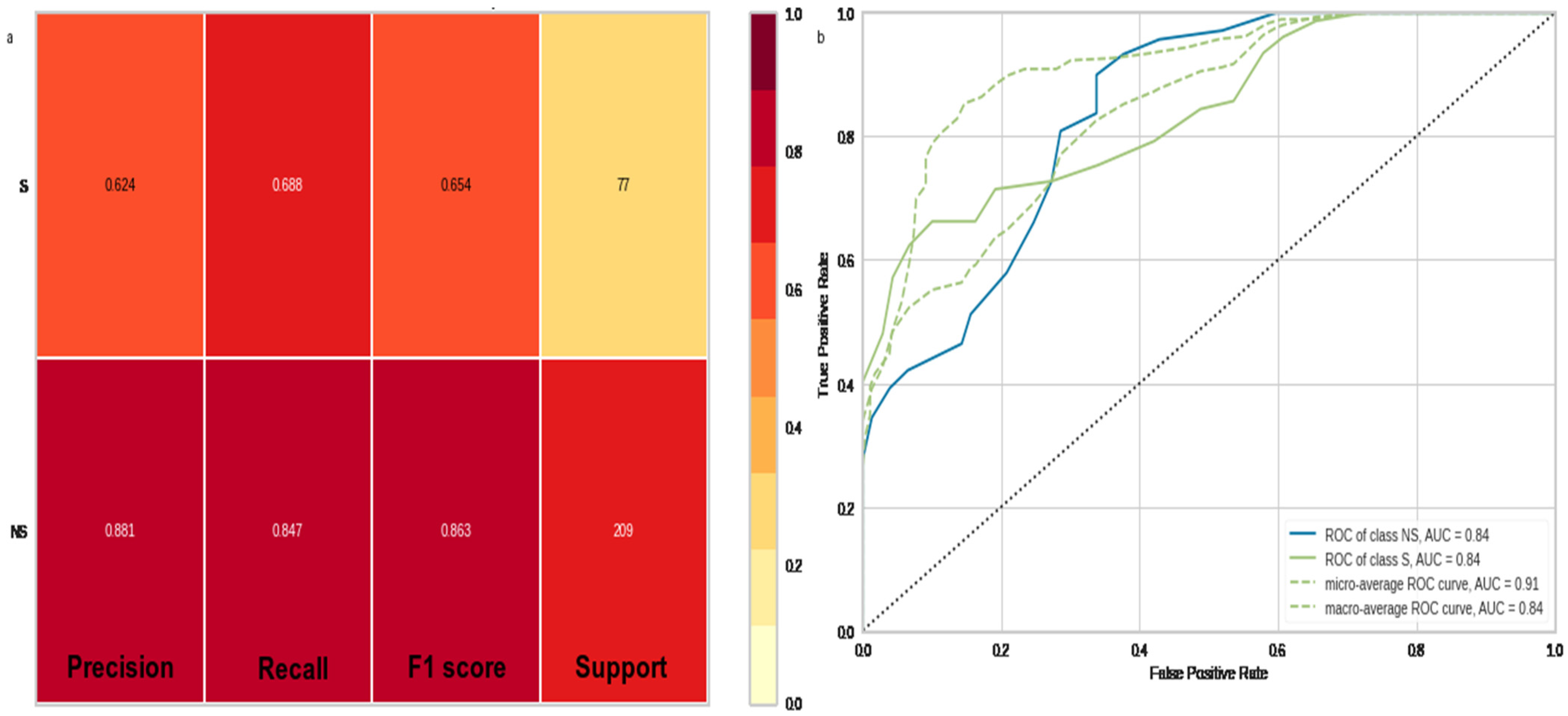
3.3. Data Mining and Machine Learning Evaluations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. 2021. Available online: https://www.fao.org/home/en/ (accessed on 11 May 2022).
- Cunicoles LGIdPAe. Produits Avicole GIPAC. 2018. Available online: http://www.gipac.tn (accessed on 11 May 2022).
- Hafez, H.M.; Attia, Y.A. Challenges to the poultry industry: Current perspectives and strategic future after the COVID-19 outbreak. Front. Vet. Sci. 2020, 7, 516. [Google Scholar] [CrossRef]
- Espinosa, R.; Tago, D.; Treich, N. Infectious diseases and meat production. Environ. Resour. Econ. 2020, 76, 1019–1044. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance: WHO. 2023. Available online: https://www.who.int/health-topics/antimicrobial-resistance (accessed on 13 June 2024).
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Redfield, R.F. Centers for Disease Control and Prevention Justification of Appropriation Estimates for Appropriations Committees Fiscal Year 2021; Communicable Disease Center: Atlanta, GA, USA, 2020.
- Pym, R. Poultry genetics and breeding in developing countries. In Poultry Development Review; FAO: Rome, Italy, 2013; pp. 80–83. [Google Scholar]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, V.; de Lurdes Enes Dapkevicius, M.; Caniça, M.; Tejedor-Junco, M.; Igrejas, G. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β-lactamase (ESBL) production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial resistance profile and ExPEC virulence potential in commensal Escherichia coli of multiple sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Agostinho, J.M.A.; Cardozo, M.V.; Borzi, M.M.; Marin, J.M. Antibiotic resistance and virulence factors among Escherichia coli isolates from avian organic fertilizer. Ciênc. Rural 2020, 50, e20180849. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, J.B.; Hayakawa, K.; Miyoshi-Akiyama, T.; Ohmagari, N.; Kirikae, T.; Nagamatsu, M.; Tojo, M.; Ohara, H.; Tandukar, S. Clinical epidemiology and molecular analysis of extended-spectrum-β-lactamase-producing Escherichia coli in Nepal: Characteristics of sequence types 131 and 648. Antimicrob. Agents Chemother. 2015, 59, 3424–3432. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–22. [Google Scholar]
- Ribeiro, L.F.; Nespolo, N.M.; Rossi, G.A.M.; Fairbrother, J.M. Exploring extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in food-producing animals and animal-derived foods. Pathogens 2024, 13, 346. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-Lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, C.M.; van der Goot, J.A.; Smith, H.E.; Kant, A.; Mevius, D.J. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: A descriptive study. PLoS ONE 2013, 8, e79005. [Google Scholar] [CrossRef]
- Gazal, L.E.d.S.; Medeiros, L.P.; Dibo, M.; Nishio, E.K.; Koga, V.L.; Gonçalves, B.C.; Grassotti, T.T.; de Camargo, T.C.L.; Pinheiro, J.J.; Vespero, E.C.; et al. Detection of ESBL/AmpC-producing and fosfomycin-resistant Escherichia coli from different sources in poultry production in Southern Brazil. Front. Microbiol. 2021, 11, 604544. [Google Scholar] [CrossRef]
- Nsubuga, M.; Galiwango, R.; Jjingo, D.; Mboowa, G. Generalizability of machine learning in predicting antimicrobial resistance in E. Coli: A multi-country case study in Africa. BMC Genom. 2024, 25, 287. [Google Scholar] [CrossRef]
- Moradigaravand, D.; Palm, M.; Farewell, A.; Mustonen, V.; Warringer, J.; Parts, L. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol. 2018, 14, e1006258. [Google Scholar] [CrossRef]
- EURL-AR. Quantification of ESBL/AmpC-Producing Escherichia coli in Caecal Content and Fresh Meat Samples. 2017. Available online: https://www.food.dtu.dk/english/-/media/institutter/foedevareinstituttet/temaer/antibiotikaresistens/eurl-ar/protocols/esbl-ampc-and-camrbapenemase-producing-e-coli/399_esbl-ampc-quantification-protocol-19-03-2018.pdf (accessed on 27 April 2025).
- CLSI. Erformance Standards for Antimicrobial Susceptibility Testing. 2020. Available online: https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf (accessed on 27 April 2025).
- EUCAST ECoAST. Clinical Breakpoints—Breakpoints and Guidance. 2021. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 27 April 2025).
- Dortet, L.; Poirel, L.; Nordmann, P. Rapid detection of ESBL-producing Enterobacteriaceae in blood cultures. Emerg. Infect. Dis. 2015, 21, 504–507. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jouy, E.; Haenni, M.; Le Devendec, L.; Le Roux, A.; Châtre, P.; Madec, J.-Y.; Kempf, I. Improvement in routine detection of colistin resistance in E. coli isolated in veterinary diagnostic laboratories. J. Microbiol. Methods 2017, 132, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Hancock, V.; Ferrieres, L.; Klemm, P. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 2008, 154, 167–175. [Google Scholar] [CrossRef]
- Tu, J.; Hue, T.; Qi, K.; Shao, Y.; Huang, B.; Wang, X.; Zhou, X. The irp2 and fyuA genes in High Pathogenicity Islands are involved in the pathogenesis of infections caused by avian pathogenic Escherichia coli (APEC). Pol. J. Vet. Sci. 2016, 19, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Kobayashi, T.; Fujimoto, F.; Okada, Y.; Higurashi, Y.; Tatsuno, K.; Okugawa, S.; Moriya, K. The prevalence of the iutA and ibeA genes in Escherichia coli isolates from severe and non-severe patients with bacteremic acute biliary tract infection is significantly different. Gut Pathog. 2021, 13, 32. [Google Scholar] [CrossRef]
- Pehlivanlar Önen, S.; Aslantaş, Ö.; Şebnem Yılmaz, E.; Kürekci, C. Prevalence of β-lactamase producing Escherichia coli from retail meat in Turkey. J. Food Sci. 2015, 80, M2023–M2029. [Google Scholar] [CrossRef]
- Hussain, A.; Shaik, S.; Ranjan, A.; Nandanwar, N.; Tiwari, S.K.; Majid, M.; Baddam, R.; Qureshi, I.A.; Semmler, T.; Wieler, L.H.; et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 2017, 8, 2120. [Google Scholar] [CrossRef]
- García-Béjar, B.; García de Blas Martín, I.; Arévalo-Villena, M.; Briones Pérez, A. High prevalence of antibiotic-resistant Escherichia coli isolates from retail poultry products in Spain. Animals 2021, 11, 3197. [Google Scholar] [CrossRef]
- Sornsenee, P.; Chimplee, S.; Arbubaker, A.; Kongchai, S.; Madimong, H.; Romyasamit, C. Occurrence, Antimicrobial Resistance Profile, and Characterization of Extended-Spectrum β-Lactamase–Producing Escherichia coli Isolates from Minced Meat at Local Markets in Thailand. Foodborne Pathog. Dis. 2022, 19, 232–240. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1216. [Google Scholar] [CrossRef]
- Ghodousi, A.; Bonura, C.; Di Carlo, P.; van Leeuwen, W.B.; Mammina, C. Extraintestinal pathogenic Escherichia coli sequence type 131 H30-R and H30-Rx subclones in retail chicken meat, Italy. Int. J. Food Microbiol. 2016, 228, 10–13. [Google Scholar] [CrossRef]
- Casella, T.; Nogueira, M.C.L.; Saras, E.; Haenni, M.; Madec, J.-Y. High prevalence of ESBLs in retail chicken meat despite reduced use of antimicrobials in chicken production, France. Int. J. Food Microbiol. 2017, 257, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef] [PubMed]
- Salah, A.M.; Mohamed, Z.; Bilel, H.; Souhir, Z.; Asma, D.; Riadh, M. ESBL/Cephalosporinase-producing Escherichia coli from retail poultry meat in Tunisia: Predominance of blaCTX-M gene and multidrug resistance. J. Microbes Microbio. Tech. 2017, 1, 101. [Google Scholar]
- Soufi, L.; Abbassi, M.S.; Sáenz, Y.; Vinué, L.; Somalo, S.; Zarazaga, M.; Abbas, A.; Dbaya, R.; Khanfir, L.; Ben Hassen, A.; et al. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog. Dis. 2009, 6, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Badi, S.; Salah Abbassi, M.; Snoussi, M.; Werheni, R.; Hammami, S.; Maal-Bared, R.; Hassen, A. High rates of antibiotic resistance and biofilm production in Escherichia coli isolates from food products of animal and vegetable origins in Tunisia: A real threat to human health. Int. J. Environ. Health Res. 2022, 32, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Dhaouadi, S.; Soufi, L.; Hamza, A.; Fedida, D.; Zied, C.; Awadhi, E.; Mtibaa, M.; Hassen, B.; Cherif, A.; Torres, C.; et al. Co-occurrence of mcr-1 mediated colistin resistance and β-lactamase-encoding genes in multidrug-resistant Escherichia coli from broiler chickens with colibacillosis in Tunisia. J. Glob. Antimicrob. Resist. 2020, 22, 538–545. [Google Scholar] [CrossRef]
- Ferjani, S.; Maamar, E.; Ferjani, A.; Meftah, K.; Battikh, H.; Mnif, B.; Hamdoun, M.; Chebbi, Y.; Kanzari, L.; Achour, W.; et al. Tunisian multicenter study on the prevalence of colistin resistance in clinical isolates of Gram negative bacilli: Emergence of Escherichia coli harbouring the mcr-1 gene. Antibiotics 2022, 11, 1390. [Google Scholar] [CrossRef]
- Devan, S.; Aklilu, E.; Lemlem, M.; Zakaria, Z.; Hamdan, R. Detection of colistin-resistant Escherichia coli isolated from broiler chickens in Kelantan, Malaysia. Trop. Biomed. 2022, 39, 197–202. [Google Scholar]
- Nath, C.; Das, T.; Islam, M.S.; Hasib, F.Y.; Singha, S.; Dutta, A.; Barua, H.; Islam, Z. Colistin Resistance in Multidrug-Resistant Escherichia coli Isolated from Retail Broiler Meat in Bangladesh. Microb. Drug Resist. 2023, 29, 523–532. [Google Scholar] [CrossRef]
- Hatrongjit, R.; Kerdsin, A.; Akeda, Y.; Hamada, S. Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX 2018, 5, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Karaiskos, I.; Giamarellou, H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: Current and emerging therapeutic approaches. Expert Opin. Pharmacother. 2014, 15, 1351–1370. [Google Scholar] [CrossRef] [PubMed]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug resistance (MDR): A widespread phenomenon in pharmacological therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef] [PubMed]
- Soufi, L.; Sáenz, Y.; Vinué, L.; Abbassi, M.S.; Ruiz, E.; Zarazaga, M.; Ben Hassen, A.; Hammami, S.; Torres, C. Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 2011, 144, 497–502. [Google Scholar] [CrossRef]
- Rafiq, K.; Sani, A.A.; Hossain, M.T.; Hossain, M.T.; Hadiuzzaman, M.; Bhuiyan, M.A.S. Assessment of the presence of multidrug-resistant Escherichia coli, Salmonella and Staphylococcus in chicken meat, eggs and faeces in Mymensingh division of Bangladesh. Heliyon 2024, 10, e36690. [Google Scholar] [CrossRef]
- Rahman, M.M.; Husna, A.; Elshabrawy, H.A.; Alam, J.; Runa, N.Y.; Badruzzaman, A.T.M.; Banu, N.A.; Al Mamun, M.; Paul, B.; Das, S.; et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep. 2020, 10, 21999. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mensah, S.; Koudandé, O.; Sanders, P.; Laurentie, M.; Mensah, G.; Abiola, F. Antimicrobial residues in foods of animal origin in Africa: Public health risks. Rev. Sci. Tech. 2014, 33, 987–996. [Google Scholar]
- White, D.G.; Hudson, C.; Maurer, J.J.; Ayers, S.; Zhao, S.; Lee, M.D.; Bolton, L.; Foley, T.; Sherwood, J. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 2000, 38, 4593–4598. [Google Scholar] [CrossRef]
- Brătfelan, D.O.; Tabaran, A.; Colobatiu, L.; Mihaiu, R.; Mihaiu, M. Prevalence and Antimicrobial Resistance of Escherichia coli Isolates from Chicken Meat in Romania. Animals 2023, 13, 3488. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Grami, R.; Mansour, W.; Dahmen, S.; Mehri, W.; Haenni, M.; Aouni, M.; Madec, J.-Y. The blaCTX-M-1 IncI1/ST3 plasmid is dominant in chickens and pets in Tunisia. J. Antimicrob. Chemother. 2013, 68, 2950–2952. [Google Scholar] [CrossRef] [PubMed]
- Nahar, A.; Awasthi, S.P.; Hatanaka, N.; Okuno, K.; Hoang, P.H.; Hassan, J.; Hinenoya, A.; Yamasaki, S. Prevalence and characteristics of extended-spectrum β-lactamase-producing Escherichia coli in domestic and imported chicken meats in Japan. J. Vet. Med. Sci. 2018, 80, 510–517. [Google Scholar] [CrossRef]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Meunier, D.; Jouy, E.; Lazizzera, C.; Kobisch, M.; Madec, J.-Y. CTX-M-1-and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 2006, 28, 402–407. [Google Scholar] [CrossRef]
- Mnif, B.; Ktari, S.; Rhimi, F.; Hammami, A. Extensive dissemination of CTX-M-1-and CMY-2-producing Escherichia coli in poultry farms in Tunisia. Lett. Appl. Microbiol. 2012, 55, 407–413. [Google Scholar] [CrossRef]
- Jouini, A.; Klibi, A.; Elarbi, I.; Chaabene, M.B.; Hamrouni, S.; Souiai, O.; Hanachi, M.; Ghram, A.; Maaroufi, A. First detection of human ST131-CTX-M-15-O25-B2 clone and high-risk clonal lineages of ESBL/pAmpC-producing E. coli isolates from diarrheic poultry in Tunisia. Antibiotics 2021, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Mesa, R.; Saco, M.; Lavilla, S.; Prats, G.; Miró, E.; Navarro, F.; Cortés, P.; Llagostera, M. ESBL-and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 2006, 118, 299–304. [Google Scholar] [CrossRef]
- Hiroi, M.; Yamazaki, F.; Harada, T.; Takahashi, N.; Iida, N.; Noda, Y.; Yagi, M.; Nishio, T.; Kanda, T.; Kawamori, F.; et al. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 2012, 74, 189–195. [Google Scholar] [CrossRef]
- Seo, K.W.; Kim, Y.B.; Jeon, H.Y.; Lim, S.-K.; Lee, Y.J. Comparative genetic characterization of third-generation cephalosporin-resistant Escherichia coli from chicken meat produced by integrated broiler operations in South Korea. Poult. Sci. 2018, 97, 2871–2879. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, Y.; Tian, Y.; Shen, Y.; Wang, S.; Zhang, Y. Clinical impact of colistin banning in food animal on mcr-1-positive Enterobacteriaceae in patients from Beijing, China, 2009–2019: A long-term longitudinal observational study. Front. Microbiol. 2022, 13, 826624. [Google Scholar] [CrossRef] [PubMed]
- Grami, R.; Mansour, W.; Mehri, W.; Bouallègue, O.; Boujaâfar, N.; Madec, J.-Y.; Haenni, M. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Eurosurveillance 2016, 21, 30144. [Google Scholar] [CrossRef] [PubMed]
- Maamar, E.; Alonso, C.A.; Hamzaoui, Z.; Dakhli, N.; Abbassi, M.S.; Ferjani, S.; Saidani, M.; Boubaker, I.B.-B.; Torres, C. Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm. Int. J. Food Microbiol. 2018, 269, 60–63. [Google Scholar] [CrossRef]
- Skov, R.L.; Monnet, D.L. Plasmid-mediated colistin resistance (mcr-1 gene): Three months later, the story unfolds. Eurosurveillance 2016, 21, 30155. [Google Scholar] [CrossRef]
- Ferjani, S.; Saidani, M.; Amine, F.S.; Boutiba-Ben Boubaker, I. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae in a Tunisian hospital. Microb. Drug Resist. 2015, 21, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Sallem, R.B.; Gharsa, H.; Slama, K.B.; Rojo-Bezares, B.; Estepa, V.; Porres-Osante, N.; Jouini, A.; Klibi, N.; Sáenz, Y.; Boudabous, A.; et al. First detection of CTX-M-1, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector-Borne Zoonotic Dis. 2013, 13, 98–102. [Google Scholar] [CrossRef]
- Cerquetti, M.; García-Fernández, A.; Giufrè, M.; Fortini, D.; Accogli, M.; Graziani, C.; Luzzi, I.; Caprioli, A.; Carattoli, A. First report of plasmid-mediated quinolone resistance determinant qnrS1 in an Escherichia coli strain of animal origin in Italy. Antimicrob. Agents Chemother. 2009, 53, 3112–3114. [Google Scholar] [CrossRef]
- Niero, G.; Bortolaia, V.; Vanni, M.; Intorre, L.; Guardabassi, L.; Piccirillo, A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018, 216, 93–98. [Google Scholar] [CrossRef]
- Fortini, D.; Fashae, K.; García-Fernández, A.; Villa, L.; Carattoli, A. Plasmid-mediated quinolone resistance and β-lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 2011, 66, 1269–1272. [Google Scholar] [CrossRef]
- Veldman, K.; van Essen-Zandbergen, A.; Kant, A.; Mevius, D. Characterization of qnr-positive Escherichia coli isolates from food-producing animals in the Netherlands. J. Antimicrob. Chemother. 2012, 67, 239–240. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M.; Gunther, N.W. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007, 4, 134–163. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Saralaya, V.; Adhikari, P.; Shenoy, S.; Baliga, S.; Hegde, A. Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann. Med. Health Sci. Res. 2015, 5, 241–246. [Google Scholar] [PubMed]
- De Carli, S.; Ikuta, N.; Lehmann, F.K.M.; da Silveira, V.P.; de Melo Predebon, G.; Fonseca, A.S.K.; Lunge, V.R. Virulence gene content in Escherichia coli isolates from poultry flocks with clinical signs of colibacillosis in Brazil. Poult. Sci. 2015, 94, 2635–2640. [Google Scholar] [CrossRef]
- Tayh, G.; Nagarjuna, D.; Sallem, R.B.; Verma, V.; Yahia, H.B.; Gharsa, H.; Yadav, M.; Ben Slama, K. Identification of Virulence Factors Among ESBL-Producing Escherichia coli Clinical Isolates from GAZA Strip, Palestine: Virulence Genes among ESBL-producing E. coli. J. Microbiol. Biotechnol. Food Sci. 2021, 11, e2865. [Google Scholar] [CrossRef]
- Messaili, C.; Messai, Y.; Bakour, R. Virulence gene profiles, antimicrobial resistance and phylogenetic groups of fecal Escherichia coli strains isolated from broiler chickens in Algeria. Vet. Ital. 2019, 55, 35–46. [Google Scholar]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef]
- Mellata, M.; Ameiss, K.; Mo, H.; Curtiss, R., III. Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coli χ7122 (O78: K80: H9). Infect. Immun. 2010, 78, 1528–1541. [Google Scholar] [CrossRef]
- Poole, N.M.; Green, S.I.; Rajan, A.; Vela, L.E.; Zeng, X.-L.; Estes, M.K.; Maresso, A.W. Role for FimH in extraintestinal pathogenic Escherichia coli invasion and translocation through the intestinal epithelium. Infect. Immun. 2017, 85, 10-1128. [Google Scholar] [CrossRef]
- Fajardo, A.; Martínez-Martín, N.; Mercadillo, M.; Galán, J.C.; Ghysels, B.; Matthijs, S.; Cornelis, P.; Wiehlmann, L.; Tümmler, B.; Baquero, F.; et al. The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 2008, 3, e1619. [Google Scholar] [CrossRef]
- Palmer, A.C.; Kishony, R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 2013, 14, 243–248. [Google Scholar] [CrossRef]
- Wozniak, R.A.; Waldor, M.K. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009, 5, e1000439. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Hammaren, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef]
- Van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140087. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Levy, S.B. The Antibiotic Paradox: How Miracle Drugs Are Destroying the Miracle; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Jackson, L.K.; Dailey, T.A.; Anderle, B.; Warren, M.J.; Bergonia, H.A.; Dailey, H.A.; Phillips, J.D. Exploiting differences in heme biosynthesis between bacterial species to screen for novel antimicrobials. Biomolecules 2023, 13, 1485. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Jouini, A.; Vinué, L.; Slama, K.B.; Sáenz, Y.; Klibi, N.; Hammami, S.; Boudabous, A.; Torres, C. Characterization of CTX-M and SHV extended-spectrum β-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J. Antimicrob. Chemother. 2007, 60, 1137–1141. [Google Scholar] [CrossRef]
- Batchelor, M.; Hopkins, K.; Threlfall, E.; Clifton-Hadley, F.; Stallwood, A.; Davies, R.; Liebana, E. bla CTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005, 49, 1319–1322. [Google Scholar] [CrossRef]
- Muzaheed Doi, Y.; Adams-Haduch, J.M.; Endimiani, A.; Sidjabat, H.E.; Gaddad, S.M.; Paterson, D.L. High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J. Antimicrob. Chemother. 2008, 61, 1393–1394. [Google Scholar] [CrossRef]
- Literacka, E.; Empel, J.; Baraniak, A.; Sadowy, E.; Hryniewicz, W.; Gniadkowski, M. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 2004, 48, 4136–4143. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Brinas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Perreten, V.; Boerlin, P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Tóth, I.; Hérault, F.; Beutin, L.; Oswald, E. Production of Cytolethal Distending Toxins by Pathogenic Escherichia coli Strains Isolated from Human and Animal Sources: Establishment of the Existence of a New cdt Variant (Type IV). J. Clin. Microbiol. 2003, 41, 4285–4291. [Google Scholar] [CrossRef]
- Ruiz, J.; Simon, K.; Horcajada, J.P.; Velasco, M.; Barranco, M.; Roig, G.; Moreno-Martínez, A.; Martínez, J.A.; de Anta, T.J.; Mensa, J.; et al. Differences in Virulence Factors among Clinical Isolates of Escherichia coli Causing Cystitis and Pyelonephritis in Women and Prostatitis in Men. J. Clin. Microbiol. 2002, 40, 4445–4449. [Google Scholar] [CrossRef]
- Vidal, M.; Kruger, E.; Durán, C.; Lagos, R.; Levine, M.; Prado, V.; Toro, C.; Vidal, R. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J. Clin. Microbiol. 2005, 43, 5362–5365. [Google Scholar] [CrossRef]
- Paton, A.W.; Paton, J.C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx 1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar]
- Yamamoto, S.; Terai, A.; Yuri, K.; Kurazono, H.; Takeda, Y.; Yoshida, O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol. 1995, 12, 85–90. [Google Scholar] [CrossRef] [PubMed]
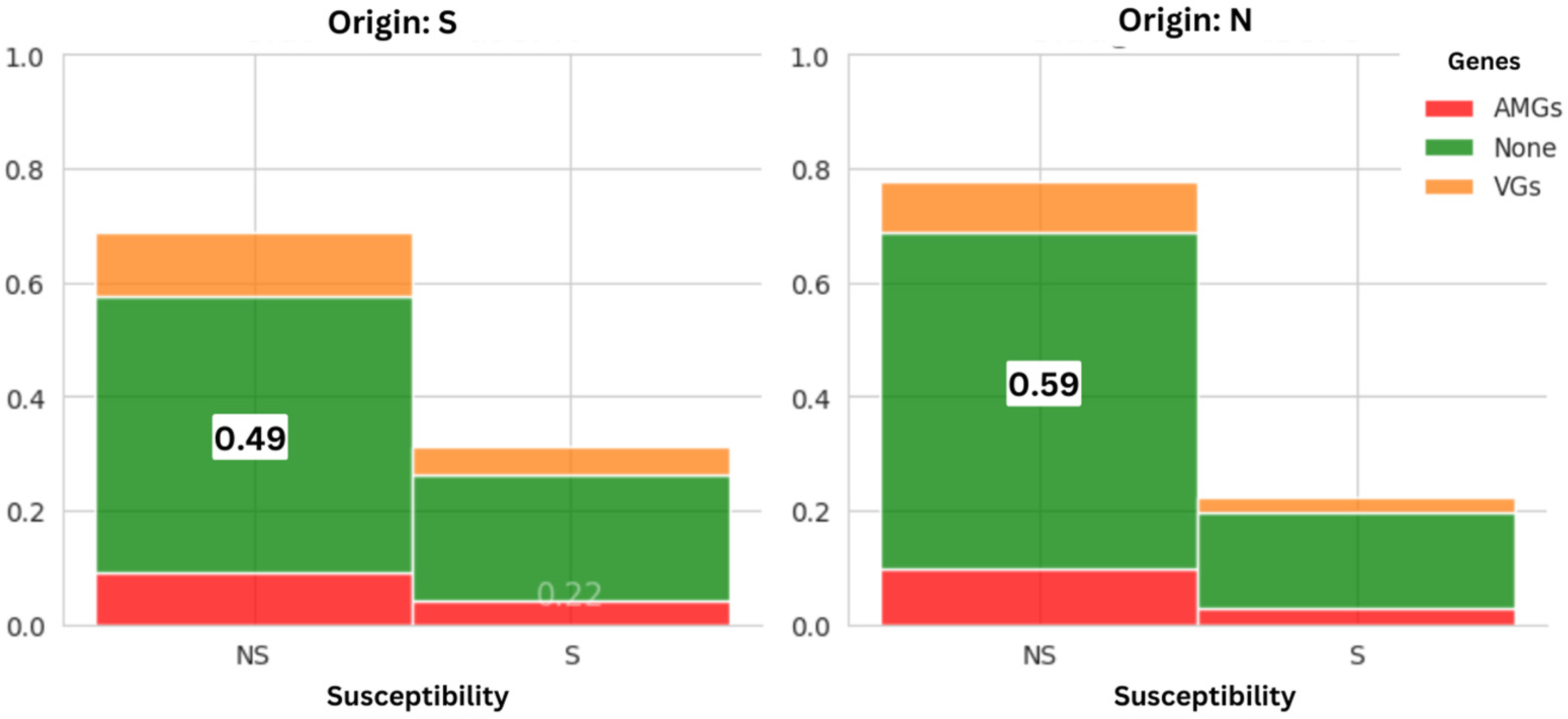

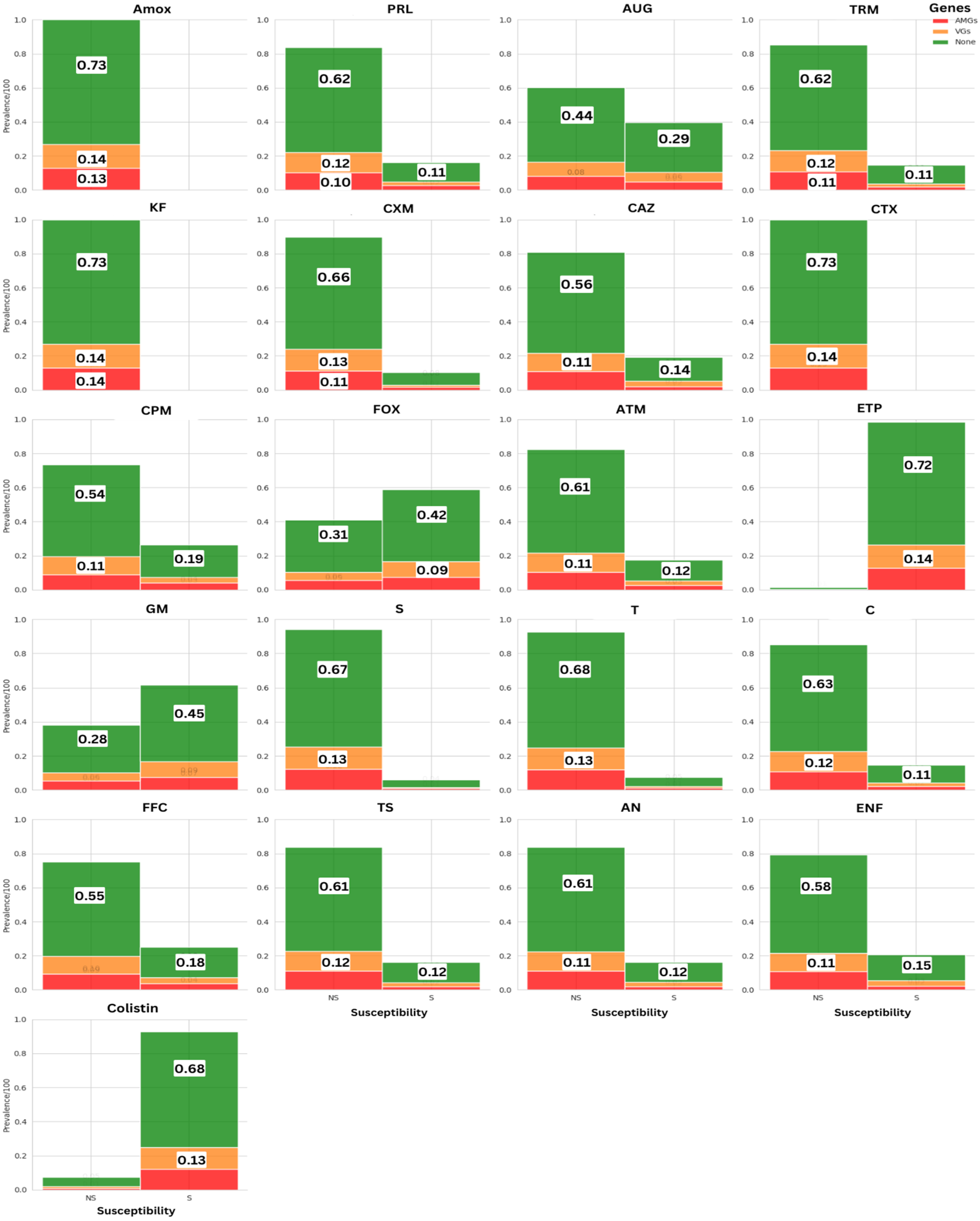

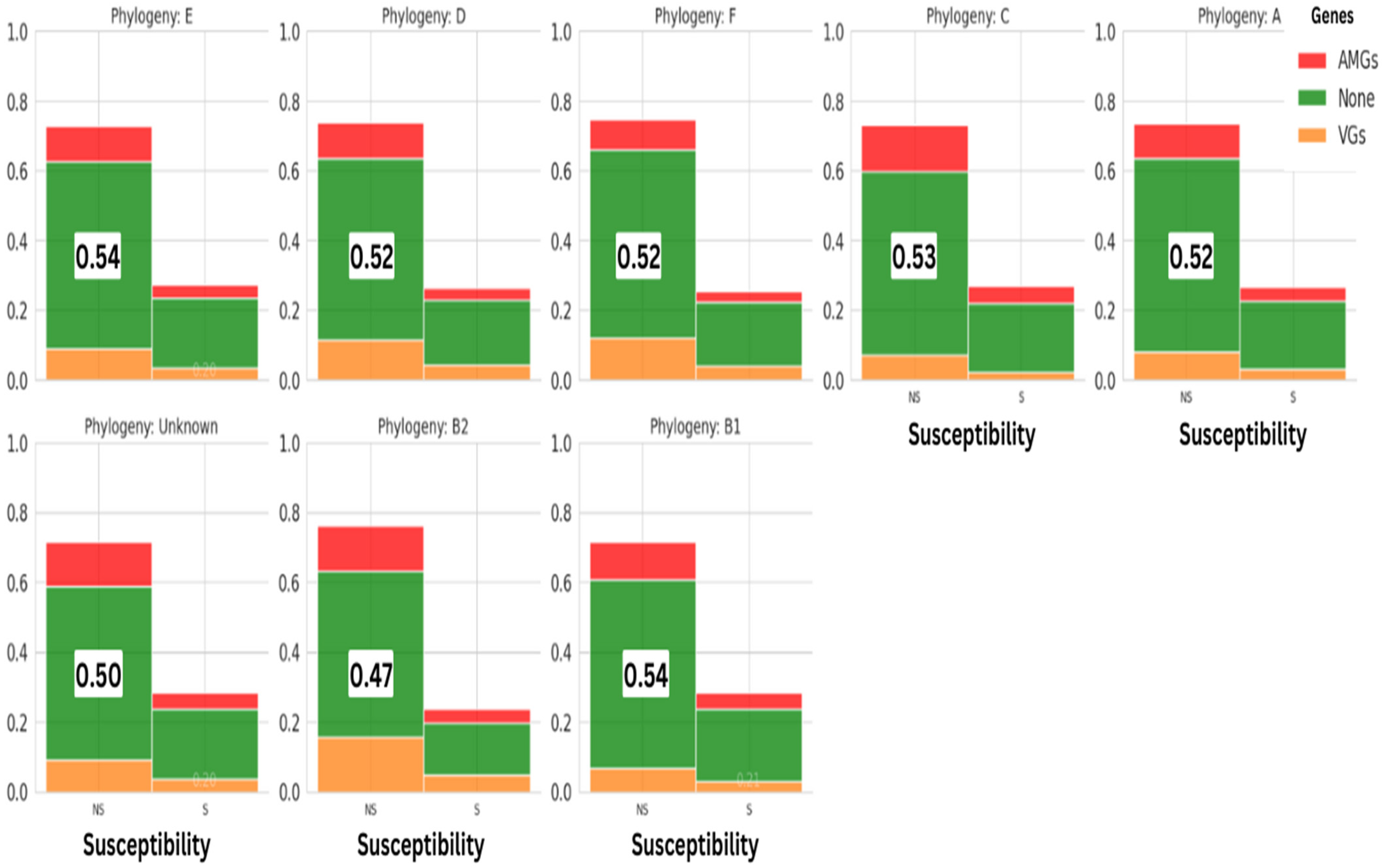
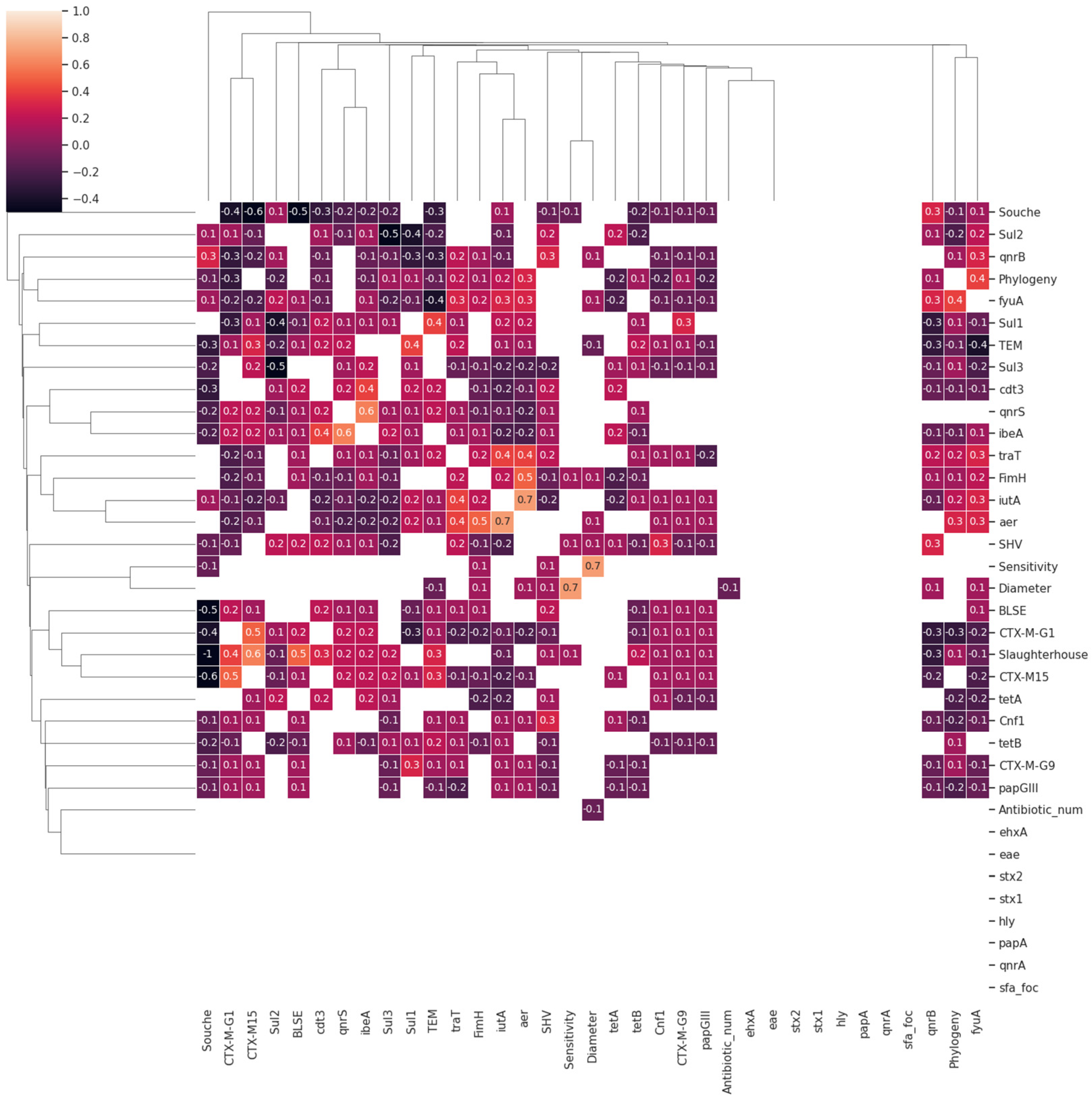


| Number of Samples | Contamination Rate E. coli (MCC) | Contamination Rate E. coli (MCC + CTX) | p-Value E. coli | |
|---|---|---|---|---|
| Supplier N | 50 | 76% (38/50) | 68% (34/50) | 0.157 |
| Supplier S | 50 | 88% (44/50) | 68% (34/50) | |
| Total | 100 | 82% (82/100) | 68% (68/100) | - |
| Strain | B14CTX | B28CTX | B44CTX | B45CTX | A10CTX |
|---|---|---|---|---|---|
| MIC (µg/mL) | 2 | <2 | 4 | 6 | 6 |
| ESBL | + | − | + | + | + |
| mcr-1 gene | + | − | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdallah, K.; Tayh, G.; Maamar, E.; Mosbah, A.; Abbes, O.; Fliss, I.; Messadi, L. Genotypic Characterisation and Risk Assessment of Virulent ESBL-Producing E. coli in Chicken Meat in Tunisia: Insights from Multi-Omics Machine Learning Perspective. Microbiol. Res. 2025, 16, 131. https://doi.org/10.3390/microbiolres16060131
Abdallah K, Tayh G, Maamar E, Mosbah A, Abbes O, Fliss I, Messadi L. Genotypic Characterisation and Risk Assessment of Virulent ESBL-Producing E. coli in Chicken Meat in Tunisia: Insights from Multi-Omics Machine Learning Perspective. Microbiology Research. 2025; 16(6):131. https://doi.org/10.3390/microbiolres16060131
Chicago/Turabian StyleAbdallah, Khaled, Ghassan Tayh, Elaa Maamar, Amine Mosbah, Omar Abbes, Ismail Fliss, and Lilia Messadi. 2025. "Genotypic Characterisation and Risk Assessment of Virulent ESBL-Producing E. coli in Chicken Meat in Tunisia: Insights from Multi-Omics Machine Learning Perspective" Microbiology Research 16, no. 6: 131. https://doi.org/10.3390/microbiolres16060131
APA StyleAbdallah, K., Tayh, G., Maamar, E., Mosbah, A., Abbes, O., Fliss, I., & Messadi, L. (2025). Genotypic Characterisation and Risk Assessment of Virulent ESBL-Producing E. coli in Chicken Meat in Tunisia: Insights from Multi-Omics Machine Learning Perspective. Microbiology Research, 16(6), 131. https://doi.org/10.3390/microbiolres16060131







