Quantitative Differences in the Human Intestinal Microbiota Through the Stages of Life: Infants, Children, Adults and the Elderly
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Permission
2.2. Study Group
2.3. Sample Collection and Preparation
2.4. Bacterial Strains and Culture Conditions
2.5. DNA Extraction from Bacterial Cultures and Feces
2.6. Primers and Probes
2.7. Quantitative Real-Time PCR (qPCR)
2.8. Statistical Analysis
3. Results
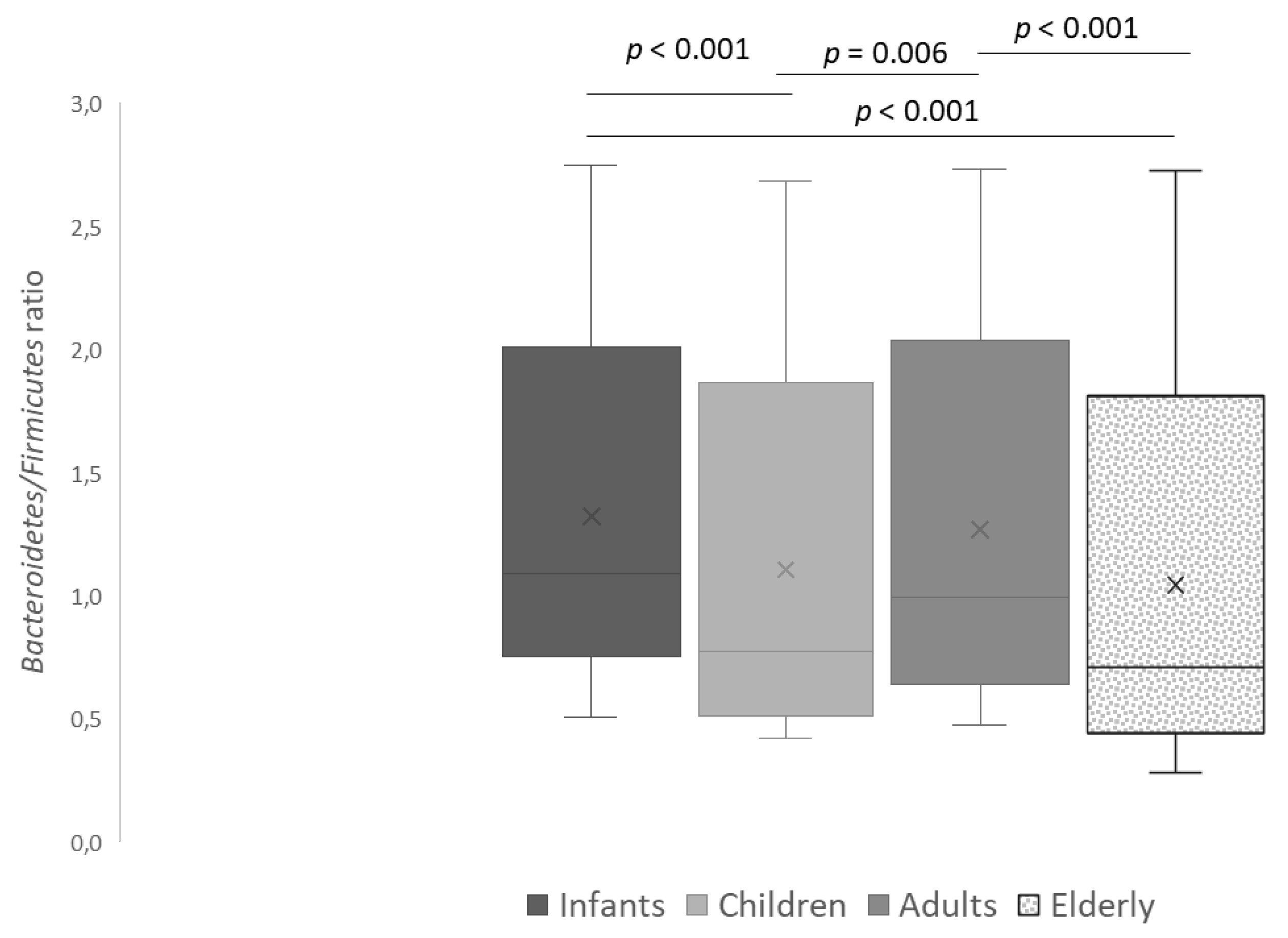
3.1. Prevalence and Counts of Intestinal Bacteria Groups in Persons of Different Age
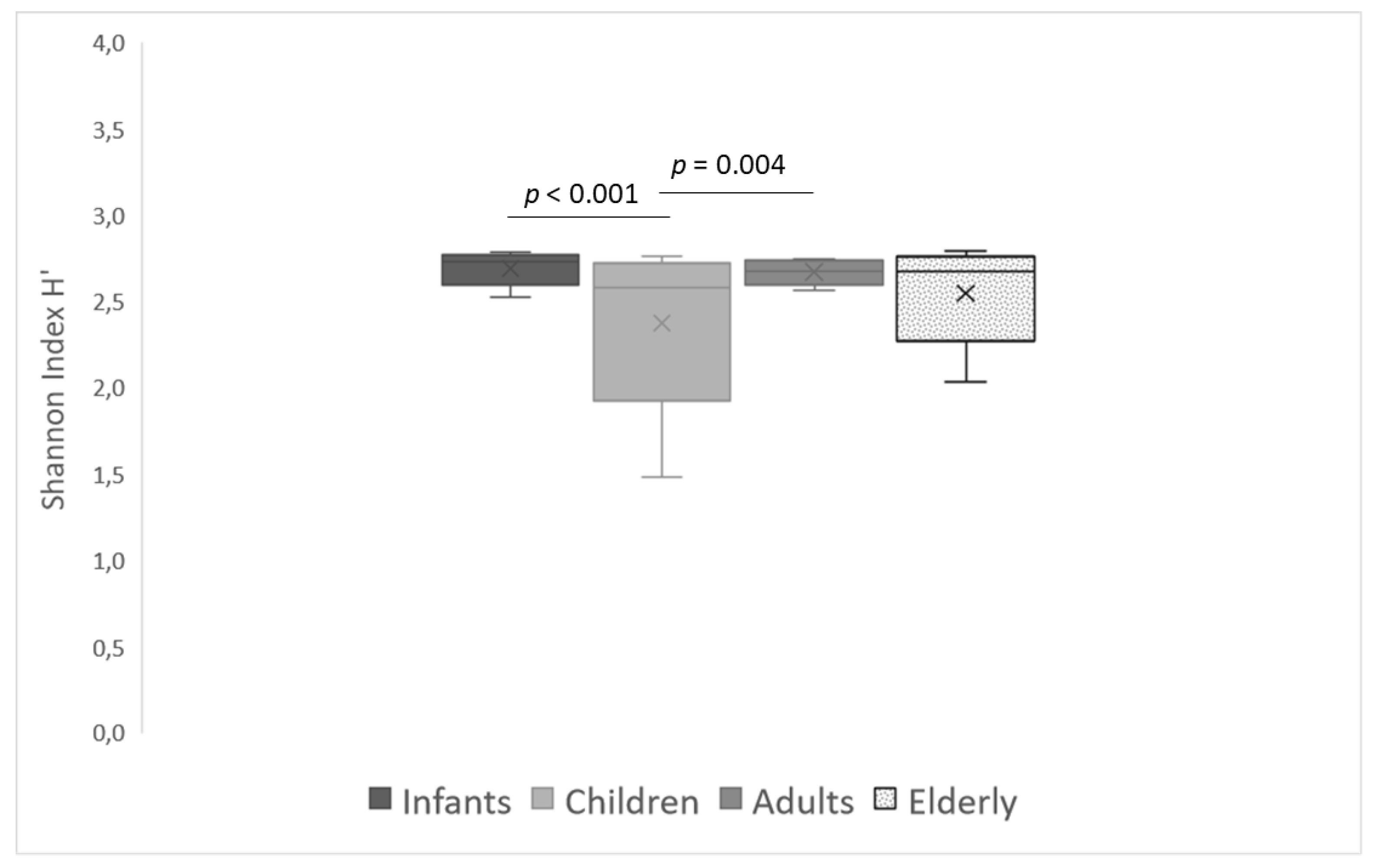
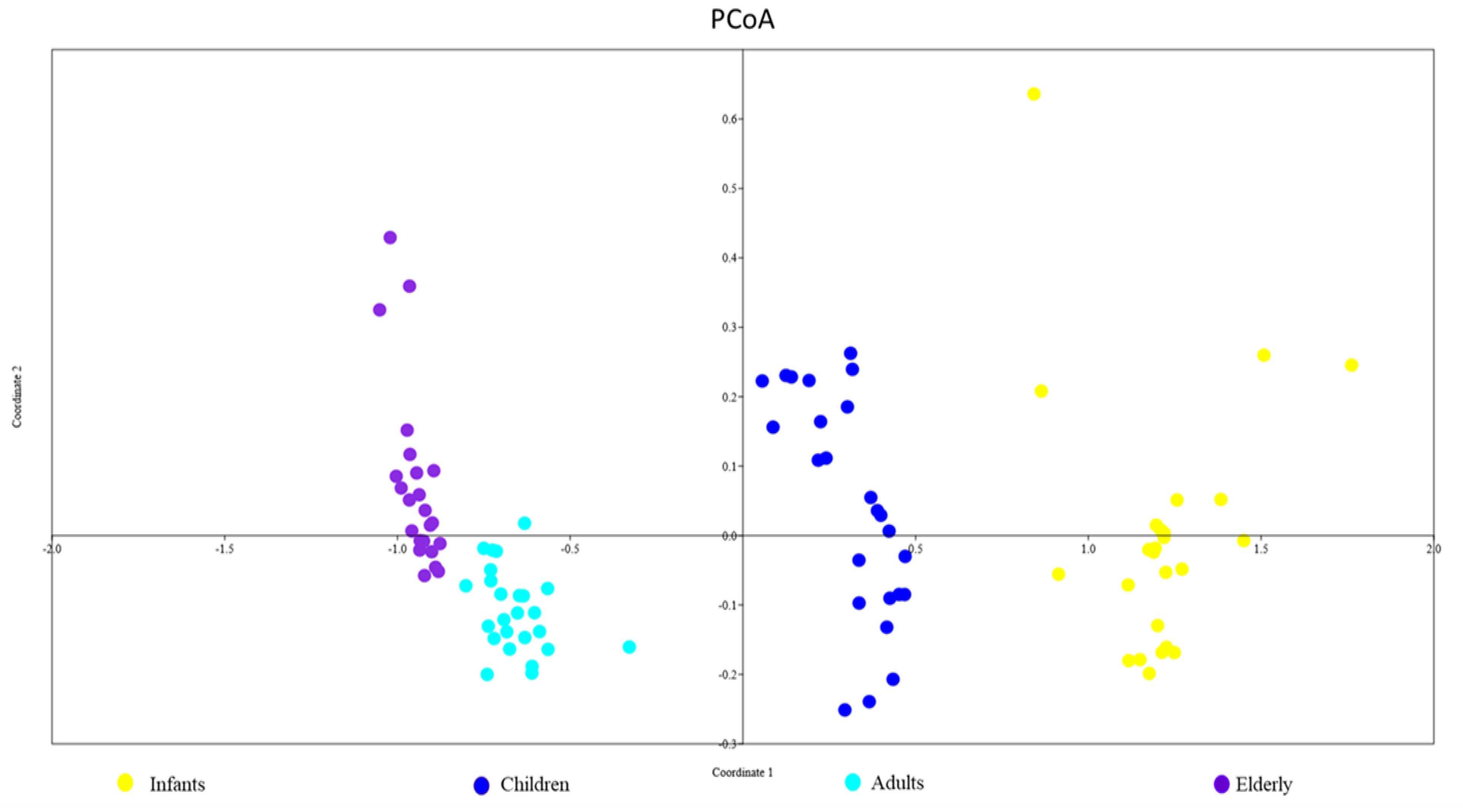
3.2. Intestinal Bacterial Diversity in Different Age Groups
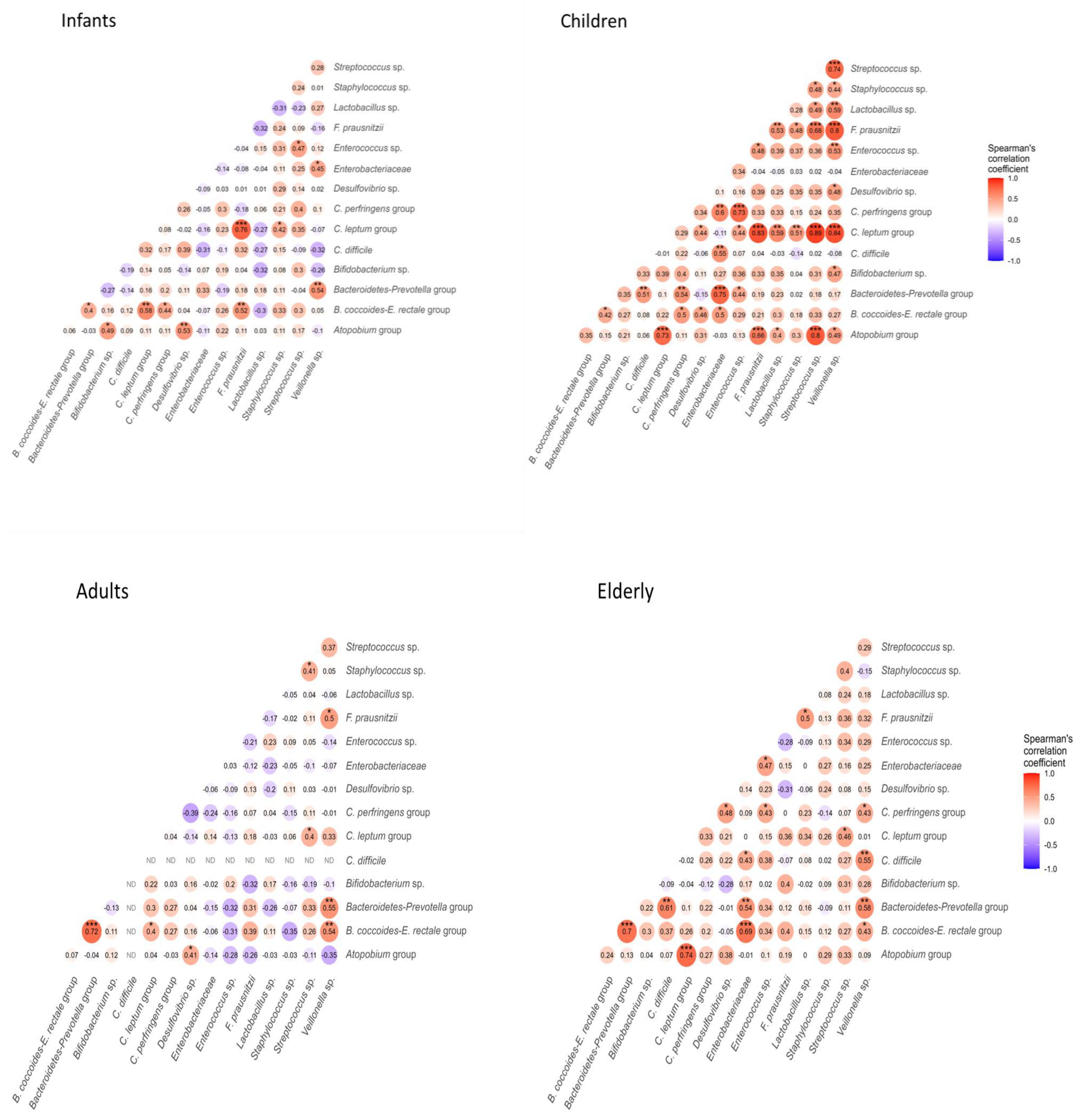
3.3. Correlation Between Age and Counts of Intestinal Bacteria Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Ladirat, S.E.; Schols, H.A.; Nauta, A.; Schoterman, M.H.; Keijser, B.J.; Montijn, R.C.; Gruppen, H.; Schuren, F.H. High-throughput analysis of the impact of antibiotics on the human intestinal microbiota composition. J. Microbiol. Methods 2013, 92, 387–397. [Google Scholar] [CrossRef]
- Nylund, L.; Satokari, R.; Salminen, S.; de Vos, W.M. Intestinal microbiota during early life—Impact on health and disease. Proc. Nutr. Soc. 2014, 73, 457–469. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Frank, D.N.; Pace, N.R. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 2008, 24, 4–10. [Google Scholar] [CrossRef]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef]
- Ohkusa, T.; Nishikawa, Y.; Sato, N. Gastrointestinal disorders and intestinal bacteria: Advances in research and applications in therapy. Front. Med. 2023, 7, 935676. [Google Scholar] [CrossRef]
- Moore, R.E.; Townsend, S.D. Temporal development of the infant gut microbiome. Open Biol. 2019, 9, 190128. [Google Scholar] [CrossRef]
- Guo, M.; Miao, M.; Wang, Y.; Duan, M.; Yang, F.; Chen, Y.; Yuan, W.; Zheng, H. Developmental differences in the intestinal microbiota of Chinese 1-year-old infants and 4-year-old children. Sci. Rep. 2020, 10, 19470. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Storro, O.; Oien, T.; Johnsen, R.; Pope, P.; Rudi, K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol. Ecol. 2014, 87, 280–290. [Google Scholar] [CrossRef]
- Bergstrom, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Molgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef]
- Cheng, J.; Ringel-Kulka, T.; Heikamp-de Jong, I.; Ringel, Y.; Carroll, I.; de Vos, W.M.; Salojarvi, J.; Satokari, R. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016, 10, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Lynch, D.B.; Jeffery, I.B.; Cusack, S.; O’Connor, E.M.; O’Toole, P.W. Diet-microbiota-health interactions in older subjects: Implications for healthy aging. Interdiscip. Top. Gerontol. 2015, 40, 141–154. [Google Scholar] [CrossRef]
- Rampelli, S.; Candela, M.; Turroni, S.; Biagi, E.; Collino, S.; Franceschi, C.; O’Toole, P.W.; Brigidi, P. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging 2013, 5, 902–912. [Google Scholar] [CrossRef]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004, 70, 3575–3581. [Google Scholar] [CrossRef] [PubMed]
- Brussow, H. Bacteriophage-host interaction: From splendid isolation into a messy reality. Curr. Opin. Microbiol. 2013, 16, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Saunier, K.; Hanisch, C.; Norin, E.; Alm, L.; Midtvedt, T.; Cresci, A.; Silvi, S.; Orpianesi, C.; Verdenelli, M.C.; et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: A cross-sectional study. Appl. Environ. Microbiol. 2006, 72, 1027–1033. [Google Scholar] [CrossRef]
- Woodmansey, E.J. Intestinal bacteria and ageing. J. Appl. Microbiol. 2007, 102, 1178–1186. [Google Scholar] [CrossRef]
- Malinen, E.; Rinttila, T.; Kajander, K.; Matto, J.; Kassinen, A.; Krogius, L.; Saarela, M.; Korpela, R.; Palva, A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am. J. Gastroenterol. 2005, 100, 373–382. [Google Scholar] [CrossRef]
- Nagpal, R.; Kumar, M.; Yadav, A.K.; Hemalatha, R.; Yadav, H.; Marotta, F.; Yamashiro, Y. Gut microbiota in health and disease: An overview focused on metabolic inflammation. Benef. Microbes 2016, 7, 181–194. [Google Scholar] [CrossRef]
- Payne, A.N.; Chassard, C.; Zimmermann, M.; Muller, P.; Stinca, S.; Lacroix, C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr. Diabetes 2011, 1, e12. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Dore, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, K.; Tanaka, R.; Urabe, T.; Ueno, Y.; Yamashiro, Y.; Nomoto, K.; Takahashi, T.; Tsuji, H.; Asahara, T.; Hattori, N. Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS ONE 2017, 12, e0171521. [Google Scholar] [CrossRef]
- Salonen, A.; Salojarvi, J.; Lahti, L.; de Vos, W.M. The adult intestinal core microbiota is determined by analysis depth and health status. Clin. Microbiol. Infect. 2012, 18 (Suppl. 4), 16–20. [Google Scholar] [CrossRef]
- Jalanka-Tuovinen, J.; Salonen, A.; Nikkila, J.; Immonen, O.; Kekkonen, R.; Lahti, L.; Palva, A.; de Vos, W.M. Intestinal microbiota in healthy adults: Temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE 2011, 6, e23035. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T.; Tanaka, R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Musfeldt, M.; Ullmann, U.; Hampe, J.; Schreiber, S. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: A global approach to the enteric flora. J. Clin. Microbiol. 2004, 42, 2566–2572. [Google Scholar] [CrossRef]
- Jian, C.; Luukkonen, P.; Yki-Jarvinen, H.; Salonen, A.; Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 2020, 15, e0227285. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Mun, S.; Oh, Y.; Cho, C.; Yun, K.; Ahn, Y.; Chung, W.; Lim, M.; Lee, K.; Hwang, T.; et al. A qRT-PCR Method Capable of Quantifying Specific Microorganisms Compared to NGS-Based Metagenome Profiling Data. Microorganisms 2022, 10, 324. [Google Scholar] [CrossRef]
- UN. World Population Prospects 2019: Highlights; UN: New York, NY, USA, 2019. [Google Scholar]
- WHO. World Health Statistics 2020: Monitoring Health for the SDGs; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Voor, T.; Julge, K.; Bottcher, M.F.; Jenmalm, M.C.; Duchen, K.; Bjorksten, B. Atopic sensitization and atopic dermatitis in Estonian and Swedish infants. Clin. Exp. Allergy 2005, 35, 153–159. [Google Scholar] [CrossRef]
- Mikelsaar, M.; Songisepp, E.; Rätsep, M.; Vallas, M.; Hütt, P.; Smidt, I.; Truusalu, K.; Sepp, E.; Mikelsaar, R.-H.; Kilk, K.; et al. Lactobacillus plantarum INDUCIA® Strain DSM 21379 as Hypocholesterolemic Agent And antimicrobial Agent Against Clostridium. Difficile. Patent Application PCT/EE2014/00007, 31 October 2014. [Google Scholar]
- Mikelsaar, M.; Sepp, E.; Štšepetova, J.; Hütt, P.; Zilmer, K.; Kullisaar, K.; Zilmer, M. Regulation of plasma lipid profile by Lactobacillus fermentum (probiotic strain ME-3 DSM14241) in a randomised controlled trial of clinically healthy adults. BMC Nutrition 2015, 1, 27. [Google Scholar] [CrossRef]
- Mikelsaar, M.; Stsepetova, J.; Hutt, P.; Kolk, H.; Sepp, E.; Loivukene, K.; Zilmer, K.; Zilmer, M. Intestinal Lactobacillus sp. is associated with some cellular and metabolic characteristics of blood in elderly people. Anaerobe 2010, 16, 240–246. [Google Scholar] [CrossRef]
- Statovci, D.; Aguilera, M.; MacSharry, J.; Melgar, S. The Impact of Western Diet and Nutrients on the Microbiota and Immune Response at Mucosal Interfaces. Front. Immunol. 2017, 8, 838. [Google Scholar] [CrossRef]
- WHO. Nutrition and Lifestyle in the Baltic Republics (EUR/ICP/LVNG 020304 E67884); WHO: Geneva, Switzerland, 1999. [Google Scholar]
- Estonian Food Infoserver. 2025. Available online: https://www.eestitoit.ee/en (accessed on 12 December 2024).
- Yang, S.; Lin, S.; Kelen, G.D.; Quinn, T.C.; Dick, J.D.; Gaydos, C.A.; Rothman, R.E. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J. Clin. Microbiol. 2002, 40, 3449–3454. [Google Scholar] [CrossRef]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef]
- van den Berg, R.J.; Vaessen, N.; Endtz, H.P.; Schulin, T.; van der Vorm, E.R.; Kuijper, E.J. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J. Med. Microbiol. 2007, 56, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Haarman, M.; Knol, J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 2005, 71, 2318–2324. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Delgado, S.; Maldonado, A.; Rodriguez, J.M. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528. [Google Scholar] [CrossRef]
- Haarman, M.; Knol, J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 2006, 72, 2359–2365. [Google Scholar] [CrossRef]
- Rinttila, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Fite, A.; Macfarlane, G.T.; Cummings, J.H.; Hopkins, M.J.; Kong, S.C.; Furrie, E.; Macfarlane, S. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 2004, 53, 523–529. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Tuikhar, N.; Keisam, S.; Labala, R.K.; Ramakrishnan, P.; Arunkuma, M.C.; Ahmed, G.; Biagi, E.; Jeyaram, K. Comparative analysis of the gut microbiota in centenarians and young adults shows a common signature across genotypically non-related populations. Mech. Ageing Dev. 2019, 179, 23–35. [Google Scholar] [CrossRef]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4578–4585. [Google Scholar] [CrossRef]
- Ringel-Kulka, T.; Cheng, J.; Ringel, Y.; Salojarvi, J.; Carroll, I.; Palva, A.; de Vos, W.M.; Satokari, R. Intestinal microbiota in healthy U.S. young children and adults—A high throughput microarray analysis. PLoS ONE 2013, 8, e64315. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Lecuyer, E.; Cerf-Bensussan, N. Role of microbiota in postnatal maturation of intestinal T-cell responses. Curr. Opin. Gastroenterol. 2011, 27, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Muncan, V.; Heijmans, J.; Krasinski, S.D.; Buller, N.V.; Wildenberg, M.E.; Meisner, S.; Radonjic, M.; Stapleton, K.A.; Lamers, W.H.; Biemond, I.; et al. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2011, 2, 452. [Google Scholar] [CrossRef] [PubMed]
- Sepp, E.; Julge, K.; Mikelsaar, M.; Bjorksten, B. Intestinal microbiota and immunoglobulin E responses in 5-year-old Estonian children. Clin. Exp. Allergy 2005, 35, 1141–1146. [Google Scholar] [CrossRef]
- Stsepetova, J.; Sepp, E.; Julge, K.; Vaughan, E.; Mikelsaar, M.; de Vos, W.M. Molecularly assessed shifts of Bifidobacterium ssp. and less diverse microbial communities are characteristic of 5-year-old allergic children. FEMS Immunol. Med. Microbiol. 2007, 51, 260–269. [Google Scholar]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Sepp, E.; Naaber, P.; Voor, T.; Mikelsaar, M.; Björksten, B. Development of intestinal micro-flora during the first month of life in Estonian and Swedish infants. Microb. Ecol. Health Dis. 2000, 12, 22–26. [Google Scholar]
- Hold, G.L.; Pryde, S.E.; Russell, V.J.; Furrie, E.; Flint, H.J. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 2002, 39, 33–39. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 2004, 186, 2099–2106. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Miquel, S.; Martin, R.; Rossi, O.; Bermudez-Humaran, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Devaux, C.A.; Million, M.; Raoult, D. The Butyrogenic and Lactic Bacteria of the Gut Microbiota Determine the Outcome of Allogenic Hematopoietic Cell Transplant. Front. Microbiol. 2020, 11, 1642. [Google Scholar] [CrossRef] [PubMed]
- Agans, R.; Rigsbee, L.; Kenche, H.; Michail, S.; Khamis, H.J.; Paliy, O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol. Ecol. 2011, 77, 404–412. [Google Scholar] [CrossRef] [PubMed]
- de Meij, T.G.; Budding, A.E.; de Groot, E.F.; Jansen, F.M.; Frank Kneepkens, C.M.; Benninga, M.A.; Penders, J.; van Bodegraven, A.A.; Savelkoul, P.H. Composition and stability of intestinal microbiota of healthy children within a Dutch population. FASEB J. 2016, 30, 1512–1522. [Google Scholar] [CrossRef]
- Enck, P.; Zimmermann, K.; Rusch, K.; Schwiertz, A.; Klosterhalfen, S.; Frick, J.S. The effects of maturation on the colonic microflora in infancy and childhood. Gastroenterol. Res. Pract. 2009, 2009, 752401. [Google Scholar] [CrossRef][Green Version]
- Gomez-Llorente, C.; Plaza-Diaz, J.; Aguilera, M.; Munoz-Quezada, S.; Bermudez-Brito, M.; Peso-Echarri, P.; Martinez-Silla, R.; Vasallo-Morillas, M.I.; Campana-Martin, L.; Vives-Pinera, I.; et al. Three main factors define changes in fecal microbiota associated with feeding modality in infants. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 461–466. [Google Scholar] [CrossRef]
- Piquer-Esteban, S.; Ruiz-Ruiz, S.; Arnau, V.; Diaz, W.; Moya, A. Exploring the universal healthy human gut microbiota around the World. Comput. Struct. Biotechnol. J. 2022, 20, 421–433. [Google Scholar] [CrossRef]
- Hayashi, H.; Sakamoto, M.; Kitahara, M.; Benno, Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol. Immunol. 2003, 47, 557–570. [Google Scholar] [CrossRef]
- Makivuokko, H.; Tiihonen, K.; Tynkkynen, S.; Paulin, L.; Rautonen, N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br. J. Nutr. 2010, 103, 227–234. [Google Scholar] [CrossRef]
- Zwielehner, J.; Liszt, K.; Handschur, M.; Lassl, C.; Lapin, A.; Haslberger, A.G. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp. Gerontol. 2009, 44, 440–446. [Google Scholar] [CrossRef]
- Samuel, S.C.; Hancock, P.; Leigh, D.A. An investigation into Clostridium perfringens enterotoxin-associated diarrhoea. J. Hosp. Infect. 1991, 18, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Echarri, P.P.; Gracia, C.M.; Berruezo, G.R.; Vives, I.; Ballesta, M.; Solis, G.; Morillas, I.V.; Reyes-Gavilan, C.G.; Margolles, A.; Gueimonde, M. Assessment of intestinal microbiota of full-term breast-fed infants from two different geographical locations. Early Hum. Dev. 2011, 87, 511–513. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Leitch, E.C.; Walker, A.W.; Duncan, S.H.; Holtrop, G.; Flint, H.J. Selective colonization of insoluble substrates by human faecal bacteria. Environ. Microbiol. 2007, 9, 667–679. [Google Scholar] [CrossRef]
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J.; Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef]
- Hold, G.L.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhya, I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 2014, 20, 1192–1210. [Google Scholar] [CrossRef]
- Hijova, E.; Chmelarova, A. Short chain fatty acids and colonic health. Bratisl. Lek. Listy 2007, 108, 354–358. [Google Scholar] [PubMed]
- Roy, J.; Denovan-Wright, E.M.; Linsdell, P.; Cowley, E.A. Exposure to sodium butyrate leads to functional downregulation of calcium-activated potassium channels in human airway epithelial cells. Pflugers Arch. 2006, 453, 167–176. [Google Scholar] [CrossRef]
- Mikelsaar, M.; Sepp, E.; Stsepetova, J.; Songisepp, E.; Mandar, R. Biodiversity of Intestinal Lactic Acid Bacteria in the Healthy Population. Adv. Exp. Med. Biol. 2016, 932, 1–64. [Google Scholar] [CrossRef]
- Chen, J.J.; He, S.; Fang, L.; Wang, B.; Bai, S.J.; Xie, J.; Zhou, C.J.; Wang, W.; Xie, P. Age-specific differential changes on gut microbiota composition in patients with major depressive disorder. Aging 2020, 12, 2764–2776. [Google Scholar] [CrossRef]
- Tiihonen, K.; Tynkkynen, S.; Ouwehand, A.; Ahlroos, T.; Rautonen, N. The effect of ageing with and without non-steroidal anti-inflammatory drugs on gastrointestinal microbiology and immunology. Br. J. Nutr. 2008, 100, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Delzenne, N.M. The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 2009, 15, 1546–1558. [Google Scholar] [CrossRef]
- Vaughan, E.E.; Heilig, H.G.; Ben-Amor, K.; de Vos, W.M. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 2005, 29, 477–490. [Google Scholar] [CrossRef]
- Thorasin, T.; Hoyles, L.; McCartney, A.L. Dynamics and diversity of the ‘Atopobium cluster’ in the human faecal microbiota, and phenotypic characterization of ‘Atopobium cluster’ isolates. Microbiology 2015, 161, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fak, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Backhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Grijpstra, J.; Knol, J.; Degener, J.E.; Welling, G.W. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 2000, 66, 4523–4527. [Google Scholar] [CrossRef]
- Moore, W.E.; Holdeman, L.V. Human fecal flora: The normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 1974, 27, 961–979. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Holtrop, G.; Anderson, S.E.; Lobley, G.E.; Flint, H.J. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl. Environ. Microbiol. 2007, 73, 6526–6533. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Coufalova, M.; Vitezova, M.; Rittmann, S.K.R. Sulfate-Reducing Bacteria of the Oral Cavity and Their Relation with Periodontitis-Recent Advances. J. Clin. Med. 2020, 9, 2347. [Google Scholar] [CrossRef] [PubMed]

| Group (No. Participants) | Age (Yrs.) | Sex (No. Male/Female) | |
|---|---|---|---|
| Mean ± SD | Range | ||
| Infants (n = 25) | 1.0 ± 0.05 | 0.8–1.1 | 17/8 |
| Children (n = 25) | 5.3 ± 0.2 | 5.1–5.6 | 13/12 |
| Adults (n = 25) | 48.2 ± 6.6 | 27–58 | 4/21 |
| Elderly (n = 23) | 72.9 ± 5.0 | 66–84 | 9/14 |
| Target Groups (Amplicon Length, Tm, Assay) | Primers/Probes | Sequence (5′–3′) | References |
|---|---|---|---|
| Total bacteria (466 bp, 60 °C, TaqMan) | Univ-f | TGGAGCATGTGGTTTAATTCGA | [45] |
| Univ-r | TGCGGGACTTAACCCAACA | ||
| Univ (Probe) | CACGAGCTGACGACA(AG)CCATGCA | ||
| Firmicutes phylum (126 bp, 60 °C, Sybr) | Firm934f | GGAGYATGTGGTTTAATTCGAAGCA | [46] |
| Firm1060r | AGCTGACGACAACCATGCAC | ||
| Bacteroides-Prevotella group (140 bp, 58 °C, Sybr) | Bact303-f | GGTGTCGGCTTAAGTGCCAT | [25] |
| Bact708-r | CGGACGTAAGGGCCGTGC | ||
| Blautia coccoides-Eubacterium rectale group (429 bp, 55 °C, Sybr) | Ccocc-r | AGTTTYATTCTTGCGAACG | [25] |
| Ccocc-f | CGGTACCTGACTAAGAAGC | ||
| C. leptum group (239 bp, 50 °C, Sybr) | Clept-f | GCACAAGCAGTGGAGT | [32] |
| Clept-r | CTTCCTCCGTTTTGTCAA | ||
| F. prausnitzii (158 bp, 61 °C, Sybr) | Fprau-f | GTCGCAGGATGTCAAGAC | [25] |
| Fprau-r | CCCTTCAGTGCCGCAGT | ||
| C. perfringens group (120 bp, 55 °C, Sybr) | Cperf-f | ATGCAAGTCGACCGAKG | [25] |
| Cperf-r | TATGCGGTATTAAATCTYCCTTT | ||
| C. difficile (177 bp, 60 °C, TaqMan) | Cdif398 | GAAAGTCCAAGTTTACGCTCAAT | [47] |
| Cdif399 | GCTGCACCTAAACTTACACCA | ||
| Cdif(Probe) | ACAGATGCAGCCAAAGTGGTTGAATT | ||
| Veillonella sp. (343 bp, 62 °C, Sybr) | Veilon-f | A(C/T) CAACCTGCCCTTCAGA | [25] |
| Veilon-r | CGTCCCGATTAACAGAGCTT | ||
| Lactobacillus sp. (92 bp, 60 °C, TaqMan) | AllLacto-f | TGGATGCCTTGGCACTAGGA | [48] |
| AllLacto-r | AAATCTCCGGATCAAAGCTTACTTAT | ||
| AllLacto (Probe) | TATTAGTTCCGTCCTTCATC | ||
| Staphylococcus sp. (560 bp, 62 °C, Sybr) | TStaG422 | GGCCGTGTTGAACGTGGTCAAATCA | [49] |
| TStaG765 | TIACCATTTCAGTACCTTCTGGTAA | ||
| Enterococcus sp. (144 bp, 61 °C Sybr) | Enteroc-f | CCCTTATTGTTAGTTGCCATCATT | [25] |
| Enteroc-r | ACTCGTTGTACTTCCCATTGT | ||
| Streptococcus sp. (343 bp, 58 °C, Sybr) | Tut-Strep-F | GAAGAATTGCTTGAATTGGTTGAA | [49] |
| Tut-Strep-R | GGACGGTAGTTGTTGAAGAATGG | ||
| Bifidobacterium sp. (231 bp, 60 °C, TaqMan) | Allbif-f | GGGATGCTGGTGTGGAAGAGA | [50] |
| Allbif-r | TGCTCGCGTCCACTATCCAGT | ||
| AllBif (Probe) | TCAAACCACCACGCGCCA | ||
| Atopobium group (120 bp, 61 °C, Sybr) | c-Atopo-F: | ACCGCTTTCAGCAGGGA | [51] |
| c-Atopo-R: | ACGCCAATGAATCCGGAT | ||
| Enterobacteriacea (195bp,58 °C, Sybr) | Eco1457-f | CATTGACGTTACCCGCAGAAGAAGC | [21] |
| Eco1652- | CTCTACGAGACTCAAGCTTGC | ||
| Desulfovibrio sp. (135 bp, 63 °C, Sybr) | Dsv691-f | CCGTAGATATCTGGAGGAACATCAG | [52] |
| Dsv826-r | ACATCTAGCATCCATCGTTTACAGC |
| Domain | Phylum | Class | Genus/Bacterial Group | Infants | Children | Adults | Elderly | p Value |
|---|---|---|---|---|---|---|---|---|
| Bacteria | Total | Total | Total bacteria | a,b 11.0 ± 0.5 | a,c 10.5 ± 0.9 | c,d 11.0 ± 0.9 | b,d 10.1 ± 0.7 | c p = 0.002 |
| a,b,d p < 0.001 | ||||||||
| Bacteroidetes | Bacteroidia | Bacteroides-Prevotella group | a,b 9.7 ± 1.6 | a,c 8.1 ± 2.0 | 9.0 ± 1.8 | b,c 6.8 ± 1.5 | a p = 0.012 | |
| b,c p < 0.001 | ||||||||
| Firmicutes | Clostridia | Firmicutes group | a,b 9.2 ± 0.3 | a,c 10.2 ± 0.7 | c,d 9.4 ± 0.4 | b,d 10.1 ± 0.1 | a,b,c,d p < 0.001 | |
| B. coccoides-E. rectale group | a 9.7 ± 1.1 | 8.9 ± 2.0 | c 9.6 ± 0.6 | a,c 7.9 ± 1.1 | a,c p < 0.001 | |||
| C. leptum group | 7.7 ± 0.9 | 7.3 ± 1.4 | 7.8 ± 0.9 | 7.8 ± 0.8 | NS | |||
| F. prausnitzii | a,b,c 8.0±0.7 | a,d 7.3 ± 1.6 | b,d,e 9.2 ± 0.9 | c,e 7.5 ± 0.5 | a,b,c p < 0.001 | |||
| d p = 0.005 | ||||||||
| C. perfringens group | a,b,c 6.0 ± 0.4 | a,d 5.0 ± 0.5 | b,e 5.1 ± 0.2 | c,d,e 4.0 ± 0.4 | a,b,c,d,e p < 0.001 | |||
| C. difficile | 2.5±1.1 | 1.0 ± 0.4 | ND | 1.9 ± 0.7 | c p < 0.001 | |||
| d p = 0.005 | ||||||||
| Bacilli | Lactobacillus sp. | a,b,c 6.3±1.2 | a,c 7.5 ± 0.6 | c,d 5.6 ± 0.9 | b,d 7.7 ± 0.5 | a,c,d p < 0.001 | ||
| b p = 0.019 | ||||||||
| Enterococcus sp. | a,b,c 7.5 ± 0.3 | a 7.1 ± 0.5 | b,d 6.8 ± 0.7 | c,d 6.1 ± 1.0 | a p = 0.002 | |||
| b,c p < 0.001 | ||||||||
| d p = 0.007 | ||||||||
| Streptococcus sp. | a,b,c 7.5 ± 0.4 | a 6.9 ± 0.8 | b 6.9 ± 0.6 | c 6.8 ± 0.9 | a p = 0.003 | |||
| c p = 0.001 | ||||||||
| b p < 0.001 | ||||||||
| Staphylococcus sp. | 4.3 ± 0.4 | 4.4 ± 0.7 | 4.3 ± 0.5 | 4.9 ± 1.5 | NS | |||
| Negativicutes | Veillonella sp. | a,b,c 7.8 ± 1.2 | a 5.8 ± 1.8 | b,d 6.8 ± 1.0 | c,d 5.8 ± 1.0 | a,c,d p < 0.001 | ||
| b p = 0.003 | ||||||||
| Actinobacteria | Actinobacteria | Bifidobacterium sp. | 7.1 ± 1.3 | a 7.6 ± 0.9 | a 6.6 ± 1.4 | 6.9 ± 1.4 | a p = 0.003 | |
| Atopobium group | 6.8 ± 1.0 | a 7.2 ± 1.1 | 6.6 ± 1.2 | a 6.5 ± 0.8 | a p = 0.003 | |||
| Proteobacteria | Proteobacteria | Enterobacteriaceae | a,b,c 8.0 ± 1.0 | a 7.1 ± 0.7 | b 6.2 ± 1.3 | c 6.3 ± 1.4 | a p = 0.009 | |
| b,c p < 0.001 | ||||||||
| Desulfovibrio sp. | 4.2 ± 1.4 | 4.7 ± 1.4 | c 5.8 ± 1.8 | c 4.5 ± 1.0 | c p = 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štšepetova, J.; Šebunova, N.; Voor, T.; Soeorg, H.; Rätsep, M.; Mändar, R.; Mikelsaar, M.; Sepp, E. Quantitative Differences in the Human Intestinal Microbiota Through the Stages of Life: Infants, Children, Adults and the Elderly. Microbiol. Res. 2025, 16, 60. https://doi.org/10.3390/microbiolres16030060
Štšepetova J, Šebunova N, Voor T, Soeorg H, Rätsep M, Mändar R, Mikelsaar M, Sepp E. Quantitative Differences in the Human Intestinal Microbiota Through the Stages of Life: Infants, Children, Adults and the Elderly. Microbiology Research. 2025; 16(3):60. https://doi.org/10.3390/microbiolres16030060
Chicago/Turabian StyleŠtšepetova, Jelena, Natalja Šebunova, Tiia Voor, Hiie Soeorg, Merle Rätsep, Reet Mändar, Marika Mikelsaar, and Epp Sepp. 2025. "Quantitative Differences in the Human Intestinal Microbiota Through the Stages of Life: Infants, Children, Adults and the Elderly" Microbiology Research 16, no. 3: 60. https://doi.org/10.3390/microbiolres16030060
APA StyleŠtšepetova, J., Šebunova, N., Voor, T., Soeorg, H., Rätsep, M., Mändar, R., Mikelsaar, M., & Sepp, E. (2025). Quantitative Differences in the Human Intestinal Microbiota Through the Stages of Life: Infants, Children, Adults and the Elderly. Microbiology Research, 16(3), 60. https://doi.org/10.3390/microbiolres16030060






