Abstract
One of the critical steps in lignocellulosic deconstruction is the hydrolysis of cellulose by cellulases. Aspergillus oryzae can produce and secrete a large amount of various extracellular enzymes, including cellulases. However, due to the lack of a comprehensive characterization of the cellulase genes in A. oryzae, the development and application of A. oryzae cellulase are greatly limited. In this study, a total of 219 glycosyl hydrolase genes were systematically identified from the A. oryzae 3.042 genome and classified into 40 glycosyl hydrolase families. Among these glycosyl hydrolase genes, 26 genes encoding the cellulases of endoglucanase, exoglucanase, and β-glucosidase were identified and functionally characterized. The chromosome localizations, gene structures, functional domains, and subcellular localizations of these 26 cellulases were analyzed by bioinformatics. In addition, analysis of the expression patterns revealed that the expression of A. oryzae cellulase genes was time-specific, and most of the cellulase genes were inhibited under low- and high-temperature stress and high salt stress, which had important guiding significance for understanding the transcription patterns of A. oryzae cellulase genes. These findings lay a foundation for our subsequent modification of cellulase activity to realize the industrial applications of A. oryzae cellulase genes in cellulose biorefineries.
1. Introduction
Cellulose, which is the major component of plant biomass, is the most abundant biomass resource on the earth and a sustainable energy source [1]. Cellulose is a macromolecular polysaccharide composed exclusively of multiple glucose molecules that are linked together by β-1,4 glycosidic bonds [2]. The efficient conversion of cellulose into fermentable sugars by hydrolysis of glycosidic bonds is a crucial process for the sustainable production of biofuels, which can be used as an alternative source of renewable energy to meet the future energy demands [3]. The degradation of cellulose is mediated by a complex of enzymes known as cellulases. Cellulase is a group of enzymes with different activities, including endoglucanase (endo-1,4-β-D-glucanase, EC 3.2.1.4), exoglucanase (exo-1,4-β-D-glucanase or cellobiohydrolase, EC 3.2.1.91), and β-glucosidase (1,4-β-D-glucosidase, EC 3.2.1.21) [4]. These cellulase enzymes synergistically degrade cellulose into glucose units, which can then be utilized in the textile, paper and pulp industries, laundry industries, biofuel production, and amino acid synthesis [2,5]. Studies have shown that these three types of cellulases belong to the glycoside hydrolase (GH) family. There are 189 GH families in the Carbohydrate-Active Enzymes (CAZy) database, and the cellulases reported so far are mainly distributed in 20 GH families.
Cellulases are widely found in various microorganisms in nature. For example, bacteria and fungi can produce cellulase by the decomposition mechanism or fermentation process [6,7]. The cellulases used in industrial production, which are mainly derived from fungi, have the characteristics of high yield and high activity. Commonly utilized fungal cellulases have been produced and identified from various filamentous fungi, including Aspergillus [8], Trichoderma [9], and Penicillium [10]. Aspergillus oryzae is a filamentous fungus known for its ability to produce and secrete a variety of extracellular enzymes, including cellulases [11]. To our knowledge, the systematic studies of A. oryzae cellulases have not been reported. In particular, the diversity, abundance, and characteristics of A. oryzae cellulase genes are still far from fully explored. Therefore, in this study, we aimed to identify and comprehensively analyze the cellulase genes in the A. oryzae genomes. Our study investigated the gene structure, chromosomal distributions, and subcellular localizations of the cellulase genes. We also studied the phylogenetic relationships of A. oryzae cellulases with the reported cellulases of Trichoderma reesei and Aspergillus nidulans. In addition, we analyzed the expression patterns of A. oryzae cellulase genes in different growth stages and under different stress conditions. Our study provided valuable insights into the potential function of A. oryzae cellulases.
2. Materials and Methods
2.1. Identification of Glycoside Hydrolase Family from A. oryzae 3.042 Genome
The A. oryzae 3.042 genome was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/?term=Aspergillus+oryzae, accessed on 16 August 2024). The BLASTP program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 16 August 2024) was used to predict A. oryzae glycoside hydrolase family with a threshold e-value of 1 × 10−10, using Aspergillus gene sequences as query sequences. All potential glycoside hydrolase families were identified using HMMER v3.1 software (http://hmmer.org/, accessed on 25 August 2024) and predicted the domain of Glyco_hydro_1, Glyco_hydro_2, Glyco_hydro_3, etc. (PF00232, PF00703, PF00933, etc.). The sequences that generated hits with glycoside hydrolase genes encoding Glyco_hydro domains were considered as glycoside hydrolase family. And then, the CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml, accessed on 3 September 2024) and PFAM databases (http://pfam-legacy.xfam.org/, accessed on 3 September 2024) were utilized to validate the potential glycoside hydrolase family in A. oryzae.
2.2. Functional Annotation of A. oryzae Glycoside Hydrolase Family
The genes of the A. oryzae glycoside hydrolase family were compared against the NCBI non-redundant protein (NR) databases (https://www.ncbi.nlm.nih.gov/refseq/about/nonredundantproteins/, accessed on 16 August 2024) to predict and classify functions using the BLASTX program with an E-value cutoff at 1 × 10−5, and the BLAST results were imported into Blast2GO v4.1 software to retrieve associated GO terms [12]. A. oryzae glycoside hydrolase genes were submitted to the KEGG Automatic Annotation Server (KAAS), and the single-directional best hit information method was selected [13]. The KEGG enrichment graph was generated online (https://www.omicshare.com/tools/Home/Soft/pathwaygseasenior, accessed on 3 September 2024).
2.3. Identify the Cellulase Genes from the Glycoside Hydrolase Family of A. oryzae
According to the Carbohydrate-Active Enzymes (CAZy) database (https://www.cazy.org/Glycoside-Hydrolases.html, accessed on 3 September 2024), functional annotation, and literature report [14], we identified glycoside hydrolase families containing cellulase genes from the glycoside hydrolase families of A. oryzae. In total, six families comprising 26 genes were found to encode β-glucosidase (EC 3.2.1.21), exoglucanase (EC 3.2.1.91), and endoglucanase (EC 3.2.1.4).
2.4. The Chromosomal Locations of A. oryzae Cellulase Genes
The chromosomal locations of the 26 identified cellulase genes were determined by downloading the locus coordinates from the A. oryzae RIB40 genomics database. The distribution of 26 cellulase genes on chromosomes was mapped using MapGene2 Chrome (http://mg2c.iask.in/mg2c_v2.0/, accessed on 3 September 2024) and visualized with MapChart v2.2 software.
2.5. Analysis of Gene Structure and Conserved Domain
The cellulase gene structures were analyzed by the Visualize Gene Structure (Basic) in the TBtools-II v2.20 software. The protein sequences of A. oryzae cellulase were submitted to the Batch CD-Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 4 September 2024) to predict the conserved domains with default parameters [15]. The result was downloaded and submitted to Visualize NCBI CDD Domain Pattern in TBtools-II v2.20 software to obtain the conserved domain graph of A. oryzae cellulase [16]. The subcellular localization of A. oryzae cellulases was predicted using an online WoLF PSORT system (https://wolfpsort.hgc.jp/, accessed on 4 September 2024). MOTIF Search was used to predict and analyze the motifs of A. oryzae cellulases (https://www.genome.jp/tools/motif/, accessed on 4 September 2024).
2.6. Multi Sequence Alignment and Phylogenetic Analysis
The protein sequences of known cellulase in representative fungi Aspergillus nidulans, Aspergillus flavus, Aspergillus niger, and Trichoderma reesei were downloaded from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/?term=, accessed on 16 August 2024). ClustalW of MEGA v6.0 software was used to align the cellulase sequences of A. oryzae, A. nidulans, A. flavus, A. niger, and T. reesei and analyze the phylogenetic relationships. Based on the alignment results in MEGA v6.0 software, the neighbor-joining (NJ) tree was constructed with bootstrap replications of 1000.
2.7. Effects of Different Growth Stages, Temperatures, and Salt Concentration on the Expression Patterns of A. oryzae Cellulase Genes
The transcriptional data of A. oryzae that were used to analyze the expression patterns of A. oryzae cellulase genes were downloaded from the NCBI/SRA database (Bioproject Accession: PRJNA407002; Biosample: SAMN07637372; Bioproject: PRJNA383095; BioSample: SAMN06759404; Bioproject: PRJNA774152; Biosample: SAMN22555497, SAMN22555498, SAMN22555499). Gene expression levels were normalized using the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) method [17]. And then, a heatmap of 26 cellulase genes in different growth stages, temperatures, and salt concentrations was generated using HemI v1.0 software (http://hemi.biocuckoo.org/down.php, accessed on 4 September 2024) [18]. In addition, the edgeR package was used to identify differentially expressed genes (DEGs) of 26 cellulases across samples. Genes with a fold change ≥1 and a false discovery rate (FDR) < 0.05 in a comparison were identified as significant differentially expressed genes [19]. A Venn diagram of A. oryzae differentially expressed cellulase genes was generated using Venny v2.1 software (https://bioinfogp.cnb.csic.es/tools/venny/index.html, accessed on 4 September 2024).
3. Results
3.1. Identification and Annotation of A. oryzae Glycoside Hydrolase Family
BLASTP analysis was used to check the predicted glycoside hydrolase (GH) family from the A. oryzae 3.042 genome. All potential A. oryzae GH families were used to identify Glyco_hydro_1, Glyco_hydro_2, Glyco_hydro_3 domain, etc., (PF00232, PF00703, PF00933, etc.) using HMMER3.1. In total, 261 A. oryzae GH domains (Table S1) were identified and classified into 52 domain types that are shown in Figure 1. All these 261 GH domains were encoded by 219 A. oryzae GH genes and distributed in 40 GH families. More details about the 219 A. oryzae glycoside hydrolase genes are shown in Table S2.
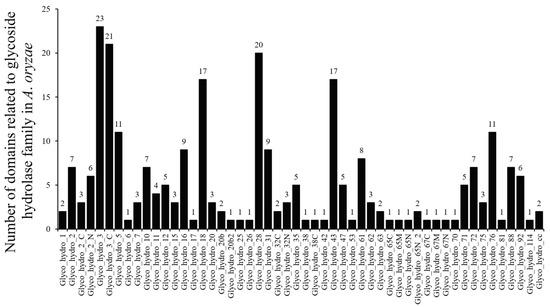
Figure 1.
The numbers of glycoside hydrolase domains in Aspergillus oryzae.
The GO functional enrichment analysis revealed that the 219 A. oryzae GH genes were primarily related to catalytic activity (GO:0003824), metabolic process (GO:0008152), binding (GO:0005488) (Figure 2A–C, Tables S2 and S3). The KEGG analysis revealed that the GH genes were mainly enriched in metabolic pathways, starch and sucrose metabolism, biosynthesis of secondary metabolites, amino sugar and nucleotide sugar metabolism, and carbon metabolism (Tables S2 and S4). The top 25 KEGG pathways are shown in Figure 2D. These analyses of GO and KEGG annotation were helpful to further understand the function of these GH genes in A. oryzae.
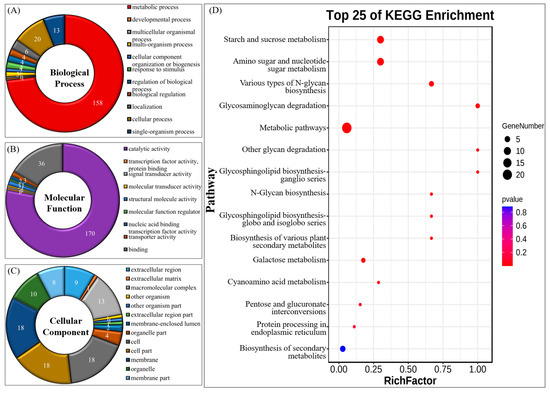
Figure 2.
The GO and KEGG annotation of glycoside hydrolase family in Aspergillus oryzae. (A–C) The GO functional enrichment analysis of 219 glycoside hydrolase genes in A. oryzae. (D) The KEGG analysis of 219 glycoside hydrolase genes in A. oryzae.
3.2. Identified Analysis of the Cellulase Genes from the A. oryzae GH Family
Based on the GO, KEGG and BLASTP annotations, we found there were 26 GH genes that were annotated as β-glucosidase (EC3.2.1.21), exoglucanase (EC.3.2.1.91), and endoglucanase (EC 3.2.1.4) (Table 1 and Table S2). These 26 genes belonged to GH1, GH3, GH5, GH7, and GH12, respectively (Table 2). There were 20 genes encoded with cellulase in GH1 and GH3 families. Two GH5 genes and one GH12 gene encoded the endoglucanase of cellulase. In addition, GH7 families possessed both exoglucanase and endoglucanase of two types of cellulase activities (Table 1 and Table 2). Therefore, 26 genes in A. oryzae containing the cellulase activities of β-glucosidase, exoglucanase, and endoglucanase were identified from the 219 GH genes. All the details of these 26 genes, such as gene IDs, sizes of deduced peptides, and Pfams, gene description, corresponding cellulase activity, are listed in Table 1.

Table 1.
Identification information of 26 cellulase genes in Aspergillus oryzae.

Table 2.
Classification of cellulase genes in glycoside hydrolase families of Aspergillus oryzae.
3.3. Distribution Analysis of A. oryzae Cellulase Genes on Chromosomal Locations
A. oryzae has a genome size of approximately 37.9 Mb, containing about 11,720 predicted genes, and is assembled into eight chromosomes [20]. Based on this genomic framework, chromosome localization analysis was performed to label the positions of cellulase gene family members and observe whether they were clustered on chromosomes. Fungal cellulase genes are usually randomly distributed throughout the genome, and each gene has its own regulatory unit. In our study, the 26 cellulase genes were randomly distributed on eight chromosomes of A. oryzae (Figure 3). In addition, the number of cellulase genes distributed on chromosome 2 is the highest, with five cellulase genes, while the number of genes distributed on chromosome 7 is the least, with only one cellulase gene. The chromosomal location revealed that the cellulase genes of the same GH family member were not clustered and scattered on different chromosomes, which demonstrated that the expression of cellulase genes in the same GH family may be regulated by multiple different transcription factors.
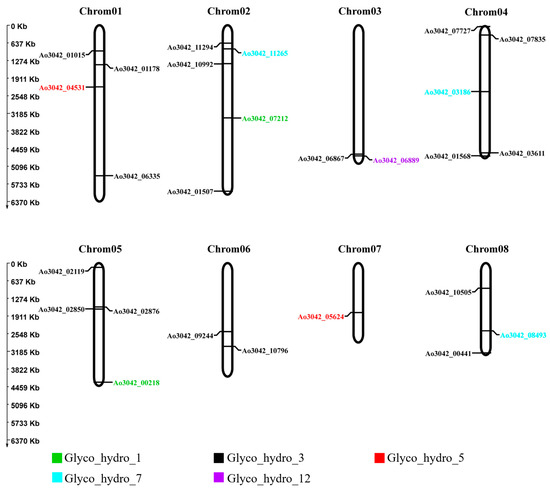
Figure 3.
The chromosome localization of 26 cellulase genes on eight Aspergillus oryzae chromosomes. The genes with the same color on the chromosomes were members of the same hydrolase family.
3.4. Analysis of Cellulase Gene Structure and Conserved Domain
The number of exons and introns in the A. oryzae cellulase genes varied greatly, with 19 cellulase genes (73%) containing two to seven exons. Six cellulase genes contained one exon, and one had 11 exons. Overall, one to three exon genes were the most common, including 16 genes (Figure 4A). The conserved domains of the A. oryzae cellulase were analyzed by using the NCBI CD-Search (Figure 4B). Except for one glucosidase (Ao3042_06867), the other glucosidases in GH1 and GH3 contained an extra fibronectin type III-like (Fn3-like, PF14310) domain, and four glucosidases had a PA14 domain (PF07691). In addition, one endoglucanase in the GH5 family contained a CBD domain (PF00734). All this information could help elucidate the function and evolutionary relationships of the A. oryzae cellulase genes.
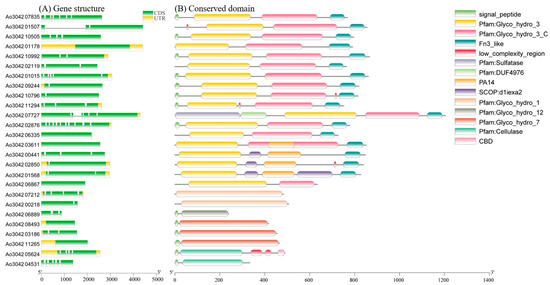
Figure 4.
The gene structure and conserved domain arrangement of 26 A. oryzae cellulases based on their phylogenetic relationships. (A) The gene structure of 26 A. oryzae cellulases. (B) The conserved domains of A. oryzae cellulase were found by using the NCBI CD-Search.
Cellulases are ubiquitously present in different life forms either as intracellular or extracellular proteins. The prediction of protein subcellular localization showed that most of the A. oryzae cellulases (17.65%) were located extracellular, and seven cellulases were in the cytoplasm. In addition, two cellulases are located in the mitochondria. We found A. oryzae exoglucanases and endoglucanases in GH5, GH7, and GH12 that belonged to extracellular proteins, while the two β-glucosidases in GH1 were located in the cytoplasm. The details of conserved domain and subcellular localization of these A. oryzae cellulases are listed in Table S5.
3.5. Phylogenetic Analysis of A. oryzae Cellulases
A. nidulans, A. flavus, A. niger, and T. reesei are the representative filamentous fungi for cellulase production, especially A. nidulans and T. reesei. A neighbor-joining (NJ) phylogenetic tree was constructed using MEGA6.0 for multiple sequence alignment of cellulases with 1000 bootstrap replications to analyze phylogenetic relationships of cellulases among A. oryzae, A. flavus, A. nidulans, Aspergillus niger, and T. reesei. The 26 A. oryzae cellulases were roughly divided into two subgroups in the NJ phylogenetic tree with the homologous cellulases of A. flavus, A. nidulans, Aspergillus niger, and T. reesei (Figure 5). All the A. flavus, A. nidulans, Aspergillus niger, and T. reesei β-glucosidases of cellulases in the GH3 and GH1 families were clustered together in one subgroup in the NJ phylogenetic tree. The endoglucanase cellulases of the GH5 and GH12 families clustered in a subgroup. In addition, one endoglucanase and two exoglucanases in GH7 were clustered with the GH5 and GH12 families. As expected, cellulases that are in the same GH family could be clustered together between different Aspergillus and Trichoderma, which suggested fungal cellulases were evolutionarily conserved and perform similar genetic functions.
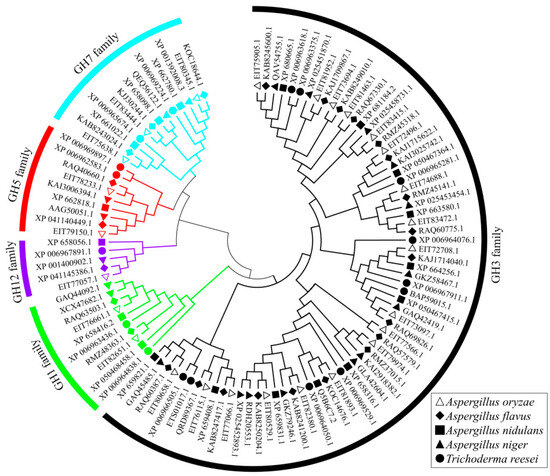
Figure 5.
Phylogenetic analysis of the cellulases among Aspergillus oryzae, Aspergillus flavus, Aspergillus nidulans, Aspergillus niger, and Trichoderma reesei. Cellulase sequences of these three fungi were aligned using ClustalW and a neighbor-joining (NJ) tree was constructed based on the alignment results in MEGA6.0 with bootstrap replications of 1000.
3.6. Expression Patterns of A. oryzae Cellulase Genes at Different Growth Stages, Temperatures, and Salt Concentrations
The expression pattern of 26 A. oryzae cellulase genes at different growth stages and under different temperatures and salt stress was performed using the data from the open dataset. With the extension of growth time, more and more cellulase genes were induced to express (Figure 6A). During the first 24 h, only six cellulase genes were significantly induced to express, while more than half of the genes were upregulated at 72 h compared with 24 h (Figure 6A). Interestingly, the expression of six cellulase genes induced at 24 h was significantly inhibited at 48 h and 72 h, including three endoglucanase genes. Comparative transcriptome analysis showed that there was little difference in A. oryzae cellulase gene expression levels between 72 h and 48 h, but there were 11 differentially expressed genes (DEGs) between 72 h and 24 h, including seven significantly upregulated cellulase genes and four significantly downregulated genes (Figure 7A). Comparative analysis of expression levels at different times suggests that the expression patterns of A. oryzae cellulase genes are obvious time-specific expressions.
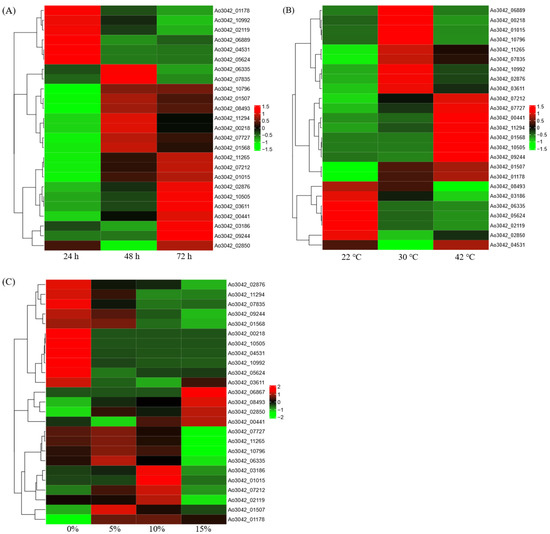
Figure 6.
Expression patterns of Aspergillus oryzae cellulase genes at different experimental treatments. (A) The heatmaps of 26 cellulase genes at different growth stages (24 h, 48 h, and 72 h); (B) The heatmaps of 26 cellulase genes in three temperature treatments (22 °C, 42 °C, and 30 °C); (C) The heatmaps of 26 cellulase genes under different salt stress (0%, 5%, 10%, 15% NaCl). Red and green colors represent the upregulation and downregulation of gene expression, respectively.
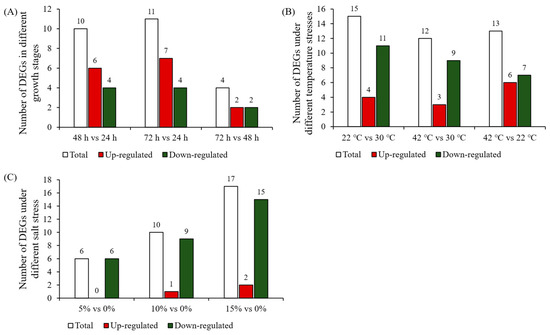
Figure 7.
The number of differentially expressed genes at different growth stages, temperatures, and salt concentrations, respectively. (A) The number of differentially expressed genes in the comparative transcriptome at different growth stages. (B) The number of differentially expressed genes in the comparative transcriptome under different temperature stresses. (C) The number of differentially expressed genes in the comparative transcriptome at different salt concentrations. The differentially expressed genes were selected by the |log2FoldChange| > 1 and Padj < 0.05.
The expression trend of cellulase genes at low temperature, high temperature, and the optimum temperature of A. oryzae growth was opposite. For example, cellulase genes that were induced at low temperatures were significantly inhibited at high temperatures, while cellulase genes that upregulated at high temperatures were downregulated at low temperatures (Figure 6B). Comparative transcriptome analysis showed that there were 15 and 12 DEGs in 22 °C vs. 30 °C and 42 vs. 30 °C comparative transcriptomes (Figure 7B). The expression patterns show that most cellulase genes were significantly inhibited under low-temperature and high-temperature stresses in A. oryzae. Three of four identified endoglucanase genes were downregulated in high-temperature stress.
In addition, salt stress also had a great influence on the expression level of some A. oryzae cellulase genes (Figure 6C). The expression profile in response to salt stress revealed that most of the differentially expressed genes were downregulated in 5% vs. 0%, 10% vs. 0%, and 15% vs. 0% comparative transcriptomes. For example, 6 of 6, 9 of 10, and 15 of 17 DEGs were markedly downregulated in the comparative transcriptomes of 5% vs. 0%, 10% vs. 0%, and 15% vs. 0%, respectively (Figure 7C), which indicated that high salt significantly inhibited the expression of cellulase genes in A. oryzae. Interestingly, 15 of 17 downregulated genes under high salt stress were β-glucosidase, except for two endoglucanase genes in the GH family.
Further analysis of differentially expressed genes by Venn diagram revealed that few cellulase genes were affected by the growth time of A. oryzae or sensitive to temperature and salt stress. The expression levels of three cellulase genes, including Ao3042_06889, Ao3042_10505, and Ao3042_00218, were significantly affected by the growth stages in A. oryzae (Figure 8A, red font, Table S6). Five differentially expressed cellulase genes existed in three comparative transcriptomes, indicating that their expressions were sensitive to low- and high-temperature stresses (Figure 8B, red font, Table S6). In addition, six cellulase genes were sensitive to salt concentration, and their expressions were significantly inhibited under 5 %, 10 % and 15 % salt stresses (Figure 8C, red font, Table S6). Interestingly, we found three cellulase genes were commonly shared among the five comparative transcriptomes, which suggested their expressions were sensitive to both temperature and salt stress (Figure 8D, Table S7). Furthermore, two cellulase gene expressions only strongly responded to high salt stress (Figure 8D, Table S7).
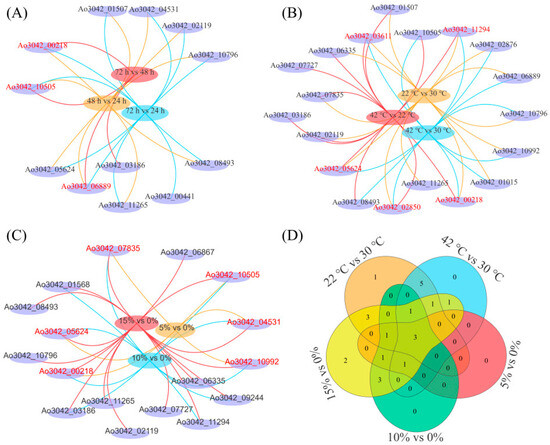
Figure 8.
The Venn analysis of differentially expressed cellulase genes at different growth stages, temperatures, and salt concentrations. (A–C) Distribution of differentially expressed genes in comparative transcriptomes. The red marker genes indicate that they were present in the three comparative transcriptomes in the same treatment. For example, the three genes with red color in (A) were differentially expressed in the comparative transcriptomes of 48 h vs. 24 h, 72 h vs. 24 h, and 72 h vs. 48 h. (D) The Venn analysis of differentially expressed cellulase genes under temperature and salt stress.
4. Discussion
Cellulose, the most abundant renewable biomass resource in the world, can be used in a variety of industrial applications, such as biofuel production, papermaking and pulping industries, bioethanol production, and animal feeds [21,22]. However, cellulosic biomass needs to be effectively degraded into fermentable sugars by cellulases before it is utilized by microorganisms, as most microorganisms are unable to directly metabolize it [23]. Cellulases are the main enzymes required for the hydrolysis of cellulosic polymer materials to produce soluble sugars [24]. At present, cellulases utilized in industry are mainly derived from bacteria and filamentous fungi [25,26]. In general, filamentous fungi exhibit higher levels of protein expression and can secrete proteins directly into the extracellular matrix, making them the preferred hosts for enzyme production [27]. Although there are a larger number of efficient cellulase-producing microorganisms, such as T. reesei and A. niger, they are unable to produce an efficient cellulase system [28]. Different fungal species have different cellulase genes, and the ability of a single cellulose-degrading fungal strain to express three types of cellulases at sufficiently high concentrations is rare. Therefore, these three types of cellulases are usually produced separately using different strains and then blended. In recent years, the research on cellulases has primarily focused on A. niger [8], Trichoderma, T. reesei [9], and Penicillium sp. [29], while the studies on the cellulase gene family of A. oryzae have been rarely reported. However, the lack of a clear understanding of structural and functional properties of A. oryzae cellulases seriously impeded its development and utilization. The exponential growth of genomic data presents a unique opportunity for the identification and analysis of cellulase in A. oryzae. Thus, the discovery of cellulase genes in A. oryzae holds great significance for the rational utilization of cellulase in industry.
Cellulase is a group of individual enzymes, including three types of endoglucanase, exoglucanase, and β-glucosidase, which are classified into the glycoside hydrolase (GH) family [4]. Currently, there are 189 GH families in the CAZy database, of which 20 GH families contain biochemically confirmed cellulase. In this study, a total of 219 GH genes in A. oryzae were identified by HMMER3.1 and classified into 40 GH families. We identified eight genes containing the Pfam of auxiliary activity family 9 (AA9, PF03443) (Table S8), which were originally classified as GH61. Recent studies have shown that lytic polysaccharide monooxygenases from the AA9 family act on cellulose through an oxidative mechanism, synergistically promoting cellulose saccharification with other cellulose-degrading enzymes [30]. Although the AA9 family is involved in improving the degradation efficiency of cellulose microfibrils [30], they lack the conserved clusters of catalytic acidic amino acids typically present in most GHs [31]. Moreover, many members of the GH61 family lack detectable cellulase activity themselves. Therefore, GH61 is unlikely to be true GHs and has been reclassified into the AA9 family [32]. In our study, if we did not classify these eight genes into the GH family, then there were 211 GH genes in A. oryzae.
So far, cellulose degradation requires three types of enzymes—β-1,4-endoglucanases, β-1,4-exoglucanases/cellobiohydrolases, and β-glucosidase—which are classified into 20 GH families in the CAZy database. In our study, 26 GH genes having cellulase activities of β-glucosidase (EC3.2.1.21), exoglucanase (EC.3.2.1.91), and endoglucanase (EC 3.2.1.4) were identified from 219 GH genes in A. oryzae 3.042. These 26 cellulase genes belonged to the GH1, GH3, GH5, GH7, and GH12 families in A. oryzae. We found that the genes in the A. oryzae 3.042 GH2, GH6, GH10, GH16, and GH26 families did not have cellulase activity as reported. In comparison, A. oryzae RIB4 contains 207 GH genes and 26 cellulases [33]. Although A. oryzae 3.042 had twelve more GH genes than RIB40, their number of cellulase genes was consistent. In addition, one of the most efficient cellulose-producing fungi, T. reesei, harbors 31 cellulase genes, including 13 endoglucanases, three exoglucanases, and 15 β-glucosidases [34]. These cellulases demonstrate high efficiency in breaking down cellulose by acting synergistically in T. reesei [34]. In our study, we found 20 of 26 cellulases possessed the activity of β-glucosidase, which indicated that A. oryzae might have the ability to express β-glucosidase at sufficiently high concentrations and could be specially used to produce β-glucosidase. In addition, A. oryzae also had two exoglucanases and four endoglucanases, so the cellulases in A. oryzae might also have been the same as T. reesei, which can effectively degrade cellulose through synergistic action. Therefore, the 26 cellulase genes identified in this study provide genetic information sources for studying the synergistic effect of A. oryzae cellulases on efficient degradation of cellulose.
Most fungal cellulases typically possess a dual-domain structure consisting of one catalytic domain and one or more carbohydrate-binding domains (CBD). However, there are also some cellulases that lack the CBD, such as the cellulases of β-glucosidase. Many carbohydrate-binding modules (CBMs) are classified as CBDs because of their strong affinity for cellulose. The first CBM_1 domain that was found appended to the exoglucanase CBH1 and CBH2 from T. reesei was used for the first discovered fungal CBDs [35,36]. It has been suggested that CBM does not have catalytic activity, but the binding characteristic of CBM can improve the catalytic function of cellulase through targeting the enzyme to the substrate and increasing the affinity of substrate–enzyme proximity [37,38,39]. Studies have shown that removing the CBM from the enzyme results in reduced enzymatic activity [40] and reduced stability [41,42]. In our study, one endoglucanase in GH5 contained a CBD domain, which might play an important role in improving the catalytic function of A. oryzae cellulase. In addition, we found 19 of 20 A. oryzae β-glucosidases contained an extra Fn3-like domain. The function of the Fn3-like domain may be similar to that of CBM, which can not only enhance the activity of cellulases but also increase the substrate proximity to the catalytic domain [43],44]. Another similarity between Fn3 and CBM is that they have a binding site for calcium ions, which plays an important role in the substrate interactions [43,44]. Therefore, the Fn3-like domain in A. oryzae β-glucosidase might play a biological role as a functional domain in enhancing the catalytic center and even represent a novel CBM-like domain. However, the domain analysis was to better understand the structure–function relationship and the cellulose degradation accomplished by the A. oryzae cellulases.
Cellulase production is based on the specific growth rate as well as the synthetic ability of the microorganisms [45]. Various fermentation conditions have important effects on cellulase production, including fermentation method, temperature, salt concentration, incubation period, carbon or nitrogen source, pH, and fungal species [46,47,48]. The incubation period, temperature, and salt concentration are of crucial significance for cellulase production, as they directly affect the growth of microorganisms and the bioactive production. Sulyman et al. suggested that cellulase production increased with the prolongation of culture time, reached the maximum production at 120 h, and then decreased at 144 h [49]. In our study, we found the expression pattern of cellulase genes in A. oryzae was time-specific expression, and more than 70% of the cellulase genes had significantly higher expression levels at 72 h than at 24 h. Unfortunately, the transcriptome data of incubation periods were only 24 h, 48 h, and 72 h. Therefore, based on the previous studies [49], we speculated that there might be more A. oryzae cellulase genes upregulated at 96 h and 120 h. In addition, we found three cellulase gene expressions were time-specific, of which two genes encoding β-glucosidase were significantly induced at 72 h. There is a possibility that the expression patterns of cellulase genes might provide a theoretical basis for us to produce specific cellulase by regulating the culture time of A. oryzae.
Temperature also plays a central role in the metabolic activities of microorganisms and affects their growth [50]. Sulyman et al. revealed that the optimal temperature for endoglucanase and β-glucosidase production is 40 °C [49]. In addition, Ali et al. reported that the maximum yield of cellulases from A. niger and Aspergillus terreus is 40 °C [51]. Unlike research reports, we analyzed the expression patterns of cellulase genes in A. oryzae under low-temperature (22 °C) and high-temperature (42 °C) conditions. We found that the expression levels of most cellulase genes in A. oryzae were significantly inhibited under low- and high-temperature stresses. However, we also find four cellulase genes at low temperature, and three genes at high temperature were significantly upregulated. In addition, the salt concentration in the medium also affects the synthesis of cellulases, and the addition of salt has a significant negative effect on the cellulase production in A. niger [52]. Consistent with this result, most of A. oryzae cellulase genes were inhibited under high salt concentration in our study. For example, 9 of 10 and 15 of 17 differentially expressed cellulase genes were downregulated at 10% and 5% salt concentration compared to the control. Furthermore, we found three cellulase genes were extremely sensitive to both temperature and salt stress. In any case, these results have important guiding significance for understanding the transcription patterns of A. oryzae cellulase genes and designing the culture conditions to produce specific cellulases.
Taken together, these findings indicate that cellulase production in A. oryzae is strongly influenced by environmental conditions. In addition, studies in filamentous fungi, such as A. nidulans [53], A. niger [54], and T. reesei [54], have demonstrated that cellulase genes are tightly controlled by both global carbon and nitrogen metabolism regulators (e.g., CreA, AreA) and substrate-specific transcription factors (e.g., XlnR, Xyr1) [55]. Some genes are co-regulated by these systems, while others are induced independently depending on the substrate and environmental conditions. These insights suggest that the 26 cellulase genes identified in A. oryzae are regulated by a combination of coordinated and gene-specific mechanisms. Nonetheless, further research is required to fully elucidate the exact regulatory network governing cellulase gene expression in this species.
5. Conclusions
Our study provides a comprehensive characterization of cellulase genes in A. oryzae. In this study, a total of 219 GH genes were identified from the A. oryzae 3.042 genome and classified into 40 GH families. And then, 26 cellulase genes belonging to the GH1, GH3, GH5, GH7, and GH10 families were identified as possessing activities of β-glucosidase, exoglucanase, and endoglucanase. We systematically analyzed gene structure, chromosome localization, conserved domain, and subcellular localization of these 26 cellulases. Additionally, phylogenetic analysis indicated that these 26 A. oryzae cellulases were roughly divided into two subgroups with the homologous cellulases of A. nidulans and T. reesei. Finally, our investigation revealed A. oryzae cellulase genes were time-specific in expression, and we identified certain genes that were inhibited under temperature and salt stress. Our results might be useful for elucidating evolutionary relationships, expression patterns, and functional divergence of cellulase genes in A. oryzae and enriching the Aspergillus cellulase gene family members. Furthermore, this study provides specific cellulase gene information for our further research on transforming A. oryzae to increase cellulase types and improve enzyme activity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres16100214/s1, Table S1: Gene information of the glycoside hydrolase family identified from Aspergillus oryzae 3.042 genome; Table S2: Information and annotation of 219 glycoside hydrolase genes in Aspergillus oryzae; Table S3: GO functional enrichment analysis of 219 glycoside hydrolase genes in Aspergillus oryzae; Table S4: KEGG enrichment analysis of 219 glycoside hydrolase genes in Aspergillus oryzae; Table S5: Motif and subcellular localization of 26 cellulase genes in Aspergillus oryzae; Table S6: Expression levels of cellulase genes affected by growth time or sensitivity to temperature and high salt stress; Table S7: Specific differentially expressed cellulase genes in response to temperature and high salt stress; Table S8: Information of eight genes containing the AA9 Pfam domain.
Author Contributions
D.H., R.Z. and Y.L., Formal Analysis; D.H. and Y.L., Writing—Original Draft Preparation; D.H. and R.Z., Investigation; C.J., Software and Methodology; C.J., Review and Editing; C.J., Supervision; C.J., Funding Acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31900063), the Jiangxi Provincial Department of Education Science and Technology Project (GJJ211103), and the Doctoral Scientific Research Foundation of Jiangxi Science and Technology Normal University (2018BSQD027).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The publicly available RNA sequencing raw data were retrieved at SRA of NCBI with accession PRJNA407002, PRJNA383095, and PRJNA774152 (https://www.ncbi.nlm.nih.gov/sra, accessed on 3 September 2024). All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zverlov, V.V.; Schwarz, W.H. Bacterial cellulose hydrolysis in anaerobic environmental subsystems—Clostridium thermocellum and Clostridium stercorarium, thermophilic plant-fiber degraders. Ann. N. Y. Acad. Sci. 2008, 1125, 298–307. [Google Scholar] [CrossRef]
- Ahmed, A.; Nasim, F.H.; Batool, K.; Bibi, A. Microbial β-glucosidase: Sources, production and applications. J. Appl. Environ. Microbiol. 2017, 5, 31–46. [Google Scholar] [CrossRef]
- Trivedi, N.; Gupta, V.; Kumar, M.; Kumari, P.; Reddy, C.R.K.; Jha, B. An alkali halotolerant cellulase from Bacillus flexus isolated from green seaweed Ulva lactuca. Carbohydr. Polym. 2011, 83, 891–897. [Google Scholar] [CrossRef]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef]
- Imran, M.; Anwar, Z.; Irshad, M.; Asad, M.J.; Ashfaq, H. Cellulase production from species of fungi and bacteria from agricultural wastes and its utilization in industry: A review. Adv. Enzym. Res. 2016, 4, 44–55. [Google Scholar] [CrossRef]
- Ahmed, A.; Bibi, A. Fungal cellulase; Production and applications: Minireview. Int. J. Health Life Sci. 2018, 4, 19–36. [Google Scholar] [CrossRef]
- Behera, B.C.; Sethi, B.K.; Mishra, R.R.; Dutta, S.K.; Thatoi, H.N. Microbial cellulases—Diversity and biotechnology with reference to mangrove environment: A review. J. Genet. Eng. Biotechnol. 2017, 15, 197–210. [Google Scholar] [CrossRef]
- Naher, L.; Fatin, S.N.; Sheikh, M.A.H.; Azeez, L.A.; Siddiquee, S.; Zain, N.M.; Karim, S.M.R. Cellulase enzyme production from filamentous fungi Trichoderma reesei and Aspergillus awamori in submerged fermentation with rice straw. J. Fungi 2021, 7, 868. [Google Scholar] [CrossRef]
- Ellilä, S.; Fonseca, L.; Uchima, C.; Cota, J.; Goldman, G.H.; Saloheimo, M.; Sacon, V.; Siika-aho, M. Development of a low-cost cellulase production process using Trichoderma reesei for Brazilian biorefineries. Biotechnol. Biofuels 2017, 10, 30. [Google Scholar] [CrossRef]
- Pachauri, P.; Aranganathan, V.; More, S.; Sullia, S.B.; Deshmukh, S. Purification and characterization of cellulase from a novel isolate of Trichoderma longibrachiatum. Biofuels 2017, 11, 85–91. [Google Scholar] [CrossRef]
- Kitamoto, K. Cell biology of the Koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2015, 79, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S. Introduction to glycoside hydrolases: Classification, identification, and occurrence. In Industrial Applications of Glycoside Hydrolases; Shrivastava, S., Ed.; Springer: Singapore, 2020; pp. 1–20. [Google Scholar]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Kolde, R. pheatmap: Pretty Heatmaps. R package Version 1.0.8. 2015. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 4 September 2024).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- You, Y.; Xu, X.; Liu, H.; Zhang, L. Locust pathogen Aspergillus oryzae XJ1 is different from Aspergillus oryzae and Aspergillus flavus based on genomics comparisons. Microorganisms 2024, 12, 2501. [Google Scholar] [CrossRef]
- Benedetti, M.; Vecchi, V.; Betterle, N.; Natali, A.; Bassi, R.; Dall’Osto, L. Design of a highly thermostable hemicellulose-degrading blend from Thermotoga neapolitana for the treatment of lignocellulosic biomass. J. Biotechnol. 2019, 296, 42–52. [Google Scholar] [CrossRef]
- Ho, M.C.; Ong, V.Z.; Wu, T.Y. Potential use of alkaline hydrogen peroxide in lignocellulosic biomass pretreatment and valorization—A review. Renew. Sustain. Energy Rev. 2019, 112, 75–86. [Google Scholar] [CrossRef]
- Jayasekara, S.; Ratnayake, R. Microbial cellulases: An overview and applications (Chapter 5). In Cellulose; IntechOpen: London, UK, 2019. [Google Scholar]
- Ariaeenejad, S.; Kavousi, K.; Mamaghani, A.S.A.; Motahar, S.F.S.; Nedaei, H.; Salekdeh, G.H. In-silico discovery of bifunctional enzymes with enhanced lignocellulose hydrolysis from microbiota big data. Int. J. Biol. Macromol. 2021, 177, 211–220. [Google Scholar] [CrossRef]
- Sethi, S.; Datta, A.; Gupta, B.L.; Gupta, S. Optimization of cellulase production from bacteria isolated from soil. Int. Sch. Res. Not. 2013, 2013, 1076–1082. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic engineering of filamentous fungi for efficient protein expression and secretion. Front. Bioeng. Biotechnol. 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Srivastava, M.; Ramteke, P.W.; Mishra, P.K. Synthetic biology strategy for microbial cellulases: An overview. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 229–238. [Google Scholar] [CrossRef]
- Prasanna, H.N.; Ramanjaneyulu, G.; Rajasekhar Reddy, B. Optimization of cellulase production by Penicillium sp. 3 Biotech 2016, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.L.; Tõlgo, M.; Olsson, L.; Nypelö, T. Carboxylation of sulfated cellulose nanocrystals by family AA9 lytic polysaccharide monooxygenases. Cellulose 2023, 30, 9331–9347. [Google Scholar] [CrossRef]
- Bauer, S.; Vasu, P.; Persson, S.; Mort, A.J.; Somerville, C.R. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 2006, 103, 11417–11422. [Google Scholar] [CrossRef]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Navarro Poulsen, J.C.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef]
- Van den Brink, J.; de Vries, R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477–1492. [Google Scholar] [CrossRef]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef]
- Van Tilbeurgh, H.; Tomme, P.; Claeyssens, M.; Bhikhabhai, R.; Pettersson, G. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei: Separation of functional domains. FEBS Lett. 1986, 204, 223–227. [Google Scholar] [CrossRef]
- Tomme, P.; Van Tilbeurgh, H.; Pettersson, G.; Van Damme, J.; Vandekerckhove, J.; Knowles, J.; Teeri, T.; Claeyssens, M. Studies of the cellulolytic system of Trichoderma reesei QM 9414: Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 1988, 170, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ortiz, V.; Heins, R.A.; Cheng, G.; Kim, E.Y.; Vernon, B.C.; Elandt, R.B.; Adams, P.D.; Sale, K.L.; Hadi, M.Z.; Simmons, B.A.; et al. Addition of a carbohydrate-binding module enhances cellulase penetration into cellulose substrates. Biotechnol. Biofuels 2013, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, A.; Pellegrini, V.O.A.; Curtolo, F.; Camilo, C.M.; Mello, B.L.; Johns, M.A.; Scott, J.L.; Guimarães, F.E.C.; Polikarpov, I. Carbohydrate binding modules enhance cellulose enzymatic hydrolysis by increasing access of cellulases to the substrate. Carbohydr. Polym. 2019, 211, 57–68. [Google Scholar] [CrossRef]
- Arai, T.; Araki, R.; Tanaka, A.; Karita, S.; Kimura, T.; Sakka, K.; Ohmiya, K. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: Importance of the CBM to cellulose hydrolysis. J. Bacteriol. 2003, 185, 504–512. [Google Scholar] [CrossRef]
- Courtade, G.; Forsberg, Z.; Heggset, E.B.; Eijsink, V.G.H.; Aachmann, F.L. The carbohydrate-binding module and linker of a modular lytic polysaccharide monooxygenase promote localized cellulose oxidation. J. Biol. Chem. 2018, 293, 13006–13015. [Google Scholar] [CrossRef]
- Teo, S.C.; Liew, K.J.; Shamsir, M.S.; Chong, C.S.; Bruce, N.C.; Chan, K.G.; Goh, K.M. Characterizing a halo-tolerant GH10 xylanase from Roseithermus sacchariphilus strain RA and its CBM-truncated variant. Int. J. Mol. Sci. 2019, 20, 2284. [Google Scholar] [CrossRef]
- Montanier, C.; Flint, J.E.; Bolam, D.N.; Xie, H.; Liu, Z.; Rogowski, A.; Weiner, D.P.; Ratnaparkhe, S.; Nurizzo, D.; Roberts, S.M.; et al. Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. J. Biol. Chem. 2010, 285, 31742–31754. [Google Scholar] [CrossRef]
- Yaniv, O.; Petkun, S.; Shimon, L.J.; Bayer, E.A.; Lamed, R.; Frolow, F. A single mutation reforms the binding activity of an adhesion-deficient family 3 carbohydrate-binding module. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 819–828. [Google Scholar] [CrossRef]
- Kunamneni, A.; Permaul, K.; Singh, S. Amylase production in solid state fermentation by the thermophilic fungus Thermomyces lanuginosus. J. Biosci. Bioeng. 2005, 100, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Norouzian, D. Effect of different factors on fermentative production of enzymes by fungi. Dyn. Biochem. Process Biotechnol. Mol. Biol. 2008, 2, 14–18. [Google Scholar]
- Okoye, I.G.; Ezugwu, A.L.; Udenwobele, D.I.; Eze, S.O.O.; Anyawu, C.U.; Chilaka, F.C. Production and partial characterization of cellulases from Aspergillus fumigatus using two distinct parts of corn cob as carbon sources. Niger. J. Biotechnol. 2013, 26, 50–59. [Google Scholar]
- Saini, A.; Aggarwal, N.K.; Yadav, A. Cost-effective cellulase production using Parthenium hysterophorus biomass as an unconventional lignocellulosic substrate. 3 Biotech 2017, 7, 12. [Google Scholar] [CrossRef]
- Sulyman, A.O.; Igunnu, A.; Malomo, S.O. Isolation, purification, and characterization of cellulase produced by Aspergillus niger cultured on Arachis hypogaea shells. Heliyon 2020, 6, e05668. [Google Scholar] [CrossRef]
- Bhanja, T.; Rout, S.; Banerjee, R.; Bhattacharya, B.C. Comparative profiles of α-amylase production in conventional tray reactor and GROWTEK bioreactor. Bioprocess Biosyst. Eng. 2007, 30, 369–376. [Google Scholar] [CrossRef]
- Ali, S.; Sayed, A.; Sarker, R.I.; Alam, R. Factors affecting cellulase production by Aspergillus terreus using water hyacinth. World J. Microbiol. Biotechnol. 1991, 7, 62–66. [Google Scholar]
- Iram, A.; Cekmecelioglu, D.; Demirci, A. Salt and nitrogen amendment and optimization for cellulase and xylanase production using dilute acid hydrolysate of distillers’ dried grains with solubles (DDGS) as the feedstock. Bioprocess Biosyst. Eng. 2022, 45, 527–540. [Google Scholar] [CrossRef]
- Lockington, R.A.; Rodbourn, L.; Barnett, S.; Carter, C.J.; Kelly, J.M. Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet. Biol. 2002, 37, 190–196. [Google Scholar] [CrossRef]
- Stricker, A.R.; Mach, R.L.; de Graaff, L.H. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 2008, 78, 211–220. [Google Scholar] [CrossRef]
- Amore, A.; Giacobbe, S.; Faraco, V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr. Genom. 2013, 14, 230–249. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
