Phylogenetic Incongruence of Cyclic di-GMP-Activated Glycosyltransferase nfrB with 16S rRNA Gene Tree Reflects In Silico-Predicted Protein Structural Divergence in Diaphorobacter nitroreducens Isolated from Estero de Paco, Manila, Philippines
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Putative D. nitroreducens Isolation
2.3. DNA Extraction
2.4. Molecular Identification and Sequencing of Putative D. nitroreducens
2.5. Phylogenetic and In Silico Protein Structure Analyses
3. Results
3.1. EMJH Media Supports D. nitroreducens Growth
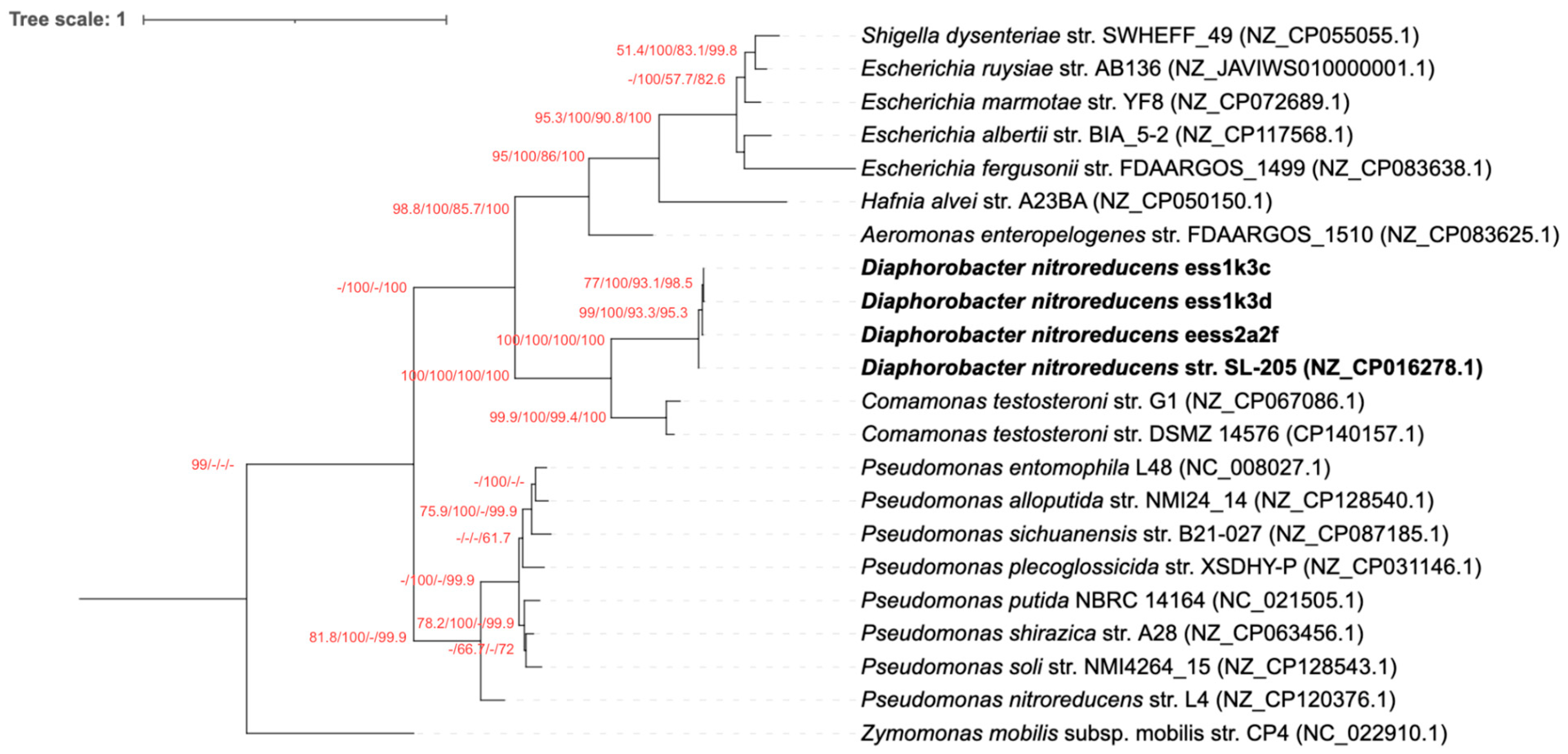
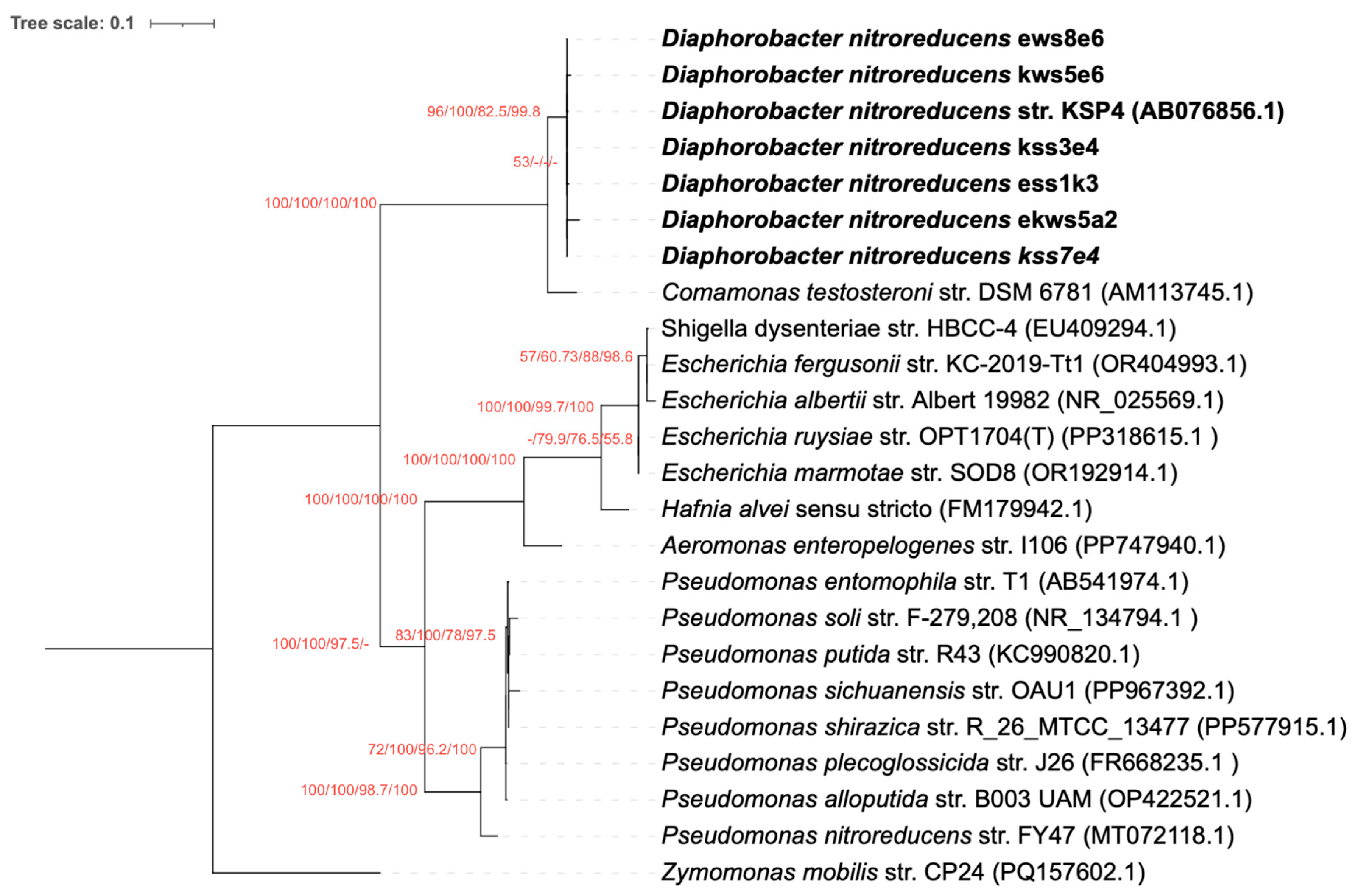
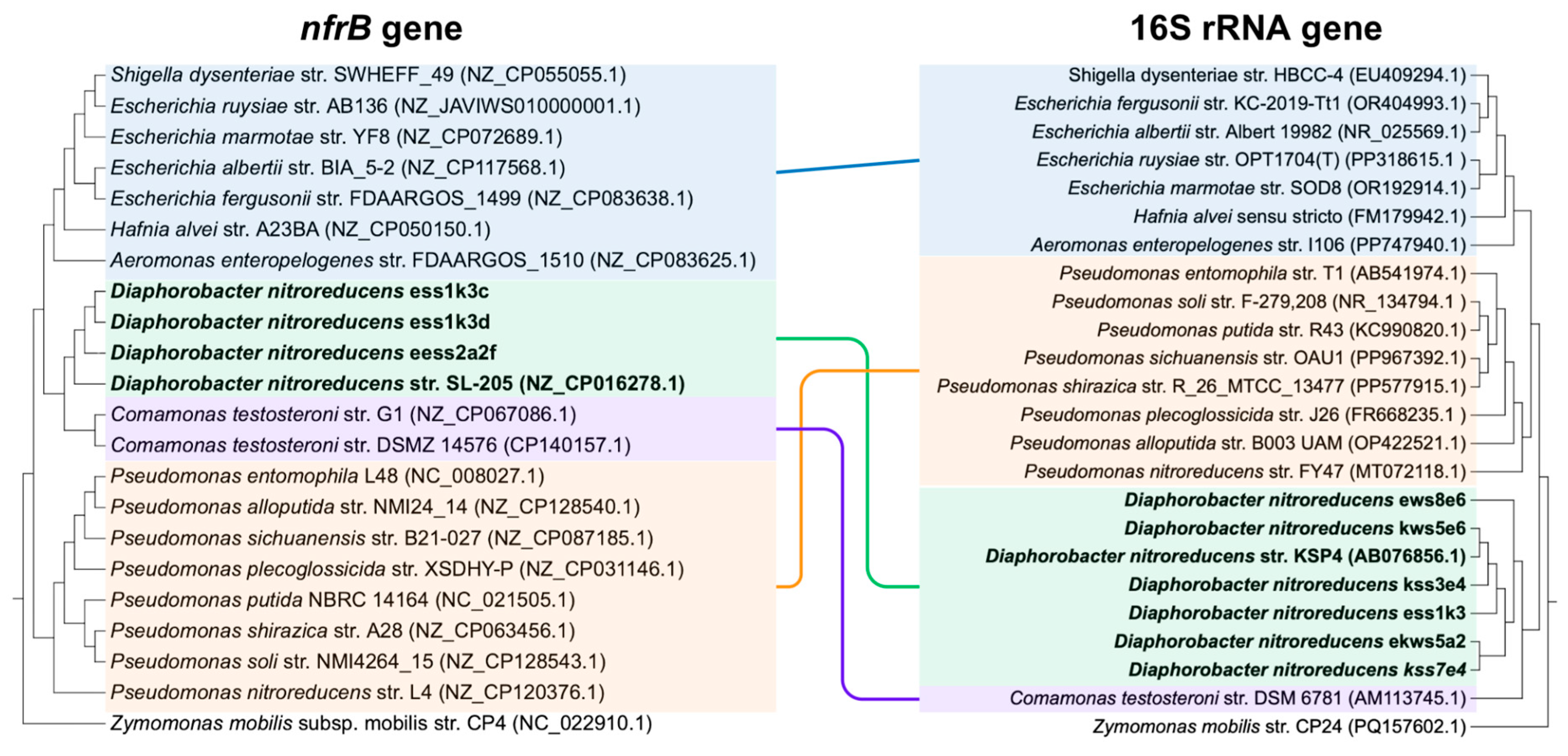
3.2. The nfrB Gene Trees Are Incongruent with the Canonical 16S rRNA Gene Phylogeny
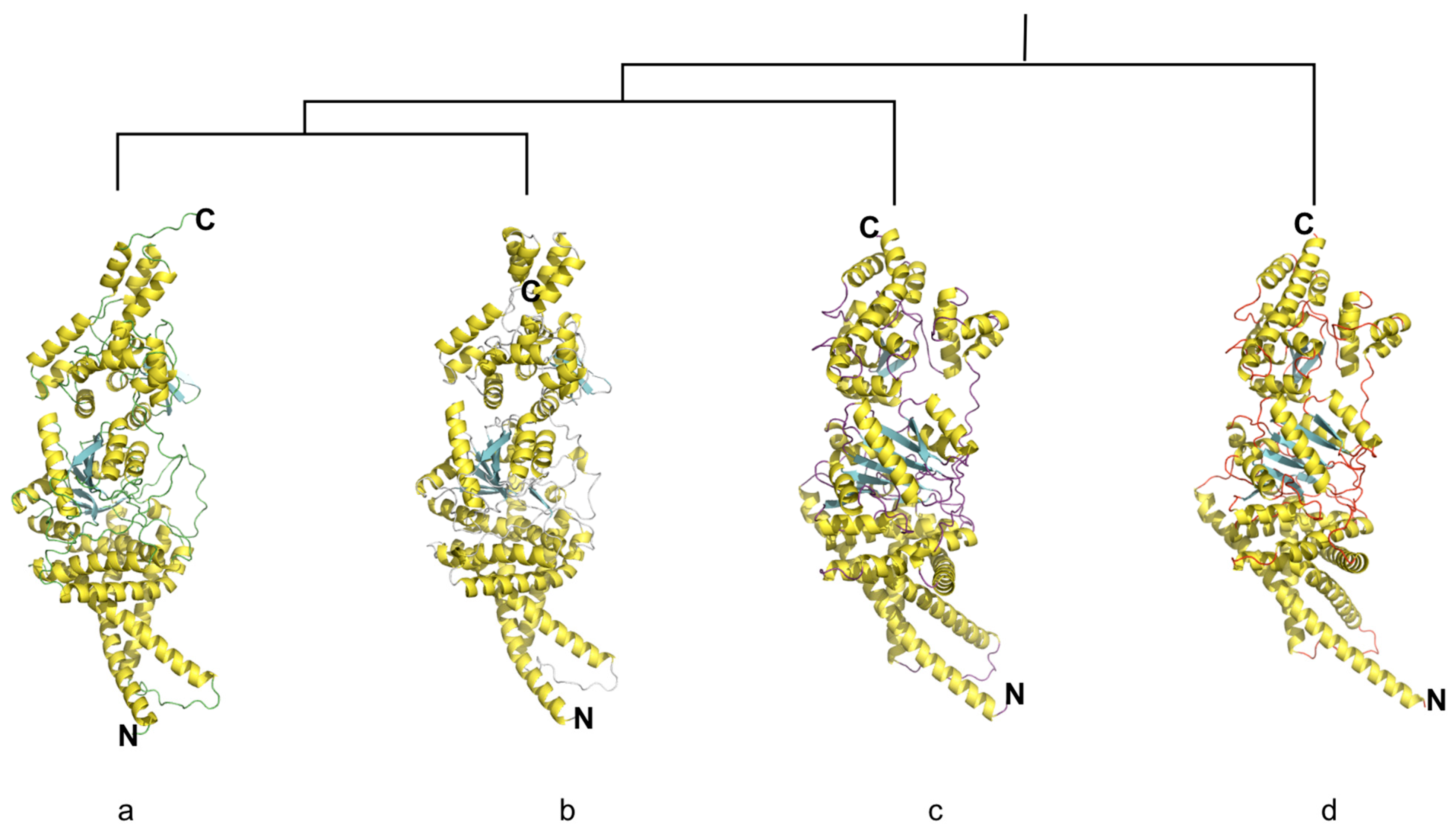
3.3. Overall Structure of Cyclic di-3′,5′-Guanylate-Activated Glycosyltransferase NfrB of D. nitroreducens
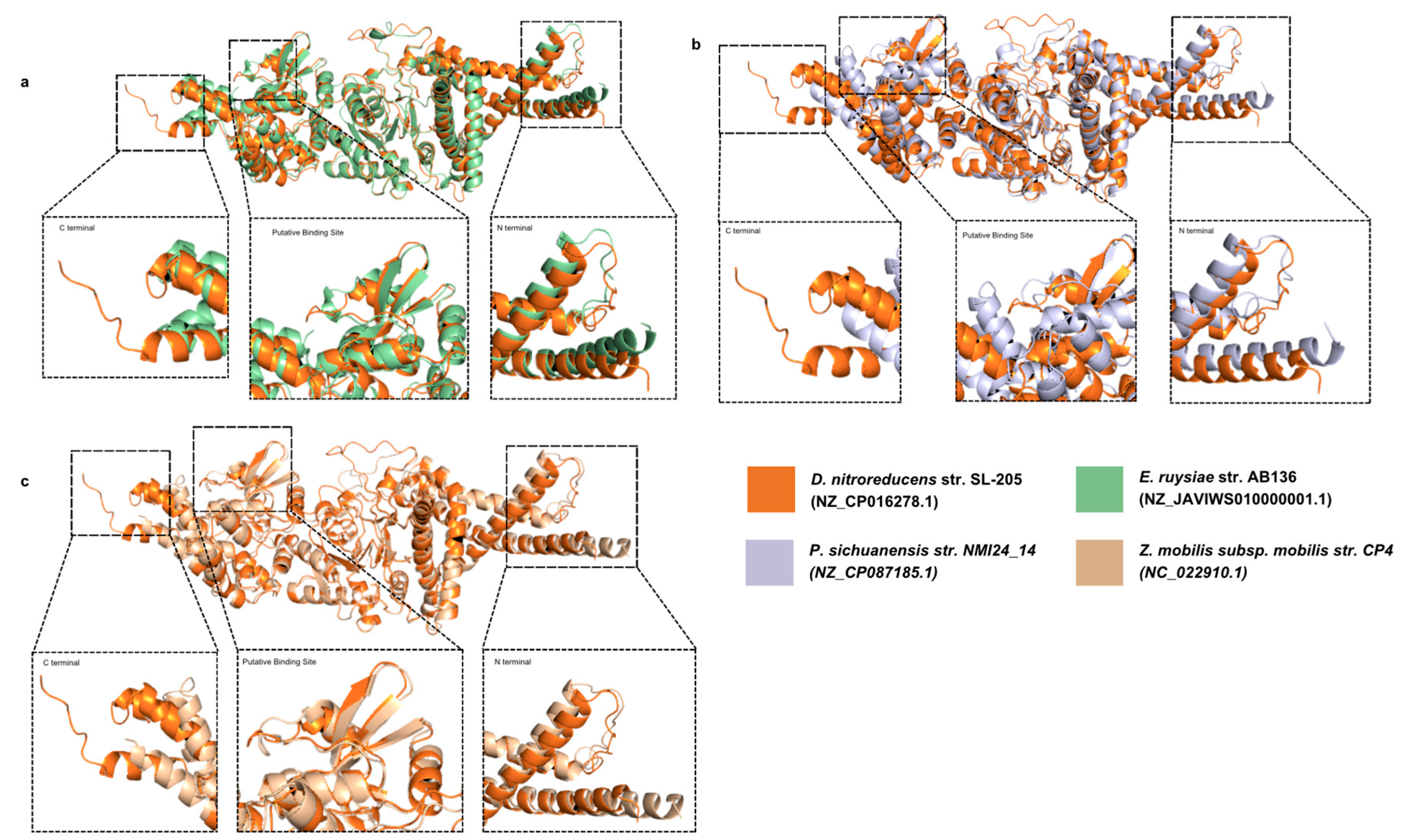
3.4. The Structure of Putative Binding Site Domains of NfrB Is Conserved Among Phyla
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, S.T.; Hiraishi, A. Diaphorobacter nitroreducens gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading denitrifying bacterium isolated from activated sludge. J. Gen. Appl. Microbiol. 2002, 48, 299–308. [Google Scholar] [CrossRef]
- Khan, S.T.; Nagao, Y.; Hiraishi, A. Nitrate removal performance of Diaphorobacter nitroreducens using biodegradable plastics as the source of reducing power. AIP Conf. Proc. 2015, 1649, 71–78. [Google Scholar] [CrossRef]
- Ni, Y.; Li, R.; Shen, X.; Yi, D.; Ren, Y.; Wang, F.; Geng, Y.; You, Q. Diaphorobacter nitroreducens synergize with oxaliplatin to reduce tumor burden in mice with lung adenocarcinoma. mSystems 2024, 9, e0132323. [Google Scholar] [CrossRef]
- Khardenavis, A.A.; Kapley, A.; Purohit, H.J. Simultaneous nitrification and denitrification by diverse Diaphorobacter sp. Appl. Microbiol. Biotechnol. 2007, 77, 403–409. [Google Scholar] [CrossRef]
- Singh, D.; Ramanathan, G. Catechol 2,3-dioxygenase isoenzymes from Diaphorabacter sp. strain DS2 demonstrate different substrate preferences. Biochem. Biophys. Res. Commun. 2025, 782, 152591. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hong, M.; Lv, J.; Tang, R.; Wang, R.; Yang, Y.; Liu, N. Enhancement on migration and biodegradation of Diaphorobacter sp. LW2 mediated by Pythium ultimum in soil with different particle sizes. Front. Microbiol. 2024, 15, 1391553. [Google Scholar] [CrossRef]
- Junkermeier, E.H.; Hengge, R. A Novel locally c-di-GMP-controlled exopolysaccharide synthase required for bacteriophage N4 infection of Escherichia coli. mBio 2021, 12, e03249-21. [Google Scholar] [CrossRef]
- Huang, L.; Jin, Y.; Zhou, D.; Liu, L.; Huang, S.; Zhao, Y.; Chen, Y. A review of the role of extracellular polymeric substances (EPS) in wastewater treatment systems. Int. J. Environ. Res. Public Health 2022, 19, 12191. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Huang, Y.; Wang, W.; Zhang, L.; Xu, J.; Li, Z.; Xu, P.; Tang, H. A novel Diaphorobacter sp. strain isolated from saponification wastewater shows highly efficient phenanthrene degradation. Environ. Res. 2022, 214, 114047. [Google Scholar] [CrossRef]
- Sellner, B.; Prakapaitė, R.; Van Berkum, M.; Heinemann, M.; Harms, A.; Jenal, U. A new sugar for an old phage: A c-di-GMP-dependent polysaccharide pathway sensitizes Escherichia coli for bacteriophage infection. mBio 2021, 12, e03246-21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Wu, R.; Zhang, H.; Wu, H. Structural and biochemical analysis of a bacterial glycosyltransferase. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; pp. 29–39. [Google Scholar] [CrossRef]
- Yakovlieva, L.; Walvoort, M.T.C. Processivity in bacterial glycosyltransferases. ACS Chem. Biol. 2019, 15, 3–16. [Google Scholar] [CrossRef]
- Schmid, J.; Heider, D.; Wendel, N.J.; Sperl, N.; Sieber, V. Bacterial glycosyltransferases: Challenges and opportunities of a highly diverse enzyme class toward tailoring natural products. Front. Microbiol. 2016, 7, 182. [Google Scholar] [CrossRef]
- Rezvani, F.; Sarrafzadeh, M.-H.; Ebrahimi, S.; Oh, H.-M. Nitrate removal from drinking water with a focus on biological methods: A review. Environ. Sci. Pollut. Res. 2017, 26, 1124–1141. [Google Scholar] [CrossRef]
- Veloso, I.; Lopes, M.; Salas, C.; Moreira, E. A comparison of three DNA extractive procedures with Leptospira for polymerase chain reaction analysis. Mem. Inst. Oswaldo Cruz 2000, 95, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Morey, R.E.; Galloway, R.L.; Bragg, S.L.; Steigerwalt, A.G.; Mayer, L.W.; Levett, P.N. Species-specific identification of Leptospiraceae by 16S RRNA gene sequencing. J. Clin. Microbiol. 2006, 44, 3510–3516. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A User-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Cummings, M.P. PAUP * (Phylogenetic analysis using parsimony (and other methods)). In Dictionary of Bioinformatics and Computational Biology; John Wiley & Sons: Hoboken, NJ, USA, 2004. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PHYML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: https://ci.nii.ac.jp/naid/10020095229/ (accessed on 14 December 2024).
- Wang, J.; Chu, L. Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 2016, 34, 1103–1112. [Google Scholar] [CrossRef]
- Guedes, I.B.; De Souza, G.O.; De Paula Castro, J.F.; Cavalini, M.B.; De Souza Filho, A.F.; Aizawa, J.; Cortez, A.; Heinemann, M.B. Improvement of the enrichment used in the EMJH medium (Ellinghausen–McCullough–Johnson–Harris) for the cultivation of Leptospira spp. Rev. Argent. Microbiol. 2021, 54, 95–99. [Google Scholar] [CrossRef]
- Heylen, K.; Vanparys, B.; Wittebolle, L.; Verstraete, W.; Boon, N.; De Vos, P. Cultivation of denitrifying bacteria: Optimization of isolation conditions and diversity study. Appl. Environ. Microbiol. 2006, 72, 2637–2643. [Google Scholar] [CrossRef]
- Loureiro, A.P.; Martins, G.; Pinto, P.; Narduche, L.; Teixeira, R.C.; Lilenbaum, W. Usage of a selective media (EMJH-STAFF) in primary culturing of pathogenic leptospires from bovine clinical samples. Lett. Appl. Microbiol. 2015, 61, 603–606. [Google Scholar] [CrossRef]
- Erşan, Y.Ç.; De Belie, N.; Boon, N. Microbially induced CaCO3 precipitation through denitrification: An optimization study in minimal nutrient environment. Biochem. Eng. J. 2015, 101, 108–118. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; Van Rensburg, M.J.J.; Bray, J.E.; Earle, S.G.; Ford, S.A.; Jolley, K.A.; McCarthy, N.D. MLST revisited: The gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 2013, 11, 728–736. [Google Scholar] [CrossRef]
- Teichman, S.; Lee, M.D.; Willis, A.D. Analyzing microbial evolution through gene and genome phylogenies. Biostatistics 2023, 25, 786–800. [Google Scholar] [CrossRef]
- Shimma, Y.-I.; Saito, F.; Oosawa, F.; Jigami, Y. Construction of a library of human glycosyltransferases immobilized in the cell wall of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006, 72, 7003–7012. [Google Scholar] [CrossRef]
- Mollicone, R.; Gibaud, A.; François, A.; Ratcliffe, M.; Oriol, R. Acceptor specificity and tissue distribution of three human α-3-fucosyltransferases. Eur. J. Biochem. 1990, 191, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Bencúrová, M.; Rendić, D.; Fabini, G.; Kopecky, E.-M.; Altmann, F.; Wilson, I.B.H. Expression of eukaryotic glycosyltransferases in the yeast Pichia Pastoris. Biochimie 2003, 85, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Shi, Y.; Wu, N.C.; Grande, G.; Douthit, L.; Wang, H.; Zhou, W.; Sharpless, K.B.; Wilson, I.A.; Xie, J.; et al. Bacterial glycosyltransferase-mediated cell-surface chemoenzymatic glycan modification. Nat. Commun. 2019, 10, 1799. [Google Scholar] [CrossRef] [PubMed]
- Anyaogu, D.C.; Mortensen, U.H. Manipulating the glycosylation pathway in bacterial and lower eukaryotes for production of therapeutic proteins. Curr. Opin. Biotechnol. 2015, 36, 122–128. [Google Scholar] [CrossRef] [PubMed]
| Tree | −ln L | Δ −ln L | SH | Wtd-SH |
|---|---|---|---|---|
| 2 | 6377.75631 | (best) | ||
| 1 | 6641.07196 | 263.31565 | 0.0000 * | 0.0000 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marababol, R.J.L.; Rivera, W.L. Phylogenetic Incongruence of Cyclic di-GMP-Activated Glycosyltransferase nfrB with 16S rRNA Gene Tree Reflects In Silico-Predicted Protein Structural Divergence in Diaphorobacter nitroreducens Isolated from Estero de Paco, Manila, Philippines. Microbiol. Res. 2025, 16, 212. https://doi.org/10.3390/microbiolres16100212
Marababol RJL, Rivera WL. Phylogenetic Incongruence of Cyclic di-GMP-Activated Glycosyltransferase nfrB with 16S rRNA Gene Tree Reflects In Silico-Predicted Protein Structural Divergence in Diaphorobacter nitroreducens Isolated from Estero de Paco, Manila, Philippines. Microbiology Research. 2025; 16(10):212. https://doi.org/10.3390/microbiolres16100212
Chicago/Turabian StyleMarababol, Ram Julius L., and Windell L. Rivera. 2025. "Phylogenetic Incongruence of Cyclic di-GMP-Activated Glycosyltransferase nfrB with 16S rRNA Gene Tree Reflects In Silico-Predicted Protein Structural Divergence in Diaphorobacter nitroreducens Isolated from Estero de Paco, Manila, Philippines" Microbiology Research 16, no. 10: 212. https://doi.org/10.3390/microbiolres16100212
APA StyleMarababol, R. J. L., & Rivera, W. L. (2025). Phylogenetic Incongruence of Cyclic di-GMP-Activated Glycosyltransferase nfrB with 16S rRNA Gene Tree Reflects In Silico-Predicted Protein Structural Divergence in Diaphorobacter nitroreducens Isolated from Estero de Paco, Manila, Philippines. Microbiology Research, 16(10), 212. https://doi.org/10.3390/microbiolres16100212







