Identification of Listeria Isolates by Using a Pragmatic Multilocus Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. DNA Extraction and Purification
2.3. MLPA Genes and Primers
2.4. PCR Amplification and Sequencing
2.5. Single Gene and Multilocus Sequence Analysis
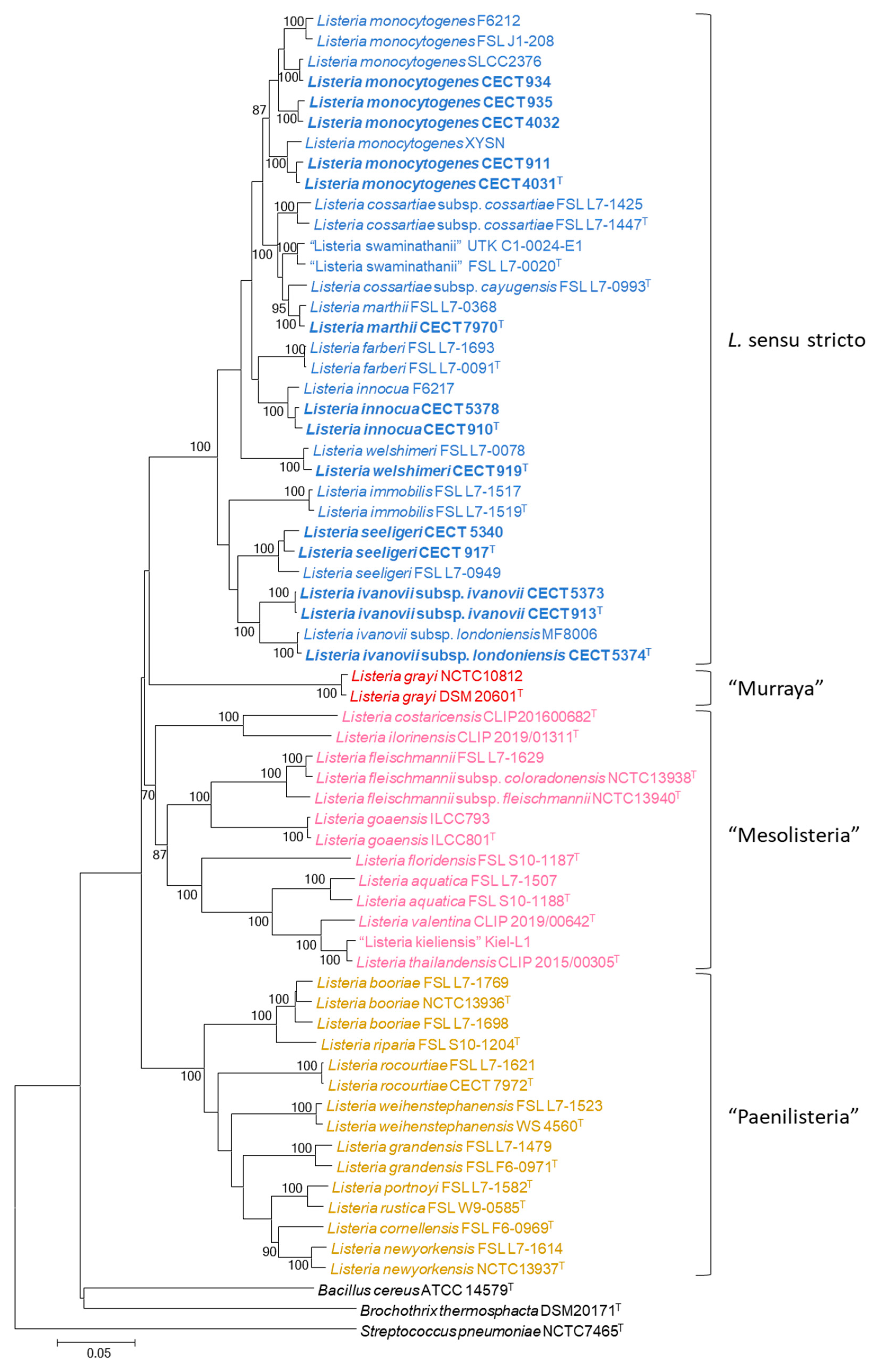
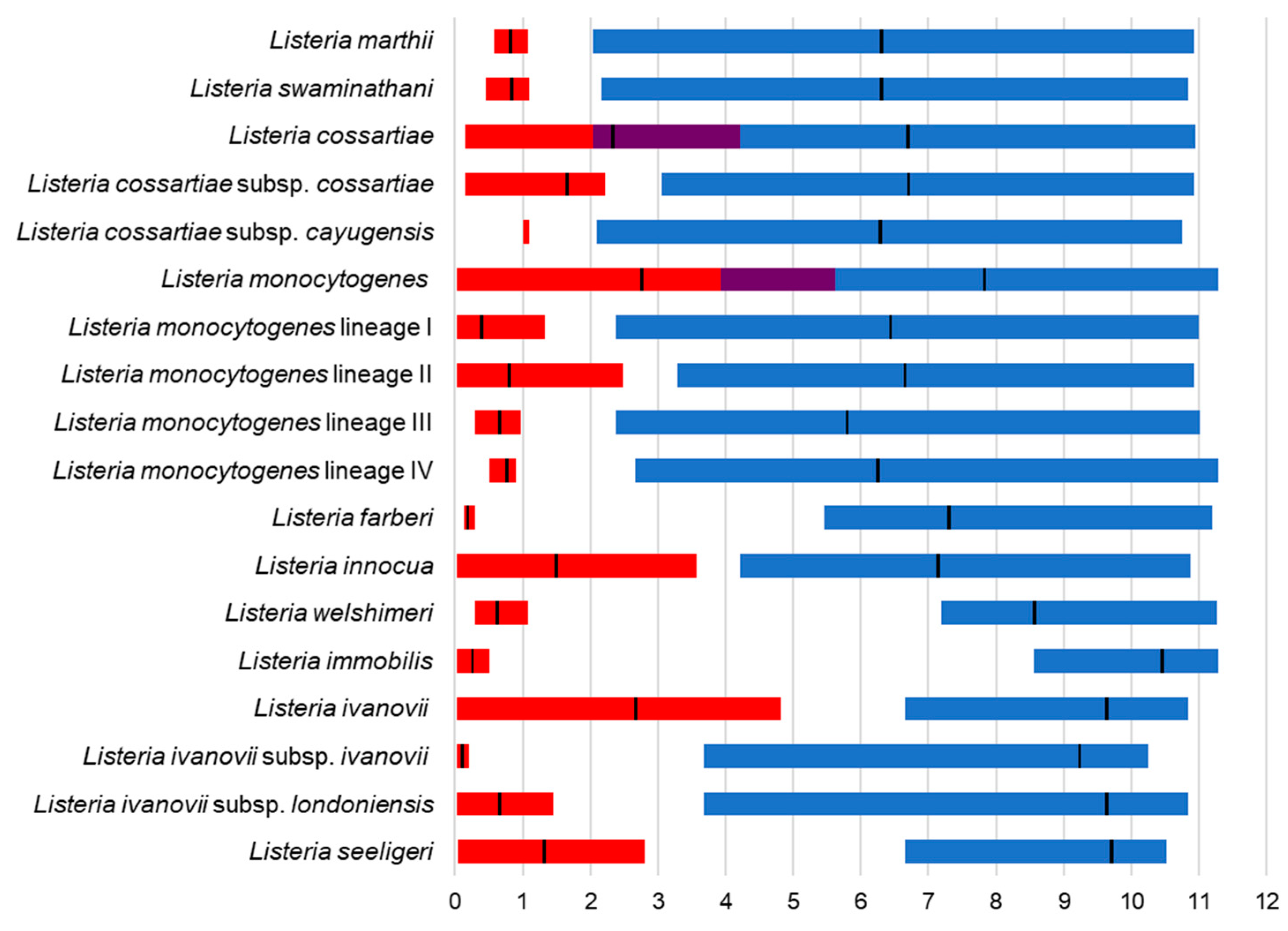
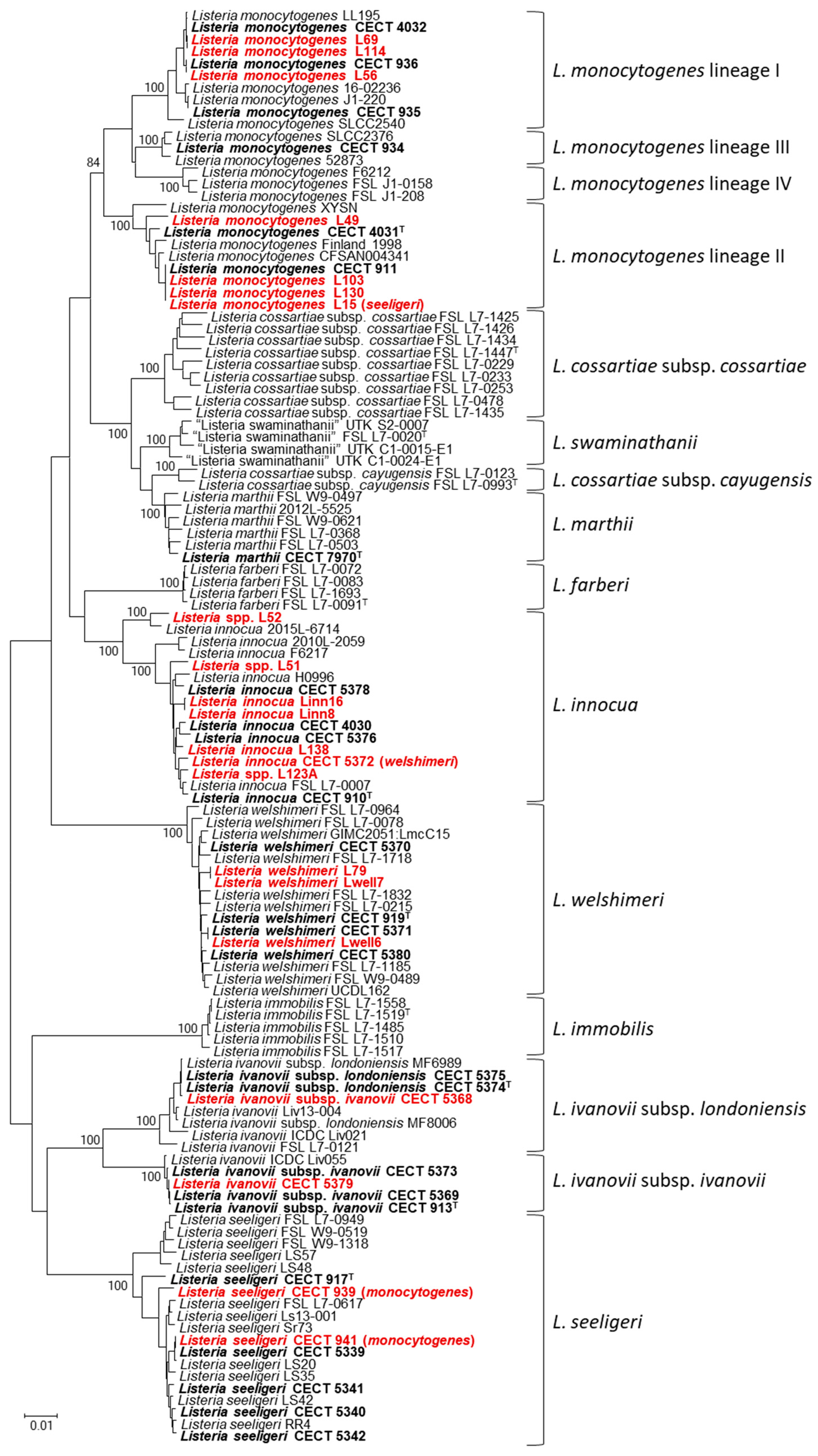
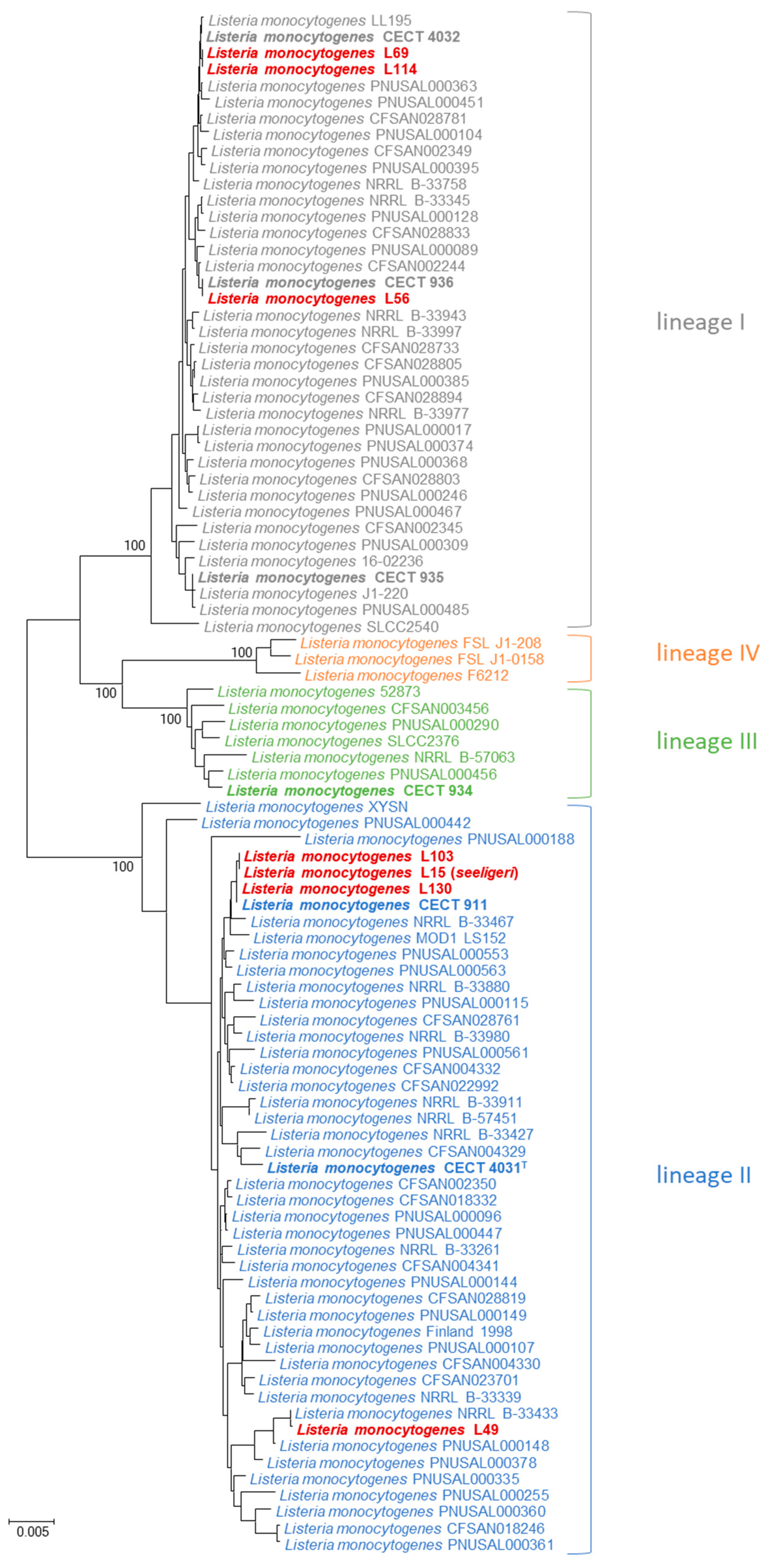
3. Results
4. Discussion
4.1. MLPA of the Genus Listeria
4.2. MLPA Clustering of Listeria Sensu Stricto
4.3. Identification of Listeria Isolates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria Monocytogenes—from Saprophyte to Intracellular Pathogen. Nat. Rev. Microbiol. 2009, 7, 623. [Google Scholar] [CrossRef] [PubMed]
- Linke, K.; Rückerl, I.; Brugger, K.; Karpiskova, R.; Walland, J.; Muri-Klinger, S.; Tichy, A.; Wagner, M.; Stessl, B. Reservoirs of Listeria Species in Three Environmental Ecosystems. Appl. Environ. Microbiol. 2014, 80, 5583–5592. [Google Scholar] [CrossRef] [PubMed]
- Hain, T.; Chatterjee, S.S.; Ghai, R.; Kuenne, C.T.; Billion, A.; Steinweg, C.; Domann, E.; Kärst, U.; Jänsch, L.; Wehland, J.; et al. Pathogenomics of Listeria spp. Int. J. Med. Microbiol. IJMM 2007, 297, 541–557. [Google Scholar] [CrossRef] [PubMed]
- McLauchlin, J.; Rees, C.E.D. Listeria. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Oxford, UK, 2015; pp. 1–29. ISBN 978-1-118-96060-8. [Google Scholar]
- Favaro, M.; Sarmati, L.; Sancesario, G.; Fontana, C. First Case of Listeria Innocua Meningitis in a Patient on Steroids and Eternecept. JMM Case Rep. 2014, 1, e003103. [Google Scholar] [CrossRef]
- Perrin, M.; Bemer, M.; Delamare, C. Fatal Case of Listeria Innocua Bacteremia. J. Clin. Microbiol. 2003, 41, 5308–5309. [Google Scholar] [CrossRef]
- Rapose, A.; Lick, S.D.; Ismail, N. Listeria Grayi Bacteremia in a Heart Transplant Recipient. Transpl. Infect. Dis. 2008, 10, 434–436. [Google Scholar] [CrossRef]
- Guillet, C.; Join-Lambert, O.; Monnier, A.L.; Leclercq, A.; Mechaï, F.; Mamzer-Bruneel, M.-F.; Bielecka, M.K.; Scortti, M.; Disson, O.; Berche, P.; et al. Human Listeriosis Caused by Listeria ivanovii. Emerg. Infect. Dis. 2010, 16, 136. [Google Scholar] [CrossRef]
- European Food Safety Authority. European Centre for Disease Prevention and Control (ECDC) The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Food and Agriculture Organization; World Health Organization. Listeria Monocytogenes in Ready-to-Eat (RTE) Foods: Attribution, Characterization and Monitoring: Meeting Report; FAO; WHO: Rome, Italy, 2022; ISBN 978-92-5-136983-8.
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria Monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria Monocytogenes Lineages: Genomics, Evolution, Ecology, and Phenotypic Characteristics. Int. J. Med. Microbiol. IJMM 2011, 301, 79–96. [Google Scholar] [CrossRef]
- den Bakker, H.C.; Didelot, X.; Fortes, E.D.; Nightingale, K.K.; Wiedmann, M. Lineage Specific Recombination Rates and Microevolution in Listeria Monocytogenes. BMC Evol. Biol. 2008, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Zadoks, R.N.; Fortes, E.D.; Dogan, B.; Cai, S.; Chen, Y.; Scott, V.N.; Gombas, D.E.; Boor, K.J.; Wiedmann, M. Listeria Monocytogenes Isolates from Foods and Humans Form Distinct but Overlapping Populations. Appl. Environ. Microbiol. 2004, 70, 5833–5841. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, K.K.; Windham, K.; Wiedmann, M. Evolution and Molecular Phylogeny of Listeria Monocytogenes Isolated from Human and Animal Listeriosis Cases and Foods. J. Bacteriol. 2005, 187, 5537–5551. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A New Perspective on Listeria Monocytogenes Evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. Worldwide Distribution of Major Clones of Listeria Monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef]
- Balandyté, L.; Brodard, I.; Frey, J.; Oevermann, A.; Abril, C. Ruminant Rhombencephalitis-Associated Listeria Monocytogenes Alleles Linked to a Multilocus Variable-Number Tandem-Repeat Analysis Complex. Appl. Environ. Microbiol. 2011, 77, 8325–8335. [Google Scholar] [CrossRef][Green Version]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria Monocytogenes Hypervirulence by Harnessing Its Biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria Monocytogenes Pan-Genome Reveals Dynamic Integration Hotspots and Mobile Genetic Elements as Major Components of the Accessory Genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef]
- Raufu, I.A.; Moura, A.; Vales, G.; Ahmed, O.A.; Aremu, A.; Thouvenot, P.; Tessaud-Rita, N.; Bracq-Dieye, H.; Krishnamurthy, R.; Leclercq, A.; et al. Listeria ilorinensis sp. Nov., Isolated from Cow Milk Cheese in Nigeria. Int. J. Syst. Evol. Microbiol. 2022, 72, 005437. [Google Scholar] [CrossRef]
- Carlin, C.R.; Liao, J.; Weller, D.; Guo, X.; Orsi, R.; Wiedmann, M. Listeria cossartiae sp. Nov., Listeria immobilis sp. Nov., Listeria portnoyi sp. Nov. and Listeria rustica sp. Nov., Isolated from Agricultural Water and Natural Environments. Int. J. Syst. Evol. Microbiol. 2021, 71, 004795. [Google Scholar] [CrossRef]
- Quereda, J.J.; Leclercq, A.; Moura, A.; Vales, G.; Gómez-Martín, Á.; García-Muñoz, Á.; Thouvenot, P.; Tessaud-Rita, N.; Bracq-Dieye, H.; Lecuit, M. Listeria valentina sp. Nov., Isolated from a Water Trough and the Faeces of Healthy Sheep. Int. J. Syst. Evol. Microbiol. 2020, 70, 5868–5879. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, A.; Moura, A.; Vales, G.; Tessaud-Rita, N.; Aguilhon, C.; Lecuit, M. Listeria thailandensis sp. Nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Doijad, S.P.; Poharkar, K.V.; Kale, S.B.; Kerkar, S.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Malik, S.V.S.; Ahmad, R.Y.; Hudel, M.; et al. Listeria goaensis sp. Nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 3285–3291. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Montero, K.; Leclercq, A.; Moura, A.; Vales, G.; Peraza, J.; Pizarro-Cerdá, J.; Lecuit, M. Listeria costaricensis sp. Nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 844–850. [Google Scholar] [CrossRef]
- Weller, D.; Andrus, A.; Wiedmann, M.; den Bakker, H.C. Listeria booriae sp. Nov. and Listeria newyorkensis sp. Nov., from Food Processing Environments in the USA. Int. J. Syst. Evol. Microbiol. 2015, 65, 286–292. [Google Scholar] [CrossRef]
- den Bakker, H.C.; Warchocki, S.; Wright, E.M.; Allred, A.F.; Ahlstrom, C.; Manuel, C.S.; Stasiewicz, M.J.; Burrell, A.; Roof, S.; Strawn, L.K.; et al. Listeria floridensis sp. Nov., Listeria Aquatica sp. Nov., Listeria cornellensis sp. Nov., Listeria Riparia sp. Nov. and Listeria grandensis sp. Nov., from Agricultural and Natural Environments. Int. J. Syst. Evol. Microbiol. 2014, 64, 1882–1889. [Google Scholar] [CrossRef]
- Lang Halter, E.; Neuhaus, K.; Scherer, S. Listeria weihenstephanensis sp. Nov., Isolated from the Water Plant Lemna Trisulca Taken from a Freshwater Pond. Int. J. Syst. Evol. Microbiol. 2013, 63, 641–647. [Google Scholar] [CrossRef]
- Bertsch, D.; Rau, J.; Eugster, M.R.; Haug, M.C.; Lawson, P.A.; Lacroix, C.; Meile, L. Listeria fleischmannii sp. Nov., Isolated from Cheese. Int. J. Syst. Evol. Microbiol. 2013, 63, 526–532. [Google Scholar] [CrossRef]
- den Bakker, H.C.; Manuel, C.S.; Fortes, E.D.; Wiedmann, M.; Nightingale, K.K. Genome Sequencing Identifies Listeria fleischmannii Subsp. Coloradonensis Subsp. Nov., Isolated from a Ranch. Int. J. Syst. Evol. Microbiol. 2013, 63, 3257–3268. [Google Scholar] [CrossRef]
- Graves, L.M.; Helsel, L.O.; Steigerwalt, A.G.; Morey, R.E.; Daneshvar, M.I.; Roof, S.E.; Orsi, R.H.; Fortes, E.D.; Milillo, S.R.; den Bakker, H.C.; et al. Listeria marthii sp. Nov., Isolated from the Natural Environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 2010, 60, 1280–1288. [Google Scholar] [CrossRef]
- Leclercq, A.; Clermont, D.; Bizet, C.; Grimont, P.A.D.; Le Flèche-Matéos, A.; Roche, S.M.; Buchrieser, C.; Cadet-Daniel, V.; Le Monnier, A.; Lecuit, M.; et al. Listeria rocourtiae sp. Nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2210–2214. [Google Scholar] [CrossRef] [PubMed]
- Carlin, C.R.; Liao, J.; Hudson, L.K.; Peters, T.L.; Denes, T.G.; Orsi, R.H.; Guo, X.; Wiedmann, M. Soil Collected in the Great Smoky Mountains National Park Yielded a Novel Listeria sensu stricto species, L. swaminathanii. Microbiol. Spectr. 2022, 10, e00442-22. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.K.; Chaggar, H.K.; Schamp, C.N.; Claxton, M.L.; Bryan, D.W.; Peters, T.L.; Song, Y.; Carlin, C.R.; den Bakker, H.C.; Denes, T.G. Phenotypic Characterization and Analysis of Complete Genomes of Two Distinct Strains of the Proposed Species “L. swaminathanii”. Sci. Rep. 2022, 12, 9137. [Google Scholar] [CrossRef]
- Chiara, M.; Caruso, M.; D’Erchia, A.M.; Manzari, C.; Fraccalvieri, R.; Goffredo, E.; Latorre, L.; Miccolupo, A.; Padalino, I.; Santagada, G.; et al. Comparative Genomics of Listeria Sensu Lato: Genus-Wide Differences in Evolutionary Dynamics and the Progressive Gain of Complex, Potentially Pathogenicity-Related Traits through Lateral Gene Transfer. Genome Biol. Evol. 2015, 7, 2154–2172. [Google Scholar] [CrossRef]
- Orsi, R.H.; Wiedmann, M. Characteristics and Distribution of Listeria spp., Including Listeria Species Newly Described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef]
- Rocourt, J.; Wehmeyer, U.; Stackebrandt, E. Transfer of Listeria dentrificans to a New Genus, Jonesia gen. Nov., as Jonesia denitrificans Comb. Nov. Int. J. Syst. Evol. Microbiol. 1987, 37, 266–270. [Google Scholar] [CrossRef]
- Rocourt, J.; Boerlin, P.; Grimont, F.; Jacquet, C.; Piffaretti, J.-C. Assignment of Listeria Grayi and Listeria Murrayi to a Single Species, Listeria Grayi, with a Revised Description of Listeria Grayi. Int. J. Syst. Evol. Microbiol. 1992, 42, 171–174. [Google Scholar] [CrossRef]
- ISO 11290-1:2017(En); Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:11290:-1:ed-2:v1:en (accessed on 11 June 2024).
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; de Peer, Y.V.; Vandamme, P.; Thompson, F.L.; et al. Re-Evaluating Prokaryotic Species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef]
- Martinez-Murcia, A.J.; Monera, A.; Saavedra, M.J.; Oncina, R.; Lopez-Alvarez, M.; Lara, E.; Figueras, M.J. Multilocus Phylogenetic Analysis of the Genus Aeromonas. Syst. Appl. Microbiol. 2011, 34, 189–199. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Kämpfer, P. Multilocus Sequence Analysis (MLSA) in Prokaryotic Taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Soler, L.; Yáñez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalán, V.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic Analysis of the Genus Aeromonas Based on Two Housekeeping Genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C.J. Multilocus Sequence Typing of Bacteria. Annu. Rev. Microbiol. 2006, 60, 561–588. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus Sequence Typing: A Portable Approach to the Identification of Clones within Populations of Pathogenic Microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef]
- Sullivan, C.B.; Diggle, M.A.; Clarke, S.C. Multilocus Sequence Typing. Mol. Biotechnol. 2005, 29, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Martínez-Murcia, A. Phylogenetic Analyses of the Genus Aeromonas Based on Housekeeping Gene Sequencing and Its Influence on Systematics. J. Appl. Microbiol. 2018, 125, 622–631. [Google Scholar] [CrossRef]
- Gadagkar, S.R.; Rosenberg, M.S.; Kumar, S. Inferring Species Phylogenies from Multiple Genes: Concatenated Sequence Tree versus Consensus Gene Tree. J. Exp. Zoolog. B Mol. Dev. Evol. 2005, 304B, 64–74. [Google Scholar] [CrossRef]
- Figueras, M.; Hidalgo, R.; Collado, L.; Martinez-Murcia, A. Recommendations for a New Bacterial Species Description Based on Analyses of the Unrelated Genera Aeromonas and Arcobacter. Bull. BISMiS 2011, 2, 1–16. [Google Scholar]
- Labeda, D.P. Multilocus Sequence Analysis of Phytopathogenic Species of the Genus Streptomyces. Int. J. Syst. Evol. Microbiol. 2011, 61, 2525–2531. [Google Scholar] [CrossRef] [PubMed]
- Margos, G.; Hojgaard, A.; Lane, R.S.; Cornet, M.; Fingerle, V.; Rudenko, N.; Ogden, N.; Aanensen, D.M.; Fish, D.; Piesman, J. Multilocus Sequence Analysis of Borrelia Bissettii Strains from North America Reveals a New Borrelia Species, Borrelia Kurtenbachii. Ticks Tick-Borne Dis. 2010, 1, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Macián, M.C.; Arahal, D.R.; Garay, E.; Pujalte, M.J. Multilocus Sequence Analysis of the Central Clade of the Genus Vibrio by Using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR Genes. Int. J. Syst. Evol. Microbiol. 2010, 60, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Richter, D.; Postic, D.; Sertour, N.; Livey, I.; Matuschka, F.-R.; Baranton, G. Delineation of Borrelia Burgdorferi Sensu Lato Species by Multilocus Sequence Analysis and Confirmation of the Delineation of Borrelia spielmanii sp. Nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, M.A.; Catalán, V.; Apráiz, D.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic Analysis of Members of the Genus Aeromonas Based on gyrB Gene Sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 875–883. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Pirone-Davies, C.; Chen, Y.; Pightling, A.; Ryan, G.; Wang, Y.; Yao, K.; Hoffmann, M.; Allard, M.W. Genes Significantly Associated with Lineage II Food Isolates of Listeria Monocytogenes. BMC Genom. 2018, 19, 708. [Google Scholar] [CrossRef]
- Yamamoto, S.; Harayama, S. PCR Amplification and Direct Sequencing of gyrB Genes with Universal Primers and Their Application to the Detection and Taxonomic Analysis of Pseudomonas Putida Strains. Appl. Environ. Microbiol. 1995, 61, 1104–1109. [Google Scholar] [CrossRef]
- Huang, W.M. Bacterial Diversity Based on Type II DNA Topoisomerase Genes. Annu. Rev. Genet. 1996, 30, 79–107. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.D.; Kämpfer, P.; Maiden, M.C.J.; Nesme, X.; Rosselló-Mora, R.; Swings, J.; Trüper, H.G.; et al. Report of the Ad Hoc Committee for the Re-Evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [CrossRef]
- Stevanović, O.; Milanov, D.; Prošić, I.; Gajdov, V.; Nedić, D.; Sladojević, Ž.; Radalj, A. Multilocus Sequence Analysis (MLSA) of a Nocardia Cyriacigeorgica Strain Causing Severe Bovine Mastitis in Bosnia and Herzegovina. Acta Vet. Hung. 2023, 71, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Currò, S.; Carraro, L.; Cardazzo, B.; Balzan, S.; Novelli, E.; Fontana, F.; Caburlotto, G.; Manfrin, A.; Fasolato, L. Retrospective Analysis of Vibrio spp. Isolated from Marketed Crustaceans Using Multilocus Sequence Analysis. Ital. J. Food Saf. 2023, 12, 11045. [Google Scholar] [CrossRef] [PubMed]
- Meghdadi, H.; Khosravi, A.D.; Hashemzadeh, M.; Tabandeh, M.R. New Design of Multilocus Sequence Analysis of rpoB, ssrA, Tuf, atpE, Ku, and dnaK for Identification of Mycobacterium Species. Mol. Biol. Rep. 2022, 49, 7967–7977. [Google Scholar] [CrossRef] [PubMed]
- Schardt, J. Characterization of Listeria Sensu Stricto Specific Genes Involved in Colonization of the Gastrointestinal Tract by Listeria monocytogenes. Doctoral Dissertation, Technische Universität München, München, Germany, 2018. [Google Scholar]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A Hybrid Sub-Lineage of Listeria Monocytogenes Comprising Hypervirulent Isolates. Nat. Commun. 2019, 10, 4283. [Google Scholar] [CrossRef] [PubMed]
- Daza Prieto, B.; Pietzka, A.; Martinovic, A.; Ruppitsch, W.; Zuber Bogdanovic, I. Surveillance and Genetic Characterization of Listeria Monocytogenes in the Food Chain in Montenegro during the Period 2014–2022. Front. Microbiol. 2024, 15, 1418333. [Google Scholar] [CrossRef] [PubMed]
- Nouws, S.; Bogaerts, B.; Verhaegen, B.; Denayer, S.; Crombé, F.; De Rauw, K.; Piérard, D.; Marchal, K.; Vanneste, K.; Roosens, N.H.C.; et al. The Benefits of Whole Genome Sequencing for Foodborne Outbreak Investigation from the Perspective of a National Reference Laboratory in a Smaller Country. Foods 2020, 9, 1030. [Google Scholar] [CrossRef]
- Mispelaere, M.; De Rop, A.-S.; Hermans, C.; De Maeseneire, S.L.; Soetaert, W.K.; De Mol, M.L.; Hulpiau, P. Whole Genome–Based Comparative Analysis of the Genus Streptomyces Reveals Many Misclassifications. Appl. Microbiol. Biotechnol. 2024, 108, 453. [Google Scholar] [CrossRef]

| Locus | Encoded Protein | Amplified Fragment | Primers Code | Sequence (5′–3′) |
|---|---|---|---|---|
| gyrA | DNA gyrase subunit A | 971 bp | Lis-gyrA-3F | GATGGACATGGTAACTTTG |
| Lis-gyrA-13R2 | CGCATATCTAAAATGGCTTG | |||
| cpn60 | chaperonin-60 | 931 bp | Lis-cpn60-2F | GAATTAGAAGACCCATTTGA |
| Lis-cpn60-11R | GTTTTGCTAAACGTTCTTGT | |||
| parE | DNA topoisomerase IV subunit B | 928 bp | Lis-parE-0F’ | AATGATGATTCTATTCAGGTGC |
| Lis-parE-9R’ | CGCGAATATCGCTACCYTC | |||
| recA | homologous recombination factor | 757 bp | Lis-recA-1F | TGAAAAACAATTCGGTAAAGG |
| Lis-recA-8R | TGARATACCTTCWCCGTACA | |||
| rpoB | DNA-directed RNA polymerase subunit beta | 1021 bp | Lis-rpoB-23F | AGGATGCGATCATCATGAG |
| Lis-rpoB-33R | GCTTCGTAAGTTTTCACACG | |||
| atpA | F0F1 ATP synthase subunit alpha | 947 bp | Lis-atpA-2F | TATGGCCCAAAACTTAGAA |
| Lis-atpA-11R | TTTTCATTGCTTTAATTTGCG | |||
| gyrB | DNA gyrase subunit B | 646 bp | Lis-gyrB-4F | GCGGCGGCGGATATAAAGTA |
| Lis-gyrB-10R | CCTTCACGAACATCTTCACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Murcia, A.; Navarro, A.; Miró-Pina, C. Identification of Listeria Isolates by Using a Pragmatic Multilocus Phylogenetic Analysis. Microbiol. Res. 2024, 15, 2114-2128. https://doi.org/10.3390/microbiolres15040142
Martínez-Murcia A, Navarro A, Miró-Pina C. Identification of Listeria Isolates by Using a Pragmatic Multilocus Phylogenetic Analysis. Microbiology Research. 2024; 15(4):2114-2128. https://doi.org/10.3390/microbiolres15040142
Chicago/Turabian StyleMartínez-Murcia, Antonio, Aaron Navarro, and Caridad Miró-Pina. 2024. "Identification of Listeria Isolates by Using a Pragmatic Multilocus Phylogenetic Analysis" Microbiology Research 15, no. 4: 2114-2128. https://doi.org/10.3390/microbiolres15040142
APA StyleMartínez-Murcia, A., Navarro, A., & Miró-Pina, C. (2024). Identification of Listeria Isolates by Using a Pragmatic Multilocus Phylogenetic Analysis. Microbiology Research, 15(4), 2114-2128. https://doi.org/10.3390/microbiolres15040142






