Probiotic Enterococcus faecium CRL 183 Inhibits Candida albicans Biofilm In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Assurance of Probiotic Strain Safety
2.2. Biofilm Formation and pH Evaluation
2.3. Quantification of Viable Cells and Determination of the Anti-Candida Activity of E. faecium CRL 183
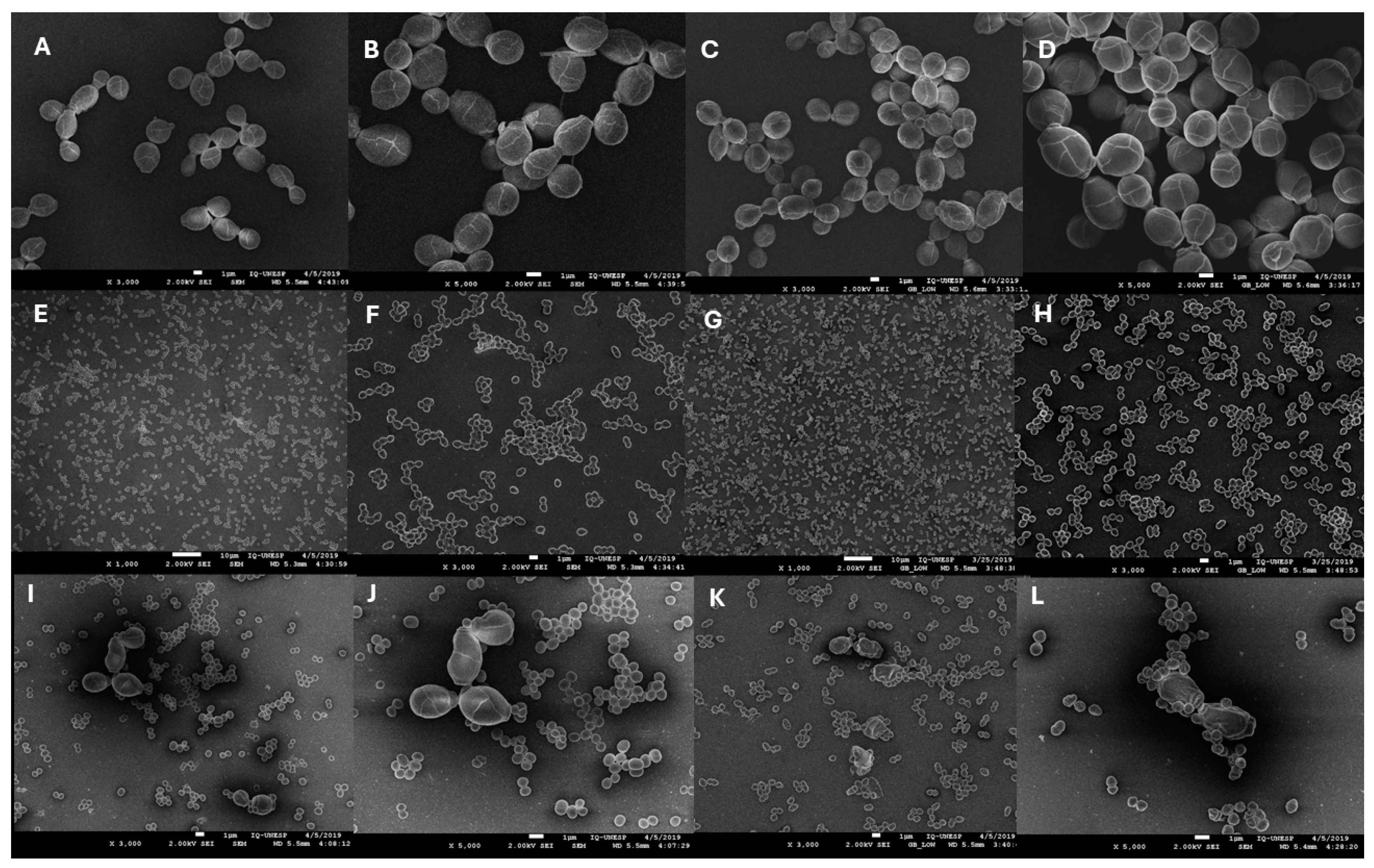
2.4. Scanning Electron Microscopy (SEM)
2.5. Statistical Analysis
3. Results
3.1. Assurance of Probiotic Strain Safety
3.2. Biofilm pH Evaluation
3.3. Anti-Candida Activity of E. faecium CRL 183
3.4. Scanning Electron Microscopy (SEM)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Park, S.J.; Choi, S.J.; Park, J.Y.; Lee, K.H. Immunological features of macrophages induced by various morphological structures of Candida albicans. J. Microbiol. Biotechnol. 2013, 23, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Mattei, A.S.; Alves, S.H.; Severo, C.B.; Da Silva Guazzelli, L.; De Mattos Oliveira, F.; Severo, L.C. Determination of germ tube, phospholipase, and proteinase production by bloodstream isolates of Candida albicans. Rev. Soc. Bras. Med. Trop. 2013, 46, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, V.H.; Wang, Y.; Bandara, H.M.H.N.; Mayer, M.P.A.; Samaranayake, L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016, 100, 6415–6426. [Google Scholar] [CrossRef]
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and characterization of an in vivo central venous catheter Candida albicans Biofilm Model. Infect. Immun. 2004, 72, 6023–6031. [Google Scholar] [CrossRef]
- Niewerth, M.; Korting, H.C. Phospholipases of Candida albicans. Phospholipasen von Candida albicans. Mycoses 2001, 367, 361–367. [Google Scholar] [CrossRef]
- Zago, C.E.; Silva, S.; Sanitá, P.V.; Barbugli, P.A.; Dias, C.M.I.; Lordello, V.B.; Vergani, C.E. Dynamics of biofilm formation and the Interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA). PLoS ONE 2015, 10, e0123206. [Google Scholar] [CrossRef]
- Fanning, S.; Mitchell, A.P. Fungal biofilms. PLoS Pathog. 2012, 8, e1002585. [Google Scholar] [CrossRef]
- Shekh, R.M.; Roy, U. Biochemical characterization of an anti-Candida factor produced by Enterococcus faecalis. BMC Microbiol. 2012, 12, 132. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Saarela, M.; Mogensen, G.; Fonden, R.; Matto, J.; Mattila-Sandholm, T. Probiotic bacteria: Safety, functional and technological properties. J. Biotechnol. 2000, 84, 197–215. [Google Scholar] [CrossRef]
- Vilela, S.F.G.; Barbosa, J.O.; Rossoni, R.D.; Santos, J.D.; Prata, M.C.A.; Anbinder, A.L.; Jorge, A.O.C.; Junqueira, J.C. Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence 2015, 6, 29–39. [Google Scholar] [CrossRef]
- Ribeiro, F.C.; de Barros, P.P.; Rossoni, R.D.; Junqueira, J.C.; Jorge, A.O.C. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 2017, 122, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Mailänder-Sánchez, D.; Braunsdorf, C.; Grumaz, C.; Müller, C.; Lorenz, S.; Stevens, P.; Wagener, J.; Hebecker, B.; Hube, B.; Bracher, F.; et al. Antifungal defense of probiotic Lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS ONE 2017, 12, e0184438. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.H.; Mayer, M.P.A.; Miyazima, T.Y.; Matsubara, V.H.; Silva, E.G.; Paula, C.R.; Campos, T.T.; Nakamae, A.E.M. A multispecies probiotic reduces oral Candida colonization in denture wearers. J. Prosthodont. 2015, 24, 194–199. [Google Scholar] [CrossRef]
- Garsin, D.A.; Lorenz, M.C. Candida albicans and Enterococcus faecalis in the gut: Synergy in commensalism? Gut Microbes 2013, 4, 409–415. [Google Scholar] [CrossRef]
- Mason, K.L.; Downward, J.R.E.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 2012, 10, 3371–3380. [Google Scholar] [CrossRef]
- Mason, K.L.; Erb Downward, J.R.; Falkowski, N.R.; Young, V.B.; Kao, J.Y.; Huffnagle, G.B. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect. Immun. 2012, 80, 150–158. [Google Scholar] [CrossRef]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef]
- Bachtiar, E.W.; Dewiyani, S.; Surono Akbar, S.M.; Bachtiar, B.M. Inhibition of Candida albicans biofilm development by unencapsulated Enterococcus faecalis cps2. J. Dent. Sci. 2016, 11, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between probiotic potential and safety concerns-an update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Pujia, A.M.; Costacurta, M.; Fortunato, L.; Merra, G.; Cascapera, S.; Calvani, M.; Gratteri, S. The probiotics in dentistry: A narrative review. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1405–1412. [Google Scholar]
- Saavedra, L.; Taranto, M.P.; Sesma, F.; De Valdez, G.F. Homemade traditional cheeses for the isolation of probiotic Enterococcus faecium strains. Int. J. Food Microbiol. 2003, 88, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, L.S.; Bedani, R.; Dejani, N.N.; Ivo De Medeiros, A.; Zuanon, J.A.S.; Spolidorio, L.C.; Adorno, M.A.T.; Varesche, M.B.A.; Galvão, F.C.; Valentini, S.R.; et al. Effect of a probiotic beverage consumption (Enterococcus faecium CRL 183 and Bifidobacterium longum ATCC 15707) in rats with chemically induced colitis. PLoS ONE 2017, 12, e0175935. [Google Scholar] [CrossRef]
- Cavallini, D.C.; Bedani, R.; Bomdespacho, L.Q.; Vendramini, R.C.; Rossi, E.A. Effects of probiotic bacteria, isoflavones and simvastatin on lipid profile and atherosclerosis in cholesterol-fed rabbits: A randomized double-blind study. Lipids Health Dis. 2009, 8, 1. [Google Scholar] [CrossRef]
- Cavallini, D.C.; Suzuki, J.Y.; Abdalla, D.S.; Vendramini, R.C.; Pauly-Silveira, N.D.; Roselino, M.N.; Pinto, R.A.; Rossi, E.A. Influence of a probiotic soy product on fecal microbiota and its association with cardiovascular risk factors in an animal model. Lipids Health Dis. 2011, 10, 126. [Google Scholar] [CrossRef]
- Kinouchi, F.L.; Maia, D.C.G.; de Abreu Ribeiro, L.C.; Placeres, M.C.P.; de Valdez, G.F.; Colombo, L.L.; Rossi, E.A.; Carlos, I.Z. A soy-based product fermented by Enterococcus faecium and Lactobacillus helveticus inhibits the development of murine breast adenocarcinoma. Food Chem. Toxicol. 2012, 50, 4144–4148. [Google Scholar] [CrossRef]
- Sivieri, K.; Spinardi-Barbisan, A.L.T.; Barbisan, L.F.; Bedani, R.; Pauly, N.D.; Carlos, I.Z.; Benzatti, F.; Vendramini, R.C.; Rossi, E.A. Probiotic Enterococcus faecium CRL 183 inhibit chemically induced colon cancer in male Wistar rats. Eur. Food Res. Technol. 2008, 228, 231–237. [Google Scholar] [CrossRef]
- Marchesin, J.C.; Celiberto, L.S.; Orlando, A.B.; de Medeiros, A.I.; Pinto, R.A.; Zuanon, J.A.S.; Spolidorio, L.C.; dos Santos, A.; Taranto, M.P.; Cavallini, D.C.U. A soy-based probiotic drink modulates the microbiota and reduces body weight gain in diet-induced obese mice. J. Funct. Foods 2018, 48, 302–313. [Google Scholar] [CrossRef]
- Cavallini, D.C.; Manzoni, M.S.J.; Bedani, R.; Roselino, M.N.; Celiberto, L.S.; Vendramini, R.C.; De Valdez, G.F.; Saes Parra Abdalla, D.; Pinto, R.A.; Rosetto, D.; et al. Probiotic soy product supplemented with isoflavones improves the lipid profile of moderately hypercholesterolemic Men: A randomized controlled trial. Nutrients 2016, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Witzler, J.J.P.; Pinto, R.A.; Font de Valdez, G.; de Castro, A.D.; Cavallini, D.C.U. Development of a potential probiotic lozenge containing Enterococcus faecium CRL 183. LWT Food Sci. Technol. 2017, 77, 193–199. [Google Scholar] [CrossRef]
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Fontana, C.R.; Abernethy, A.D.; Som, S.; Ruggiero, K.; Doucette, S.; Marcantonio, R.C.; Boussios, C.I.; Kent, R.; Goodson, J.M.; Tanner, A.C.R.; et al. The antibacterial effect of photodynamic therapy in dental plaque-derived biofilms. J. Periodontal Res. 2009, 44, 751–759. [Google Scholar] [CrossRef]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef]
- Jørgensen, M.R.; Kragelund, C.; Jensen, P.Ø.; Keller, M.K.; Twetman, S. Probiotic Lactobacillus reuteri has antifungal effects on oral Candida species in vitro. J. Oral Microbiol. 2017, 9, 1274582. [Google Scholar] [CrossRef]
- Davis, D Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 2003, 44, 1–7. [CrossRef]
- Gittings, S.; Turnbull, N.; Henry, B.; Roberts, C.J.; Gershkovich, P. Characterisation of human saliva as a platform for oral dissolution medium development. Eur. J. Pharm. Biopharm. 2015, 91, 16–24. [Google Scholar] [CrossRef]
- Chandra, J.; Mukherjee, P.K. Candida biofilms: Development, architecture, and resistance. In Microbial Biofilms; Ghannoum, M., Parsek, M., Whiteley, M., Mukherjee, P.K., Eds.; ASM Press: Washington, DC, USA, 2015; pp. 115–134. [Google Scholar] [CrossRef]
- Cavalheiro, M.; Teixeira, M.C. Candida biofilms: Threats, challenges, and promising strategies. Front. Med. 2018, 5, 28. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, X.; Zhou, P. In vitro Antifungal Effects of Berberine Against Candida spp. In Planktonic and Biofilm Conditions. Drug Des. Dev. Ther. 2020, 14, 87–101. [Google Scholar] [CrossRef]
- Tan, Y.; Leonhard, M.; Moser, D.; Ma, S.; Schneider-Stickler, B. Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm. Arch. Oral Biol. 2018, 85, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Weerasekera, M.M.; Wijesinghe, G.K.; Jayarathna, T.A.; Gunasekara, C.P.; Fernando, N.; Kottegoda, N.; Samaranayake, L.P. Culture media profoundly affect Candida albicans and Candida tropicalis growth, adhesion and biofilm development. Mem. Inst. Oswaldo Cruz 2016, 115, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010, 6, 1000828. [Google Scholar] [CrossRef]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2017, 16, 19–31. [Google Scholar] [CrossRef] [PubMed]


| Antimicrobials | Inhibition Zone Diameter (mm) | Interpretation (mm) * |
|---|---|---|
| Ciprofloxacin 5 mg | 30 | Sensitive (≥21) |
| Chloramphenicol 30 mg | 27 | Sensitive (≥18) |
| Erythromycin 15 mg | 22 | Sensitive (≥23) |
| Nitrofurantoin 300 mg | 30 | Sensitive (≥17) |
| Norfloxacin 10 mg | 25 | Sensitive (≥17) |
| Tetracycline 30 mg | 24 | Sensitive (≥19) |
| Vancomycin 30 mg | 19 | Sensitive (≥17) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lordello, V.B.; de Annunzio, S.R.; da Silva, E.V.; Taranto, M.P.; Fontana, C.R.; Cavallini, D.C.U. Probiotic Enterococcus faecium CRL 183 Inhibits Candida albicans Biofilm In Vitro. Microbiol. Res. 2024, 15, 2102-2113. https://doi.org/10.3390/microbiolres15040141
Lordello VB, de Annunzio SR, da Silva EV, Taranto MP, Fontana CR, Cavallini DCU. Probiotic Enterococcus faecium CRL 183 Inhibits Candida albicans Biofilm In Vitro. Microbiology Research. 2024; 15(4):2102-2113. https://doi.org/10.3390/microbiolres15040141
Chicago/Turabian StyleLordello, Virgínia Barreto, Sarah Raquel de Annunzio, Eliane Vale da Silva, Maria Pía Taranto, Carla Raquel Fontana, and Daniela Cardoso Umbelino Cavallini. 2024. "Probiotic Enterococcus faecium CRL 183 Inhibits Candida albicans Biofilm In Vitro" Microbiology Research 15, no. 4: 2102-2113. https://doi.org/10.3390/microbiolres15040141
APA StyleLordello, V. B., de Annunzio, S. R., da Silva, E. V., Taranto, M. P., Fontana, C. R., & Cavallini, D. C. U. (2024). Probiotic Enterococcus faecium CRL 183 Inhibits Candida albicans Biofilm In Vitro. Microbiology Research, 15(4), 2102-2113. https://doi.org/10.3390/microbiolres15040141






