Exploring Local Reservoirs for Bacteriophages with Therapeutic Potential against ESKAPE Pathogens
Abstract
1. Introduction
Bacteriophages
2. Materials and Methods
2.1. ESKAPE Strains and Growth Conditions
2.2. Water Sample Collection
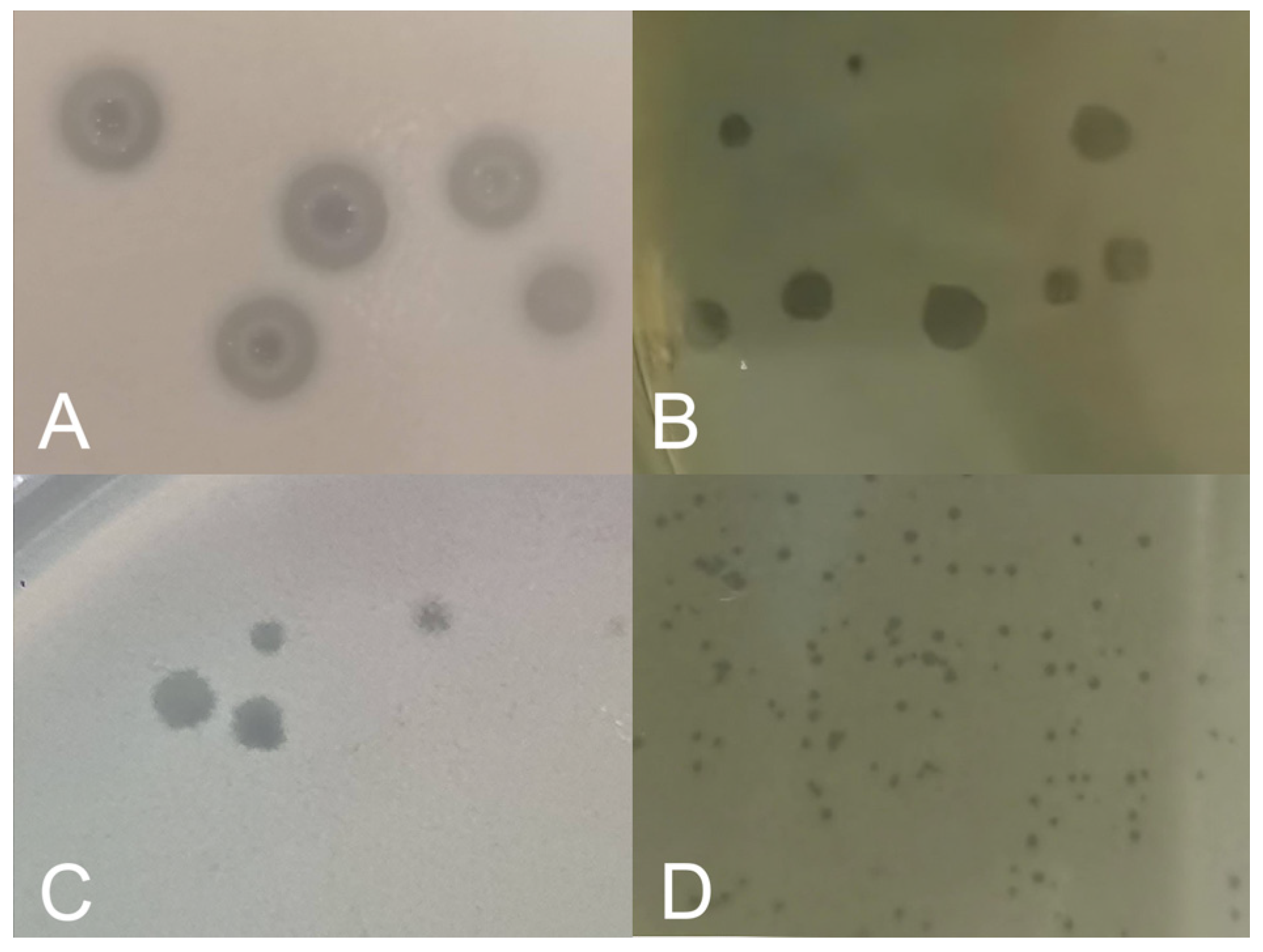
2.3. Phage Enrichment, Isolation, and Purification
2.4. Preliminary Lytic Activity Evaluation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Rosa, T.F.; Coelho, S.S.; Foletto, V.S.; Bottega, A.; Serafin, M.B.; Machado, C.S.; Franco, L.N.; de Paula, B.R.; Hörner, R. Alternatives for the treatment of infections caused by ESKAPE pathogens. J. Clin. Pharm. Ther. 2020, 45, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Moghadam, S.S.; Hosseini Doust, R.; Majidpour, A.; Adabi, M.; Minaeian, S. Antibacterial Activity and Toxicity of Zinc Oxide Nanoparticles Combined with Supernatants of Lactobacillus spp. Against ESKAPE Bacteria: A Novel Mixture. Iran. J. Pharm. Res. 2023, 22, e139222. [Google Scholar] [CrossRef]
- Gerace, E.; Mancuso, G.; Midiri, A.; Poidomani, S.; Zummo, S.; Biondo, C. Recent Advances in the Use of Molecular Methods for the Diagnosis of Bacterial Infections. Pathogens 2022, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Alam, M. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Evid. Based Nurs. 2023, 27, 103540. [Google Scholar] [CrossRef]
- Boletín Infecciones Asociadas a la Atención de la Salud (IAAS) Red Hospitalaria de Vigilancia Epidemiológica (RHOVE). Available online: https://www.gob.mx/salud/documentos/boletin-epidemiologico-rhove-2023 (accessed on 24 May 2024).
- Dávila-López, E.C.; Berumen-Lechuga, M.G.; Molina-Pérez, C.J.; Jimenez-Juarez, R.N.; Leaños-Miranda, A.; Robles-Ordoñez, N.; Peña-Cano, M.I.; Venegas-Esquivel, G.A. Antimicrobial Resistance and Antibiotic Consumption in a Secondary Care Hospital in Mexico. Antibiotics 2024, 13, 178. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181–e00219. [Google Scholar] [CrossRef] [PubMed]
- Machowska, A.; Stålsby Lundborg, C. Drivers of Irrational Use of Antibiotics in Europe. Int. J. Environ. Res. Public Health 2018, 16, 27. [Google Scholar] [CrossRef]
- Ene, A.; Miller-Ensminger, T.; Mores, C.R.; Giannattasio-Ferraz, S.; Wolfe, A.J.; Abouelfetouh, A.; Putonti, C. Examination of Staphylococcus aureus Prophages Circulating in Egypt. Viruses 2021, 13, 337. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Isaev, A.B.; Musharova, O.S.; Severinov, K.V. Microbial Arsenal of Antiviral Defenses—Part I. Biochemistry 2021, 86, 319–337. [Google Scholar] [CrossRef]
- Isaev, A.B.; Musharova, O.S.; Severinov, K.V. Microbial Arsenal of Antiviral Defenses. Part II. Biochemistry 2021, 86, 449–470. [Google Scholar] [CrossRef]
- Mann, N.H. The third age of phage. PLoS Biol. 2005, 3, e182. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Ramesh, N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS ONE 2018, 13, e0206278. [Google Scholar] [CrossRef] [PubMed]
- Batinovic, S.; Wassef, F.; Knowler, S.A.; Rice, D.T.F.; Stanton, C.R.; Rose, J.; Tucci, J.; Nittami, T.; Vinh, A.; Drummond, G.R.; et al. Bacteriophages in Natural and Artificial Environments. Pathogens 2019, 8, 100. [Google Scholar] [CrossRef]
- Modin, O.; Fuad, N.; Abadikhah, M.; I’Ons, D.; Ossiansson, E.; Gustavsson, D.J.I.; Edefell, E.; Suarez, C.; Persson, F.; Wilén, B.M. A relationship between phages and organic carbon in wastewater treatment plant effluents. Water Res. X 2022, 16, 100146. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; Latka, A.; Roszniowski, B.; Valvano, M.A.; Drulis-Kawa, Z. Phage Life Cycles Behind Bacterial Biodiversity. Curr. Med. Chem. 2017, 24, 3987–4001. [Google Scholar] [CrossRef]
- Edwards, R.A.; McNair, K.; Faust, K.; Raes, J.; Dutilh, B.E. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 2016, 40, 258–272. [Google Scholar] [CrossRef]
- Patil, A.; Banerji, R.; Kanojiya, P.; Koratkar, S.; Saroj, S. Bacteriophages for ESKAPE: Role in pathogenicity and measures of control. Expert Rev. Anti Infect. Ther. 2021, 19, 845–865. [Google Scholar] [CrossRef] [PubMed]
- Doub, J.B.; Shishido, A.; Srikumaran, U.; Haskoor, J.; Tran-Nguyen, P.; Lee, M.; Würstle, S.; Lee, A.; Kortright, K.; Chan, B.K. Salphage: Salvage bacteriophage therapy for a recalcitrant Klebsiella pneumoniae prosthetic shoulder infection—A case report. Acta Orthop. 2022, 93, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Eales, B.M.; Tam, V.H. Case Commentary: Novel Therapy for Multidrug-Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2022, 66, e0199621. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Luscher, A.; Falconnet, L.; Resch, G.; McBride, R.; Mai, Q.A.; Simonin, J.L.; Chanson, M.; Maco, B.; Galiotto, R.; et al. Personalized aerosolised bacteriophage treatment of a chronic lung infection due to multidrug-resistant Pseudomonas aeruginosa. Nat. Commun. 2023, 14, 4443. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gallardo, M.J.; Villicaña, C.; Yocupicio-Monroy, M.; Alcaraz-Estrada, S.L.; León-Félix, J. Current knowledge in the use of bacteriophages to combat infections caused by Pseudomonas aeruginosa in cystic fibrosis. Folia Microbiol. 2023, 68, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rubalskii, E.; Ruemke, S.; Salmoukas, C.; Boyle, E.C.; Warnecke, G.; Tudorache, I.; Shrestha, M.; Schmitto, J.D.; Martens, A.; Rojas, S.V.; et al. Bacteriophage Therapy for Critical Infections Related to Cardiothoracic Surgery. Antibiotics 2020, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Intralytix Safety by Nature. Available online: https://www.intralytix.com/patents (accessed on 29 June 2024).
- Clinical Trials.gov. Available online: https://clinicaltrials.gov/search?term=phage%20therapy (accessed on 29 June 2024).
- Qin, J.; Wu, N.; Bao, J.; Shi, X.; Ou, H.; Ye, S.; Zhao, W.; Wei, Z.; Cai, J.; Li, L.; et al. Heterogeneous Klebsiella pneumoniae Co-infections Complicate Personalized Bacteriophage Therapy. Front. Cell Infect. Microbiol. 2021, 10, 608402. [Google Scholar] [CrossRef] [PubMed]
- Levêque, M.; Cassir, N.; Mathias, F.; Fevre, C.; Daviet, F.; Bermudez, J.; Brioude, G.; Peyron, F.; Reynaud-Gaubert, M.; Coiffard, B. Refractory Pseudomonas aeruginosa Bronchopulmonary Infection After Lung Transplantation for Common Variable Immunodeficiency Despite Maximal Treatment Including IgM/IgA-Enriched Immunoglobulins and Bacteriophage Therapy. Infect. Drug Resist. 2023, 16, 4265–4271. [Google Scholar] [CrossRef] [PubMed]
- Blasco, L.; López-Hernández, I.; Rodríguez-Fernández, M.; Pérez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.P.; Rodríguez-Baño, J.; Tomás, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front. Med. 2023, 10, 1199657. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Romano, J.; Mastache-Estrada, L.A.; Molina-Sánchez, D.A.; Serrano-Plancarte, R.; Peña-Barrera, C.; Chávez-Bejar, M.I.; Romero-Martínez, N.; Lecona-Valera, A.N. Estabilidad y capacidad inhibitoria del bacteriófago ϕrsp, agente potencial para el biocontrol de Ralstonia solanacearum. Rev. Fitotec. Mex. 2019, 42, 13–19. [Google Scholar]
- González-Villalobos, E.; Ribas-Aparicio, R.M.; Montealegre, G.E.R.; Belmont-Monroy, L.; Ortega-García, Y.; Aparicio-Ozores, G.; Balcázar, J.L.; Eslava-Campos, C.A.; Hernández-Chiñas, U.; Molina-López, J. Isolation and characterization of novel bacteriophages as a potential therapeutic option for Escherichia coli urinary tract infections. Appl. Microbiol. Biotechnol. 2021, 105, 5617–5629. [Google Scholar] [CrossRef] [PubMed]
- Lomelí-Ortega, C.O.; Balcázar, J.L.; Quiroz-Guzmán, E. Phage therapy and aquaculture: Progress and challenges. Int. Microbiol. 2023, 26, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, X.; Li, S.; Yin, H.; Zhao, Z. Characterization and genomic analysis of a broad-spectrum lytic phage PG288: A potential natural therapy candidate for Vibrio infections. Virus Res. 2024, 341, 199320. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Saffari, M.; Siadat, S.D.; Hejazi, S.H.; Shayestehpour, M.; Motallebi, M.; Eidi, M. Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB_KshKPC-M against carbapenemase-producing Klebsiella pneumoniae strains (CRKP) isolated from Ventilator-associated pneumoniae (VAP) infection of COVID-19 patients. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 18. [Google Scholar] [CrossRef]
- Van-Twest, R.; Kropinski, A.M. Bacteriophage Enrichmnent from Water and Soil. In Bacteriophages Methods and Protocols, 1st ed.; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; pp. 15–20. [Google Scholar]
- Myers, J.; Davis, J., II; Lollo, M.; Hudec, G.; Hyman, P. More’s the Same-Multiple Hosts Do Not Select for Broader Host Range Phages. Viruses 2023, 15, 518. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, P.; Lin, Z.; Wang, T. Characterization of Two Pseudomonas aeruginosa Viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses 2019, 11, 318. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Dong, Y.; An, X.; Song, L.; Li, M.; Fan, H.; Tong, Y. Potential application of a newly isolated phage BUCT609 infecting Stenotrophomonas maltophilia. Front. Microbiol. 2022, 13, 1001237. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, C.; Medina, M.; Bonhomme, M.; Laumay, F.; Roussel-Gaillard, T.; Martins-Simoes, P.; Tristan, A.; Pirot, F.; Ferry, T.; Laurent, F.; et al. Phage Therapy against Staphylococcus aureus: Selection and Optimization of Production Protocols of Novel Broad Spectrum Silviavirus Phages. Pharmaceutics 2022, 14, 1885. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Latz, S.; Wahida, A.; Arif, A.; Häfner, H.; Hoß, M.; Ritter, K.; Horz, H.P. Preliminary survey of local bacteriophages with lytic activity against multi-drug resistant bacteria. J. Basic Microbiol. 2016, 56, 1117–1123. [Google Scholar] [CrossRef]
- Miller, J.J.; Weimer, B.C.; Timme, R.; Lüdeke, C.H.M.; Pettengill, J.B.; Bandoy, D.D.; Weis, A.M.; Kaufman, J.; Huang, B.C.; Payne, J.; et al. Phylogenetic and Biogeographic Patterns of Vibrio parahaemolyticus Strains from North America Inferred from Whole-Genome Sequence Data. Appl. Environ. Microbiol. 2021, 87, e0069321. [Google Scholar] [CrossRef] [PubMed]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.-J.; Fung, K.-M.; Lee, I.-M.; Ko, T.-P.; Lin, C.-Y.; Wong, C.-L.; Tu, I.-F.; Huang, T.-Y.; Yang, F.-L.; Chang, Y.-P.; et al. Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation. mBio 2024, 15, e0351923. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Zimmer-Faust, A.G.; Griffith, J.F.; Steele, J.A.; Santos, B.; Cao, Y.; Asato, L.; Chiem, T.; Choi, S.; Diaz, A.; Guzman, J.; et al. Relationship between coliphage and Enterococcus at southern California beaches and implications for beach water quality management. Water Res. 2023, 230, 119383. [Google Scholar] [CrossRef] [PubMed]
- Nappier, S.P.; Hong, T.; Ichida, A.; Goldstone, A.; Eftim, S.E. Occurrence of coliphage in raw wastewater and in ambient water: A meta-analysis. Water Res. 2019, 153, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef]
- Dahal, U.; Paul, K.; Gupta, S. The multifaceted genus Acinetobacter: From infection to bioremediation. J. Appl. Microbiol. 2023, 134, lxad145. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 2019, 32, e00002–e00019. [Google Scholar] [CrossRef] [PubMed]
- Ariyadasa, S.; Taylor, W.; Weaver, L.; McGill, E.; Billington, C.; Pattis, I. Nonbacterial Microflora in Wastewater Treatment Plants: An Underappreciated Potential Source of Pathogens. Microbiol. Spectr. 2023, 11, e0048123. [Google Scholar] [CrossRef] [PubMed]
- Podlacha, M.; Węgrzyn, G.; Węgrzyn, A. Bacteriophages-Dangerous Viruses Acting Incognito or Underestimated Saviors in the Fight against Bacteria? Int. J. Mol. Sci. 2024, 25, 2107. [Google Scholar] [CrossRef] [PubMed]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Lusiak-Szelachowska, M.; Zaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 100. [Google Scholar] [CrossRef]
- Stevens, R.H.; Zhang, H.; Sedgley, C.; Bergman, A.; Manda, A.R. The prevalence and impact of lysogeny among oral isolates of Enterococcus faecalis. J. Oral Microbiol. 2019, 11, 1643207. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.R. Criteria for Selecting Suitable Infectious Diseases for Phage Therapy. Viruses 2018, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.L.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef] [PubMed]
- Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of Bacteriophages with Animal and Human Organisms-Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 2021, 22, 8937. [Google Scholar] [CrossRef]
- Verbeken, G.; Pirnay, J.P.; De Vos, D.; Jennes, S.; Zizi, M.; Lavigne, R.; Casteels, M.; Huys, I. Optimizing the European regulatory framework for sustainable bacteriophage therapy in human medicine. Arch. Immunol. Ther. Exp. (Warsz). 2012, 60, 161–172. [Google Scholar] [CrossRef]
| Host Species | Strain | Source Isolation | Antimicrobial Profile | |
|---|---|---|---|---|
| E. faecium | IIH-109.3 | Catheter tip | β-LacR, CEP-2 gR, CEP-3 gR, NITR | MDR |
| IIH-160 | Sore skin | β-LacR, CEP-2 gR, CEP-3 gR, GLPR | MDR | |
| IIH-41.5 | Catheter tip | β-LacR, CEP-2 gR, CEP-3 gR, GLPR, NITR | MDR | |
| IIH-168.5 | Catheter tip | β-LacR, AMGR, CEP-2 gR, CEP-3 gR, NITR | MDR | |
| IIH-168.2 | Catheter tip | β-LacR, AMGR, CEP-2 gR, CEP-3 gR, NITR | MDR | |
| IIH-132 | Catheter tip | β-LacR, CEP-2 gR, CEP-3 gR, GLPR, NITR | MDR | |
| IIH-98 | Dialysis fluid | β-LacR, AMGR, CEP-2 gR, CEP-3 gR, GLPR, NITR | MDR | |
| IIH-140 | Catheter tip | β-LacR, CEP-2 gR, CEP-3 gR, GLPR, NITR | MDR | |
| S. aureus | IIH-107 | Catheter tip | β-LacR, FQR, MCLR, LICR | MDR |
| IIH-6 | Dialysis fluid | FQR | S | |
| IIH-145 | Ulcer | FQR, MCLR, LICR | MDR | |
| IIH-108 | Wound | β-LacR, FQR, MCLR, LICR | MDR | |
| K. pneumoniae | IIH-126 | Wound infection | PENR, β-LacR | S |
| IIH-155 | Bronchial secretion | PENR, β-LacR | S | |
| IIH-97.2 | Catheter tip | PENR | S | |
| ATCC BAA1705 | Urine | PENR, β-LacR, β-Lac/InhR, CEP-1 gR, CEP-2 gR, CEP-3 gR, CEP-4 gR, CARR, AMGR, FQR, NITR | MDR | |
| A. baumannii | AB 21/432 | Data not provided | PENR, CEP-2 gR, CEP-3 gR, CEP-4 gR, MONOR, CARR, FQR | MDR |
| AB 20/403 | Pedriatric neurology | PENR, CEP-2 gR, CEP-3 gR, CEP-4 gR, MONOR, CARR | MDR | |
| AB BAA747 | Ear pus | PENR, CEP-1 gR, CEP-2 gR | MDR | |
| AB 10/289 | Coronary Care Unit | PENR, CEP-1 gR, CEP-2 gR, CEP-3 gR, CEP-4 gR, CARR | MDR | |
| P. aeruginosa | PA IIH-97 | Bronchial secretion | AMGR, CEP-3 gR, CEP-4 gR | MDR |
| ATCC27853 | Blood culture | AMGR | S | |
| PA IIH-78 | Catheter tip | AMGR, CEP-3 gR, CEP-4 gR | MDR | |
| PA IIH-66 | Catheter tip | AMGR, CEP-3 gR, CEP-4 gR | MDR | |
| PA1 | Surgery service | AMGR, CARR, CEP-3 gR, FQR, β-Lac/InhR, MCLR, FOSR | MDR | |
| PA4 | Urgency | AMGR, CARR, CEP-3 gR, FQR, β-Lac/InhR, MCLR, FOSR | MDR | |
| PA7 | Pediatric service | AMGR, CARR, CEP-3 gR, FQR, MCLR, FOSR | MDR | |
| PA10 | Cardiology | AMGR, CARR, CEP-3 gR, FQR, MCLR, FOSR | MDR | |
| E. cloacae | IIH-41 | Blood culture | AMGR, CEP-2 gR | S |
| IIH-42 | Blood culture | AMGR, CEP-2 gR | S | |
| 04 | Pediatric service | AMGR, CEP-2 gR, CEP-3 gR, CARR | MDR | |
| 08 | Pediatric service | AMGR, CEP-2 gR, CEP-3 gR, SFNR | MDR | |
| Sample | Staphylococcus aureus | Klebsiella pneumoniae | Acinetobacter baumannii | Pseudomonas aeruginosa | Enterobacter cloacae | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IIH-145 | IIH-108 | IIH-97.2 | IIH-126 | IIH-155 | ATCC BAA 1705 | 20/403 | 21/432 | ATCC 27853 | PA10 | IIH-141 | IIH-142 | 04 | 08 | |
| M1 | 2 | 2 | 3 | 2 | 2 | |||||||||
| M2 | 3 | 3 | 2 | 1 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | ||
| M3 | 2 | 3 | 2 | 4 | ||||||||||
| M4 | 2 | 3 | 2 | 2 | ||||||||||
| M5 | 2 | 2 | 3 | |||||||||||
| M6 | 2 | 3 | 2 | 2 | ||||||||||
| M7 | 2 | 3 | 3 | 2 | ||||||||||
| M8 | 1 | 1 | 2 | 2 | ||||||||||
| M9 | 2 | 3 | 2 | 1 | ||||||||||
| M12 | 2 | 3 | 3 | 2 | ||||||||||
| Total | 15 | 23 | 3 | 3 | 5 | 1 | 22 | 23 | 2 | 2 | 2 | 2 | 2 | 2 |
| Sample | Staphylococcus aureus | Klebsiella pneumoniae | Acinetobacter baumannii | Pseudomonas aeruginosa | Enterobacter cloacae | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IIH-145 | IIH-108 | IIH-155 | ATCC 1705 | 20/403 | 21/432 | Pa7 | IIH-141 | IIH-142 | 04 | 08 | |
| M10 | 2 | 2 | 1 | 2 | 3 | 4 | 3 | 2 | 1 | 2 | |
| M11 | 1 | 1 | 3 | 3 | 2 | 4 | |||||
| M13 | 1 | 2 | 3 | 2 | |||||||
| Total | 2 | 5 | 5 | 1 | 8 | 7 | 8 | ||||
| Sample | Staphylococcus aureus | Acinetobacter baumannii | ||
|---|---|---|---|---|
| IIH-145 | IIH-108 | 20/403 | 21/432 | |
| M14 | 1 | 2 | 2 | 2 |
| M15 | 1 | 2 | 2 | 2 |
| M16 | 1 | 3 | 2 | 3 |
| M17 | 1 | 2 | 2 | 2 |
| M18 | 2 | 2 | ||
| M19 | 2 | 2 | 2 | |
| M20 | 1 | 1 | 3 | 2 |
| M21 | 1 | 1 | 2 | 3 |
| M22 | 1 | 1 | 2 | 2 |
| Total | 7 | 14 | 19 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loera Piedra, A.A.; Becerra Mejía, I.L.; Luna Galicia, B.; Martínez Díaz, S.F.; Luna Herrera, J.; Aguilera Arreola, M.G. Exploring Local Reservoirs for Bacteriophages with Therapeutic Potential against ESKAPE Pathogens. Microbiol. Res. 2024, 15, 1459-1470. https://doi.org/10.3390/microbiolres15030098
Loera Piedra AA, Becerra Mejía IL, Luna Galicia B, Martínez Díaz SF, Luna Herrera J, Aguilera Arreola MG. Exploring Local Reservoirs for Bacteriophages with Therapeutic Potential against ESKAPE Pathogens. Microbiology Research. 2024; 15(3):1459-1470. https://doi.org/10.3390/microbiolres15030098
Chicago/Turabian StyleLoera Piedra, Alejandra Aidee, Isamar Leticia Becerra Mejía, Brenda Luna Galicia, Sergio Francisco Martínez Díaz, Julieta Luna Herrera, and Ma. Guadalupe Aguilera Arreola. 2024. "Exploring Local Reservoirs for Bacteriophages with Therapeutic Potential against ESKAPE Pathogens" Microbiology Research 15, no. 3: 1459-1470. https://doi.org/10.3390/microbiolres15030098
APA StyleLoera Piedra, A. A., Becerra Mejía, I. L., Luna Galicia, B., Martínez Díaz, S. F., Luna Herrera, J., & Aguilera Arreola, M. G. (2024). Exploring Local Reservoirs for Bacteriophages with Therapeutic Potential against ESKAPE Pathogens. Microbiology Research, 15(3), 1459-1470. https://doi.org/10.3390/microbiolres15030098







