Transcriptional Regulation of the Genes Encoding Branched-Chain Aminotransferases in Kluyveromyces lactis and Lachancea kluyveri Is Independent of Chromatin Remodeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Strains
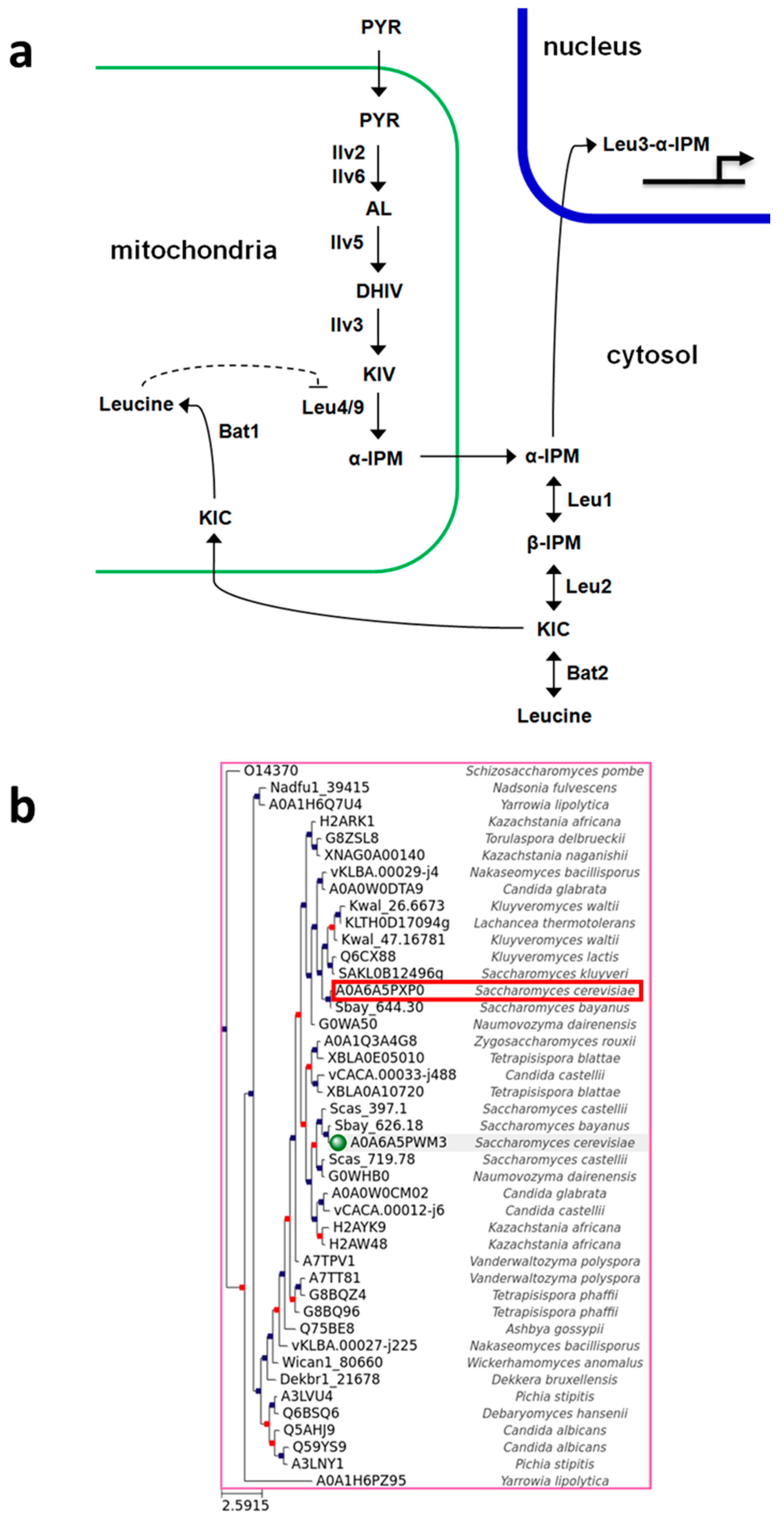
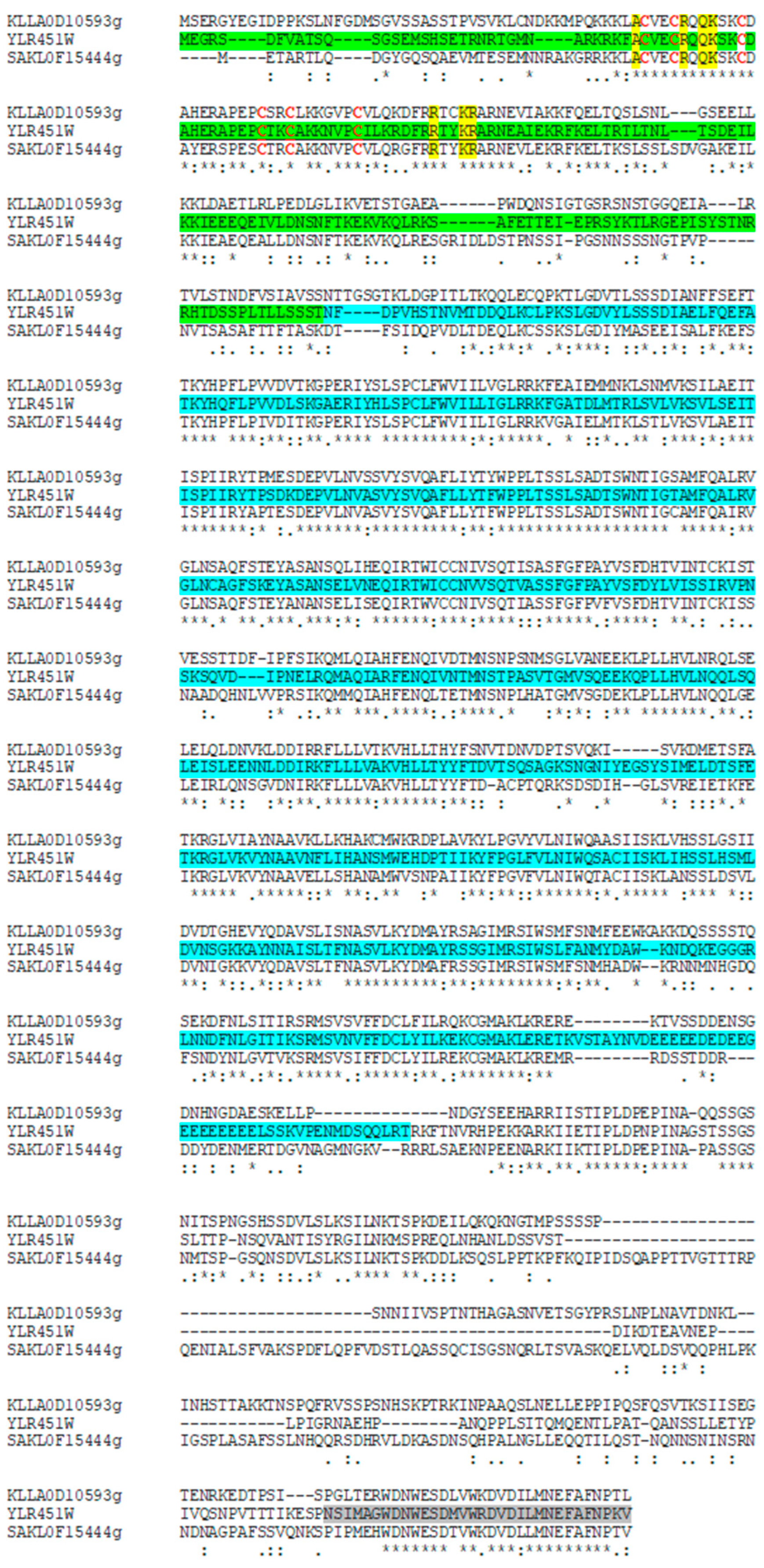
2.3. Phylogeny of Yeast Proteins
2.4. Quantitative Polymerase Chain Reaction for Differential Transcript Expression Analysis
2.5. Nucleosome Scanning Assay
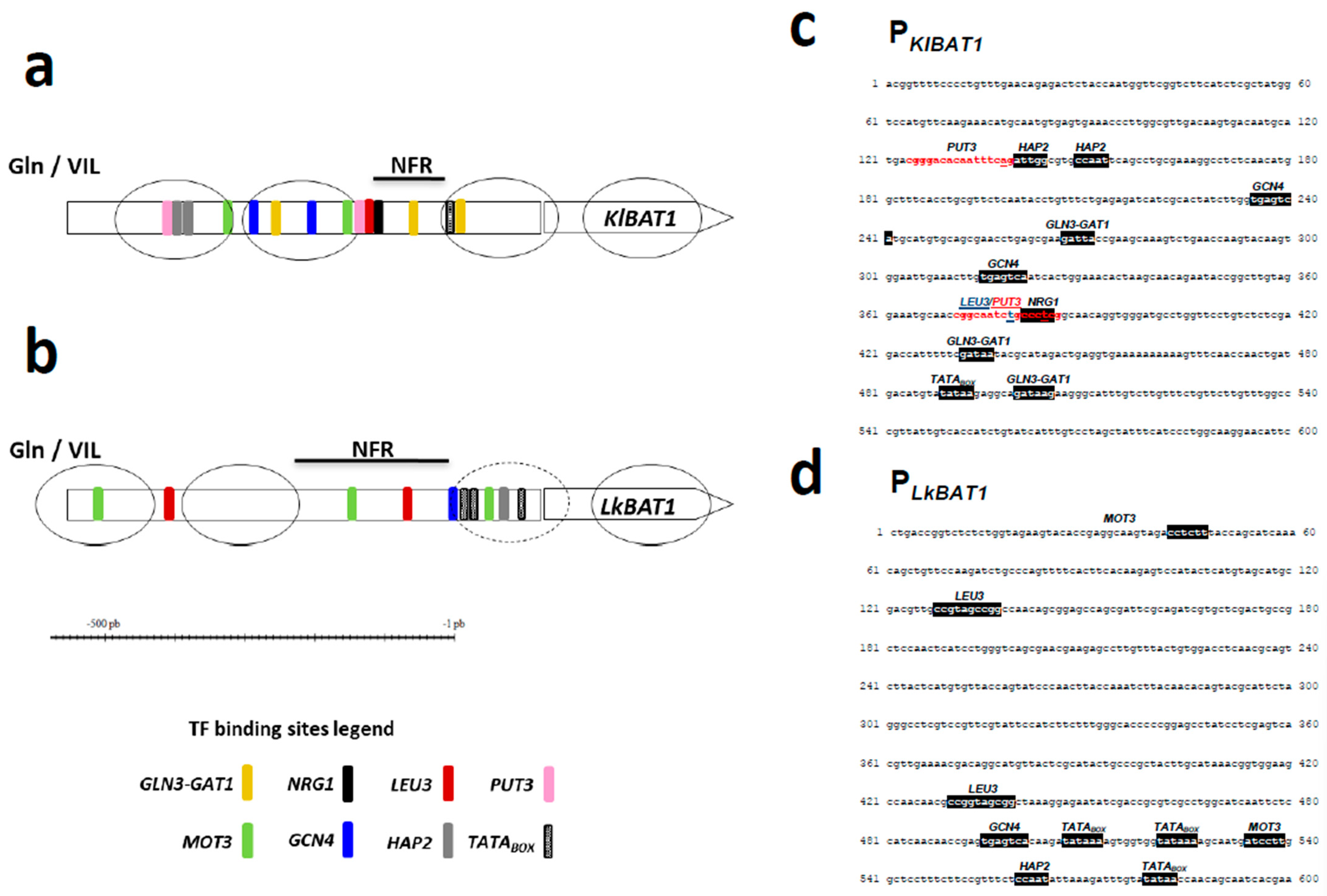
2.6. In Silico Promoter Analysis
2.7. α-IPMS Enzyme Assay and IC50 Determination
3. Results
3.1. Phylogeny of Branched-Chain Aminotransferases in the Saccharomycotina
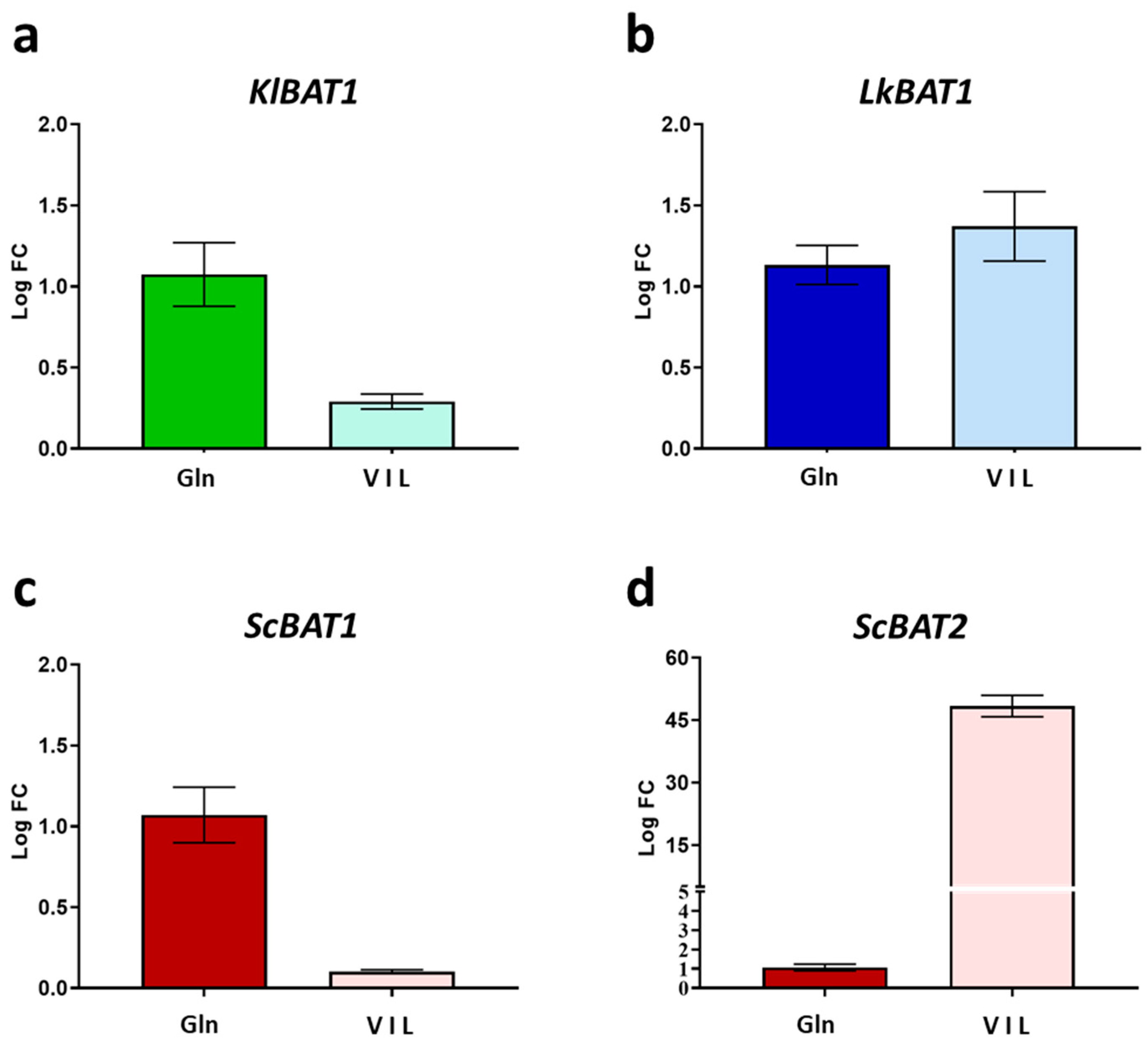
3.2. KlBAT1 and LkBAT1 Expression Profiles Are Not Dependent on Chromatin Remodeling
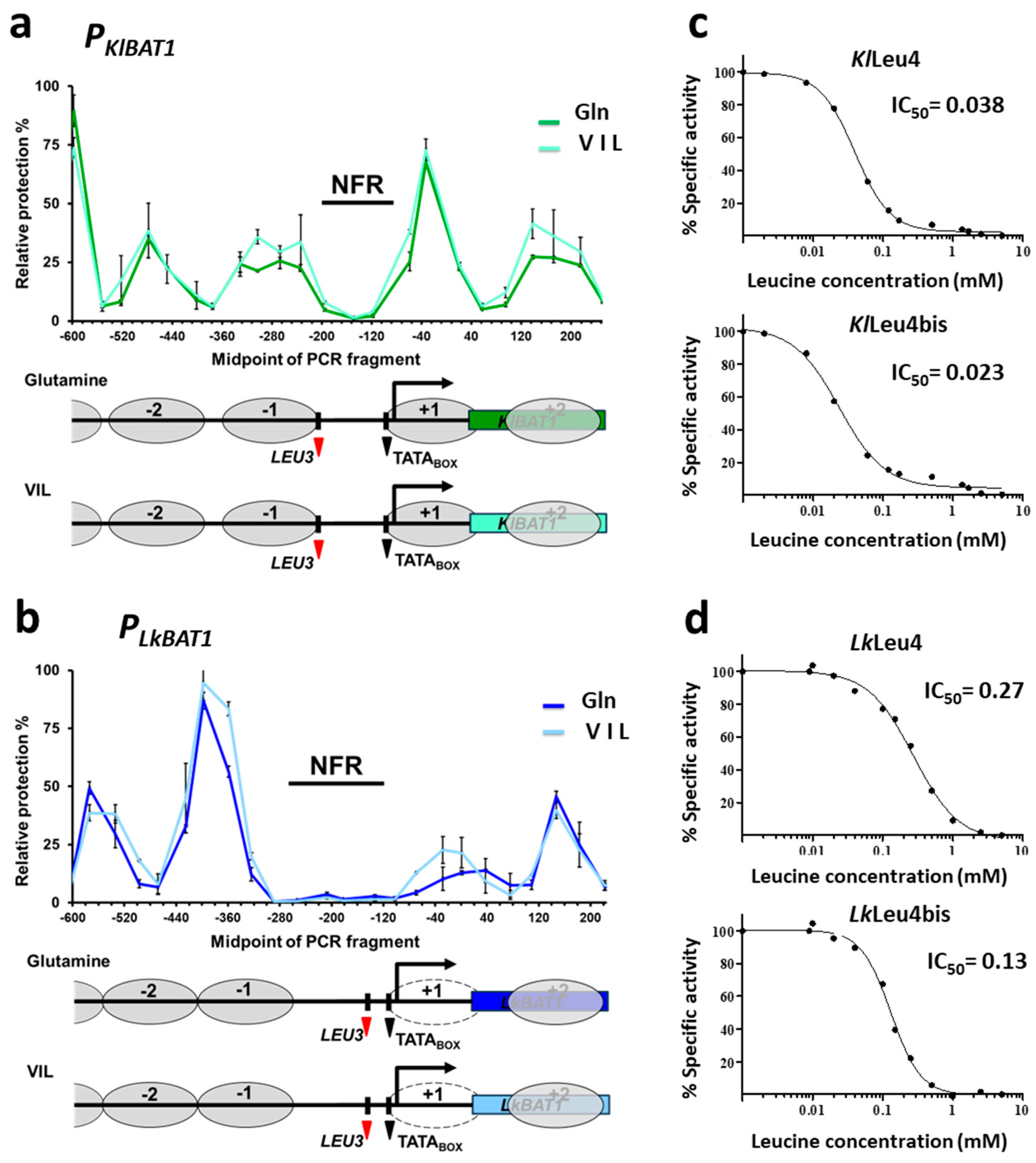
3.3. The Putative LEU3-Binding Site Is Located within the NFR of KlBAT1 and LkBAT1 Promoters
3.4. Leucine Feedback Sensitivity of the α-IPMS Enzymes Inversely Correlates with KlBAT1 and LkBAT1 Expression Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Knaap, J.A.; Verrijzer, C.P. Undercover: Gene control by metabolites and metabolic enzymes. Genes Dev. 2016, 30, 2345–2369. [Google Scholar] [CrossRef]
- Zhou, K.M.; Bai, Y.L.; Kohlhaw, G.B. Yeast regulatory protein LEU3: A structure-function analysis. Nucleic Acids Res. 1990, 18, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.M.; Kohlhaw, G.B. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J. Biol. Chem. 1990, 265, 17409–17412. [Google Scholar] [CrossRef] [PubMed]
- Friden, P.; Schimmel, P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol. Cell. Biol. 1988, 8, 2690–2697. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zheng, F.; Holmberg, S.; Kohlhaw, G.B. Yeast transcriptional regulator Leu3p: Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J. Biol. Chem. 1999, 274, 19017–19024. [Google Scholar] [CrossRef]
- Hu, Y.; Cooper, T.G.; Kohlhaw, G.B. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol. Cell. Biol. 1995, 15, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.D.; Tracy, J.W.; Kohlhaw, G.B. Subcellular localization of the leucine biosynthetic enzymes in yeast. J. Bacteriol. 1973, 116, 222–225. [Google Scholar] [CrossRef] [PubMed]
- González, J.; López, G.; Argueta, S.; Escalera-Fanjul, X.; El Hafidi, M.; Campero-Basaldua, C.; Strauss, J.; Riego-Ruiz, L.; González, A. Diversification of transcriptional regulation determines subfunctionalization of paralogous branched chain aminotransferases in the yeast Saccharomyces cerevisiae. Genetics 2017, 207, 975–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, L.-F.L.; Cunningham, T.S.; Gatzek, P.R.; Chen, W.-J.; Kohlhaw, G.B. Cloning and characterization of yeast Leu4, one of two genes responsible for α-isopropylmalate synthesis. Genetics 1984, 108, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Kohlhaw, G.B. Leucine biosynthesis in fungi: Entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 2003, 67, 1–15. [Google Scholar] [CrossRef]
- López, G.; Quezada, H.; Duhne, M.; González, J.; Lezama, M.; El-Hafidi, M.; Colón, M.; Escalera, X.M.d.l.; Flores-Villegas, M.C.; Scazzocchio, C.; et al. Diversification of paralogous alpha-isopropylmalate synthases by modulation of feedback control and hetero-oligomerization in Saccharomyces cerevisiae. Eukaryot. Cell 2015, 14, 564–577. [Google Scholar] [CrossRef]
- Colón, M.; Hernández, F.; López, K.; Quezada, H.; González, J.; López, G.; Aranda, C.; González, A. Saccharomyces cerevisiae Bat1 and Bat2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme. PLoS ONE 2011, 6, e16099. [Google Scholar] [CrossRef] [PubMed]
- Kispal, G.; Steiner, H.; Court, D.A.; Rolinski, B.; Lill, R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 Protein. J. Biol. Chem. 1996, 271, 24458–24464. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Van Nedervelde, L.; Drukker, M.; Benvenisty, N.; Debourg, A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl. Microbiol. Biotechnol. 2001, 55, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Marcet-Houben, M.; Gabaldón, T. Beyond the whole-genome duplication: Phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast lineage. PLoS Biol. 2015, 13, e1002220. [Google Scholar] [CrossRef] [PubMed]
- Montalvo-Arredondo, J.; Jiménez-Benítez, Á.; Colón-González, M.; González-Flores, J.; Flores-Villegas, M.; González, A.; Riego-Ruiz, L. Functional roles of a predicted branched chain aminotransferase encoded by the LkBAT1 gene of the yeast Lachancea kluyveri. Fungal Genet. Biol. 2015, 85, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, K.G.; Strathern, J.N. Molecular genetics in Saccharomyces kluyveri: The HIS3 homolog and its use as a selectable marker gene in S. kluyveri and Saccharomyces cerevisiae. Yeast 1993, 9, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-López, B.; Escalera-Fanjul, X.; Hersch-González, J.; Rojas-Ortega, E.; El-Hafidi, M.; Lezama, M.; González, J.; Bianchi, M.M.; López, G.; Márquez, D.; et al. In Kluyveromyces lactis a pair of paralogous isozymes catalyze the first committed step of leucine biosynthesis in either the mitochondria or the cytosol. Front. Microbiol. 2020, 11, 1843. [Google Scholar] [CrossRef]
- Vigueras-Meneses, L.G.; Escalera-Fanjul, X.; El-Hafidi, M.; Montalvo-Arredondo, J.; Gómez-Hernández, N.; Colón, M.; Granados, E.; Campero-Basaldua, C.; Riego-Ruiz, L.; Scazzocchio, C.; et al. Two alpha isopropylmalate synthase isozymes with similar kinetic properties are extant in the yeast Lachancea kluyveri. FEMS Yeast Res. 2022, 22, foac016. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, D.; Molina, M.; Chorostecki, U.; Capella-Gutiérrez, S.; Marcet-Houben, M.; Gabaldón, T. PhylomeDB V5: An expanding repository for genome-wide catalogues of annotated gene phylogenies. Nucleic Acids Res. 2021, 50, D1062–D1068. [Google Scholar] [CrossRef]
- Campero-Basaldua, C.; González, J.; García, J.A.; Ramírez, E.; Hernández, H.; Aguirre, B.; Torres-Ramírez, N.; Márquez, D.; Sánchez, N.S.; Gómez-Hernández, N.; et al. Neo-functionalization in Saccharomyces cerevisiae: A novel Nrg1-Rtg3 chimeric transcriptional modulator is essential to maintain mitochondrial DNA integrity. R. Soc. Open Sci. 2023, 10, 231209. [Google Scholar] [CrossRef]
- Byrne, K.P.; Wolfe, K.H. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005, 15, 1456–1461. [Google Scholar] [CrossRef]
- Thomas-Chollier, M.; Defrance, M.; Medina-Rivera, A.; Sand, O.; Herrmann, C.; Thieffry, D.; van Helden, J. RSAT 2011: Regulatory sequence analysis tools. Nucleic Acids Res. 2011, 39, W86–W91. [Google Scholar] [CrossRef]
- de Boer, C.G.; Hughes, T.R. YeTFaSCo: A database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res. 2011, 40, D169–D179. [Google Scholar] [CrossRef]
- Fitzgerald, M.X.; Rojas, J.R.; Kim, J.M.; Kohlhaw, G.B.; Marmorstein, R. Structure of a Leu3-DNA complex: Recognition of everted CGG half-sites by a Zn2Cys6 binuclear cluster protein. Structure 2006, 14, 725–735. [Google Scholar] [CrossRef]
- Steyer, J.T.; Todd, R.B. Branched-chain amino acid biosynthesis in fungi. Essays Biochem. 2023, 67, 865–876. [Google Scholar] [CrossRef]
- Bateman, J.M.; Iacovino, M.; Perlman, P.S.; Butow, R.A. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. J. Biol. Chem. 2002, 277, 47946–47953. [Google Scholar] [CrossRef]
- Kingsbury, J.M.; Sen, N.D.; Cardenas, M.E. Branched-chain aminotransferases control TORC1 signaling in Saccharomyces cerevisiae. PLoS Genet. 2015, 11, e1005714. [Google Scholar] [CrossRef]
- Alvers, A.L.; Fishwick, L.K.; Wood, M.S.; Hu, D.; Chung, H.S.; Dunn, W.A., Jr.; Aris, J.P. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 2009, 8, 353–369. [Google Scholar] [CrossRef]
- Ziegler, C.A.; Freddolino, P.L. The leucine-responsive regulatory proteins/feast-famine regulatory proteins: An ancient and complex class of transcriptional regulators in bacteria and archaea. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 373–400. [Google Scholar] [CrossRef]
| Species/Strain | Genotype | Source |
|---|---|---|
| K. lactis/Y155 | MATα ade2 his3 ura3 | [12] |
| L. kluyveri/Y156 | MATαura3 | [17] |
| K. lactis/Y155-1 Klleu4Δ | MATαade2 his3 ura3 KlLEU4::KanMX4 | [18] |
| K. lactis/Y155-2 Klleu4bisΔ | MATαade2 his3 ura3 KlLEU4bis::KanMX4 | [18] |
| L. kluyveri/Y156-3 Lkleu4Δ | MATα ura3 LkLEU4::NatMX | [19] |
| L. kluyveri/Y156-4 Lkleu4bisΔ | MATαura3 LkLEU4bis::KanMX4 | [19] |
| S. cerevisiae/CLA11-700 | MATα ura3 | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, J.; Quezada, H.; Campero-Basaldua, J.C.; Ramirez-González, É.; Riego-Ruiz, L.; González, A. Transcriptional Regulation of the Genes Encoding Branched-Chain Aminotransferases in Kluyveromyces lactis and Lachancea kluyveri Is Independent of Chromatin Remodeling. Microbiol. Res. 2024, 15, 1225-1238. https://doi.org/10.3390/microbiolres15030082
González J, Quezada H, Campero-Basaldua JC, Ramirez-González É, Riego-Ruiz L, González A. Transcriptional Regulation of the Genes Encoding Branched-Chain Aminotransferases in Kluyveromyces lactis and Lachancea kluyveri Is Independent of Chromatin Remodeling. Microbiology Research. 2024; 15(3):1225-1238. https://doi.org/10.3390/microbiolres15030082
Chicago/Turabian StyleGonzález, James, Héctor Quezada, Jose Carlos Campero-Basaldua, Édgar Ramirez-González, Lina Riego-Ruiz, and Alicia González. 2024. "Transcriptional Regulation of the Genes Encoding Branched-Chain Aminotransferases in Kluyveromyces lactis and Lachancea kluyveri Is Independent of Chromatin Remodeling" Microbiology Research 15, no. 3: 1225-1238. https://doi.org/10.3390/microbiolres15030082
APA StyleGonzález, J., Quezada, H., Campero-Basaldua, J. C., Ramirez-González, É., Riego-Ruiz, L., & González, A. (2024). Transcriptional Regulation of the Genes Encoding Branched-Chain Aminotransferases in Kluyveromyces lactis and Lachancea kluyveri Is Independent of Chromatin Remodeling. Microbiology Research, 15(3), 1225-1238. https://doi.org/10.3390/microbiolres15030082






