Multidrug-Resistant Enterococcus faecium and Enterococcus faecalis Isolated from Dogs and Cats in Southern Brazil
Abstract
1. Introduction
2. Materials and Methods
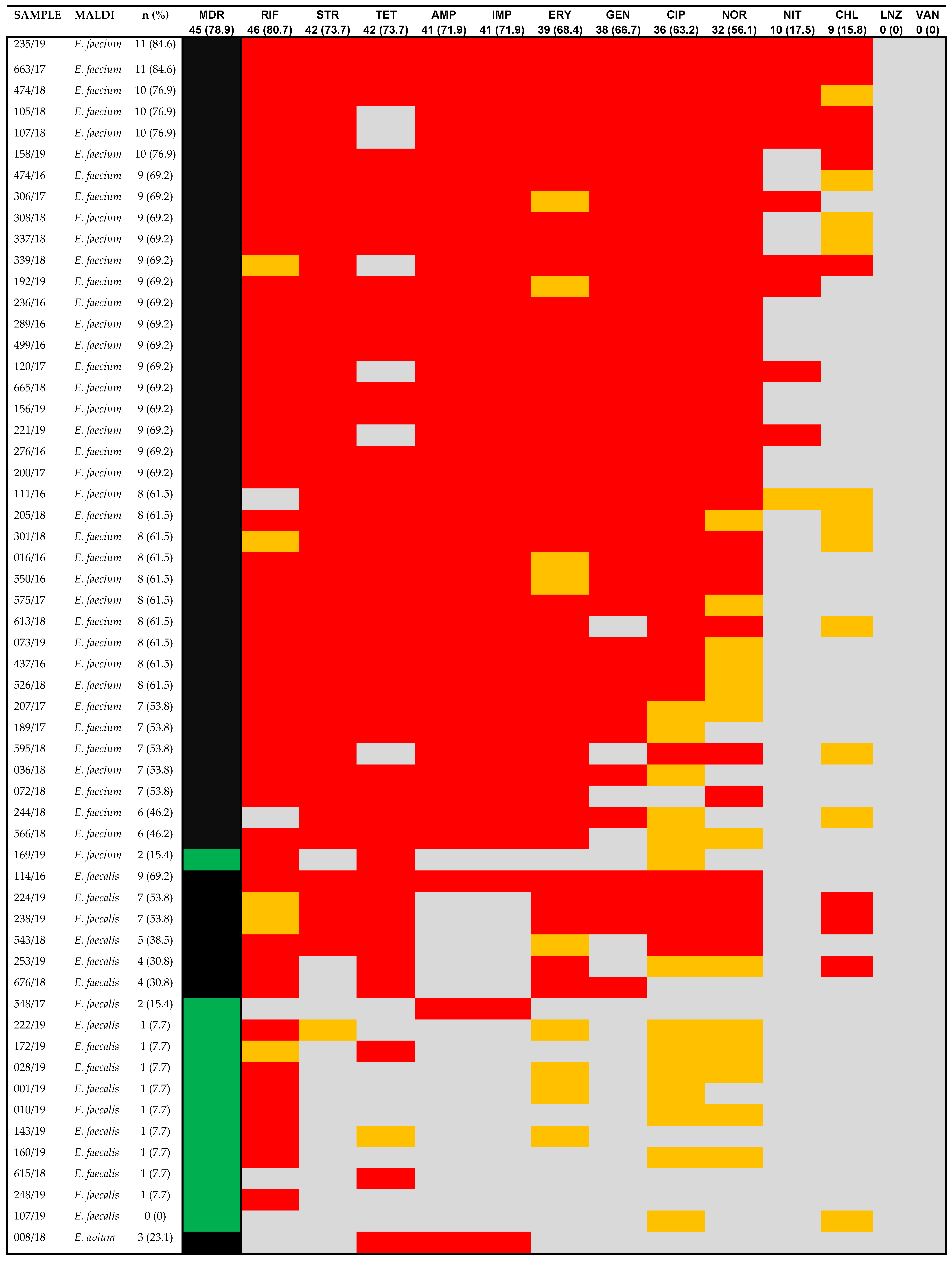
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stull, J.W.; Weese, J.S. Hospital-associated infections in small animal practice. Vet. Clin. N. Am. Small Anim. Pract. 2015, 45, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Fedorka-Cray, P.J.; Davis, J.A.; Barrett, J.B.; Frye, J.G. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 2009, 107, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Dowd, S.E.; Zurek, L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS ONE 2011, 6, e22451. [Google Scholar] [CrossRef] [PubMed]
- Damborg, P.; Top, J.; Hendrickx, A.P.A.; Dawson, S.; Willems, R.J.; Guardabassi, L. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl. Environ. Microbiol. 2009, 75, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Jackson, C.; Fedorka-Cray, P.; Davis, J.; Barrett, J.; Brousse, J.; Gustafson, J.; Kucher, M. Mechanisms of antimicrobial resistance and genetic relatedness among enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 2010, 108, 2171–2179. [Google Scholar] [CrossRef]
- Bertelloni, F.; Salvadori, C.; Lotti, G.; Cerri, D.; Ebani, V.V. Antimicrobial resistance in Enterococcus strains isolated from healthy domestic dogs. Acta Microbiol. Immunol. Hung. 2017, 64, 301–312. [Google Scholar] [CrossRef]
- Miranda, C.; Silva, V.; Igrejas, G.; Poeta, P. Impact of European pet antibiotic use on enterococci and staphylococci antimicrobial resistance and human health. Future Microbiol. 2021, 16, 185–201. [Google Scholar] [CrossRef]
- Leite-Martins, L.; Meireles, D.; Bessa, L.J.; de Matos, A.J.; da Costa, P.M. Spread of multidrug-resistant Enterococcus faecalis within the household setting. Microb. Drug Resist. 2014, 20, 501–507. [Google Scholar] [CrossRef]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef]
- Ben Said, L.; Dziri, R.; Sassi, N.; Lozano, C.; Ben Slama, K.; Ouzari, I.; Torres, C.; Klibi, N. Species distribution, antibiotic resistance and virulence traits in canine and feline enterococci in Tunisia. Acta Vet. Hung. 2017, 65, 173–184. [Google Scholar] [CrossRef] [PubMed]
- van den Bunt, G.; Top, J.; Hordijk, J.; de Greeff, S.C.; Mughini-Gras, L.; Corander, J. Intestinal carriage of ampicillin-and vancomycin-resistant Enterococcus faecium in humans, dogs and cats in the Netherlands. J. Antimicrob. Chemother. 2018, 73, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. (Eds.) Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Kim, S.H.; Chon, J.W.; Jeong, H.W.; Song, K.Y.; Kim, D.H.; Bae, D.; Kim, H.; Seo, K.-H. Identification and phylogenetic analysis of Enterococcus isolates using MALDI-TOF MS and VITEK 2. AMB Express 2023, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- CLSI—Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing 2016, 26th ed.; 100S; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.; Lahti, E.; Kuhalampi, J.; Pesonen, S.; Järvinen, A.-K.; Saijonmaa-Koulumies, L.; Honkanen-Buzalski, T. Antimicrobial resistance in Staphylococcus spp., Escherichia coli and Enterococcus spp. in dogs given antibiotics for chronic dermatological disorders, compared with non-treated control dogs. Acta Vet. Scand 2004, 45, 37–45. [Google Scholar] [CrossRef] [PubMed]
- KuKanich, K.S.; Lubbers, B.V. Review of enterococci isolated from canine and feline urine specimens from 2006 to 2011. J. Am. Anim. Hosp. Assoc. 2015, 51, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Isaiah, A.; Hoffmann, A.R.; Kelley, R.; Mundell, P.; Steiner, J.M.; Suchodolski, J.S. Characterization of the nasal and oral microbiota of detection dogs. PLoS ONE 2017, 12, e0184899. [Google Scholar] [CrossRef]
- Kataoka, Y.; Umino, Y.; Ochi, H.; Harada, K.; Sawada, T. Antimicrobial susceptibility of Enterococcal species isolated from antibiotic-treated dogs and cats. J. Vet. Med. Sci. 2014, 76, 1399–1402. [Google Scholar] [CrossRef] [PubMed][Green Version]
- da Costa, L.B.; Corá, L.F.; Correa, F.E.L.; Gabrielli, L.C.; de Oliveira, M.R.; Conceição, N.; Oliveira, A.G. High Prevalence of the aac(6′)-Ie-aph(2″)-Ia gene in hospital isolates of Enterococcus faecalis co-resistant to gentamicin and penicillin. Microb. Drug Resist. 2019, 25, 1275–1281. [Google Scholar] [CrossRef]
- Bang, K.; An, J.-U.; Kim, W.; Dong, H.-J.; Kim, J.; Cho, S. Antibiotic resistance patterns and genetic relatedness of Enterococcus faecalis and Enterococcus faecium isolated from military working dogs in Korea. J. Vet. Sci. 2017, 18, 229–236. [Google Scholar] [CrossRef]
- Cheng, C.-C.; Wang, H.-C.; Lee, W.-M.; Shyu, C.-L.; Lin, C.-C.; Chen, K.-S.; Chang, A.-C. Emphysematous pyometra secondary to Enterococcus avium infection in a dog. Tieraerztliche Prax. Ausg. Kleintiere Heimtiere 2016, 44, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; KuKanich, K.; Brown, C.E.; Zurek, L. Resident cats in small animal veterinary hospitals carry multi drug resistant enterococci and are likely involved in cross-contamination of the hospital environment. Front. Microbiol. 2012, 3, 62. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Lebreton, F.; van Schaik, W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 2013, 16, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Jara, M.C.; Frediani, A.V.; Zehetmeyer, F.K.; Bruhn, F.R.P.; Müller, M.R.; Miller, R.G.; Nascente, P.d.S. Multidrug-resistant hospital bacteria: Epidemiological factors and susceptibility profile. Microb. Drug Resist. 2021, 27, 433–440. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; WHO: Geneva, Switzerland, 2024; Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 14 June 2024).
- Abbott, Y.; Kirby, B.M.; Karczmarczyk, M.; Markey, B.K.; Leonard, F.C.; Fitzgerald, S. High-level gentamicin-resistant and vancomycin-resistant Enterococcus faecium isolated from a wound in a dog. J. Small Anim. Pract. 2009, 50, 194–197. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Tenorio, C.; Torres, C. Study of vancomycin resistance in faecal enterococci from healthy humans and dogs in Spain a decade after the avoparcin ban in Europe. Zoonoses Public Health 2013, 60, 160–167. [Google Scholar] [CrossRef]
- Gerace, E.; Mancuso, G.; Midiri, A.; Poidomani, S.; Zummo, S.; Biondo, C. Recent Advances in the Use of Molecular Methods for the Diagnosis of Bacterial Infections. Pathogens 2022, 11, 663. [Google Scholar] [CrossRef]
| Year | Total Bacterial n | Enterococcus spp. n (%) | Dogs n (%) | Cats n (%) | Horses n (%) | Cows n (%) | Missing n (%) |
|---|---|---|---|---|---|---|---|
| 2016 | 485 | 59 (12.2) | 48 (81.3) | 5 (8.5) | 5 (8.5) | 1 (1.7) | 0 (0) |
| 2017 | 646 | 76 (11.8) | 59 (77.6) | 10 (13.1) | 6 (7.9) | 0 (0) | 1 (1.3) |
| 2018 | 778 | 68 (8.7) | 53 (77.9) | 9 (13.2) | 3 (4.4) | 0 (0) | 3 (4.4) |
| 2019 | 314 | 29 (9.2) | 20 (68.9) | 6 (20.7) | 2 (5.8) | 1 (3.4) | 1 (3.4) |
| Total | 2223 | 232 (10.4) | 180 (77.6) | 30 (12.9) | 16 (6.9) | 2 (0.9) | 5 (2.1) |
| E. faecium | E. faecalis | E. avium | Total | |
|---|---|---|---|---|
| Dogs | 32 (68.1%) | 14 (29.8%) | 1 (2.1%) | 47 |
| Cats | 7 (70%) | 3 (30%) | 0 (0%) | 10 |
| Total | 39 (68.4%) | 17 (29.8%) | 1 (1.8%) | 57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, L.; Grecellé, C.Z.; Frazzon, A.P.G.; Streck, A.F.; Kipper, D.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Multidrug-Resistant Enterococcus faecium and Enterococcus faecalis Isolated from Dogs and Cats in Southern Brazil. Microbiol. Res. 2024, 15, 1083-1090. https://doi.org/10.3390/microbiolres15030071
da Silva L, Grecellé CZ, Frazzon APG, Streck AF, Kipper D, Fonseca ASK, Ikuta N, Lunge VR. Multidrug-Resistant Enterococcus faecium and Enterococcus faecalis Isolated from Dogs and Cats in Southern Brazil. Microbiology Research. 2024; 15(3):1083-1090. https://doi.org/10.3390/microbiolres15030071
Chicago/Turabian Styleda Silva, Letícia, Cristina Zaffari Grecellé, Ana Paula Guedes Frazzon, André Felipe Streck, Diéssy Kipper, André Salvador Kazantzi Fonseca, Nilo Ikuta, and Vagner Ricardo Lunge. 2024. "Multidrug-Resistant Enterococcus faecium and Enterococcus faecalis Isolated from Dogs and Cats in Southern Brazil" Microbiology Research 15, no. 3: 1083-1090. https://doi.org/10.3390/microbiolres15030071
APA Styleda Silva, L., Grecellé, C. Z., Frazzon, A. P. G., Streck, A. F., Kipper, D., Fonseca, A. S. K., Ikuta, N., & Lunge, V. R. (2024). Multidrug-Resistant Enterococcus faecium and Enterococcus faecalis Isolated from Dogs and Cats in Southern Brazil. Microbiology Research, 15(3), 1083-1090. https://doi.org/10.3390/microbiolres15030071







